Abstract
Plants have evolved intricate immune mechanisms to combat pathogen infection. Upon perception of pathogen-derived signals, plants accumulate defense hormones such as ethylene (ET), jasmonate, salicylate, and damage-associated molecular patterns to amplify immune responses. In particular, the Arabidopsis peptide Pep1 and its family members are thought to be damage-associated molecular patterns that trigger immunity through Pep1 receptor kinases PEPR1 and PEPR2. Here we show that PEPR1 specifically interacts with receptor-like cytoplasmic kinases botrytis-induced kinase 1 (BIK1) and PBS1-like 1 (PBL1) to mediate Pep1-induced defenses. In vitro and in vivo studies suggested that PEPR1, and likely PEPR2, directly phosphorylates BIK1 in response to Pep1 treatment. Surprisingly, the pepr1/pepr2 double-mutant seedlings displayed reduced in sensitivity to ET, as indicated by the elongated hypocotyls. ET-induced expression of defense genes and resistance to Botrytis cinerea were compromised in pepr1/pepr2 and bik1 mutants, reenforcing an important role of PEPRs and BIK1 in ET-mediated defense signaling. Pep treatment partially mimicked ET-induced seedling growth inhibition in a PEPR- and BIK1-dependent manner. Furthermore, both ET and Pep1 treatments induced BIK1 phosphorylation in a PEPR-dependent manner. However, the Pep1-induced BIK1 phosphorylation, seedling growth inhibition, and defense gene expression were independent of canonical ET signaling components. Together our results illustrate a mechanism by which ET and PEPR signaling pathways act in concert to amplify immune responses.
Keywords: PAMPs, DAMPs, disease resistance, plant immunity, phytohormone
Plants have evolved a sophisticated innate immune system to cope with attacks by diverse pathogenic microbes. At the core are cytoplasmic immune receptors and cell-surface immune receptors detecting various danger signals during pathogen infection. Plant cell surface-localized immune receptors, also called pattern-recognition receptors (PRRs), consist of a variety of receptor-like kinases and receptor-like proteins. Many PRRs, including FLS2, EFR, Xa21, CERK1, CEBiP, LYM1, LYM3, LYP4, and LYP6 (1–8), directly sense pathogen/microbe-associated molecular patterns (PAMPs), such as flagellin, elongation factor, quorum-sensing protein, or peptidoglycans from bacteria or chitin from fungal cell wall. In addition to PAMPs, PRRs also perceive endogenous damage-associated molecular patterns (DAMPs), such as plant cell wall fragments released by pathogen lytic enzymes or plant peptides synthesized de novo during pathogen infection. The Arabidopsis Pep1, a 23-aa peptide processed from PROPEP1 (9, 10), is thought to be a DAMP perceived by two closely related LRR receptor kinases, PEPR1 and PEPR2, to trigger immune responses (11–13). Members of the PROPEP family are transcriptionally induced by defense hormones jasmonates (JA), ethylene (ET), and salicylate (SA), or wounding, and this is thought to amplify danger signals during pathogen infection. Pep1 seems to be conserved in both dicots and monocots, because ZmPep1 has also been shown to regulate defense gene expression in maize (14).
PRRs interact with other components in a highly dynamic manner. The ligand-binding to FLS2 and EFR is known to recruit BAK1, a receptor-like kinase, forming active receptor complexes (15–18). Downstream, the receptor-like cytoplasmic kinase BIK1 and its closely related protein PBL1 interact directly with FLS2, EFR, and CERK1. The activation of these PRRs results in a rapid phosphorylation of BIK1 and PBL1, which then dissociate from the receptors to activate downstream signaling (19, 20).
In addition to immune receptors, the delicate control of plant innate immunity also involves plant hormones, among which SA, ET, and JA play key roles in regulating defense responses (21). In particular, increasing evidence indicates that ET is intimately associated with PTI signaling pathways. For example, the activation of MPK6 by flg22 stabilizes 1-aminocyclopropane-1-carboxylate (ACC) synthases ACS2 and ACS6, which are rate-limiting enzymes for ET biosynthesis (22). ET exerts its regulation on defense responses through EIN3 and EIL1, two closely related transcription factors (23, 24). For example, FLS2 transcription is positively regulated by EIN3 and EIL1 (25, 26). EIN3/EIL1 also negatively regulate SA-dependent immunity by binding to the promoter of SID2, which controls SA biosynthesis (27). Thus, a linear intracellular signaling pathway from ER-localized ET receptors to the nuclear-localized transcription factors is thought to directly execute defense gene regulation.
Interestingly, a recent report shows that etiolated bik1 seedlings are partially insensitive to ET and display elongated hypocotyl (28). Furthermore, BIK1 is phosphorylated upon ET treatment. However, the molecular mechanism by which BIK1 regulates ET signaling remains unknown. Here we show that BIK1 directly interacts with and is phosphorylated by PEPR1. Pep1-induced defenses were diminished in the bik1 mutant plants. Like bik1, the pepr1/pepr2 double mutant was partially insensitive to seedling growth inhibition by ET. Pep peptides can mimic ET-induced seedling growth inhibition in a PEPR- and BIK1-dependent but EIN3/EIL1- and/or EIN2-independent manner. PEPR1, and likely PEPR2, are required for ET- and Pep1-induced phosphorylation of BIK1. Furthermore, the ET-induced expression of several defense genes and resistance to Botrytis cinerea were compromised in pepr1/pepr2 and bik1. Our findings indicate that Pep peptides, PEPRs, and BIK1 form an extension of the canonical ET signaling pathway to regulate plant immunity.
Results
BIK1 Interacts with PEPR1 in Vitro and in Vivo.
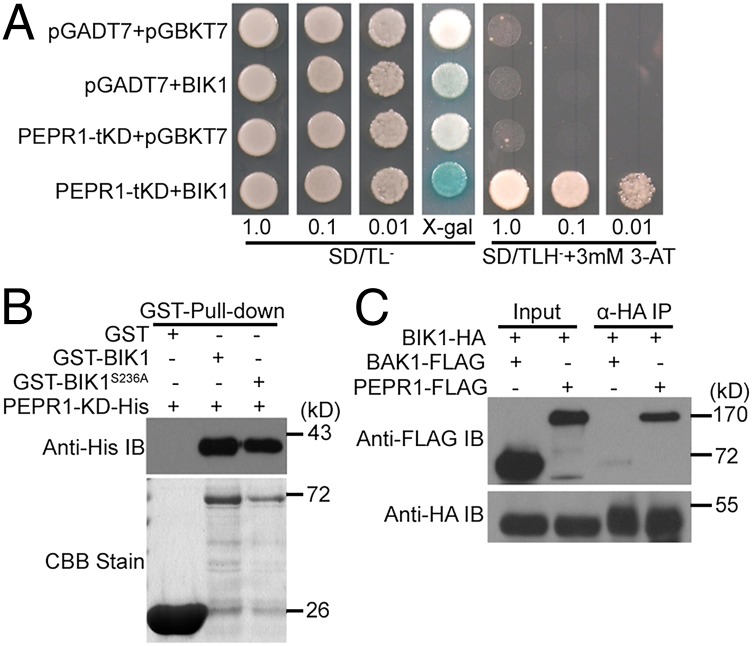
To understand mechanisms by which BIK1 regulates ET signaling and plant immunity, we performed a yeast two-hybrid screen by using BIK1 as bait. Two independent clones containing amino acids 1022–1123 and 1025–1123 of the C terminus of PEPR1 kinase domain (KD) showed a strong interaction with BIK1 (Fig. 1A). GST pull-down assay showed that a GST-tagged BIK1 also interacted with a His-tagged PEPR1 KD (amino acids 827–1123) in vitro (Fig. 1B). The BIK1S236A mutant carrying an amino acid substitution at an autophosphorylation site (20) also interacted with the PEPR1 KD, suggesting that this interaction is independent of the phosphorylation on S236 (Fig. 1B). Coimmunoprecipitation assay indicated that the full-length PEPR1-FLAG, but not BAK1-FLAG, specifically interacted with BIK1-HA in Arabidopsis protoplasts (Fig. 1C). The lack of BIK1–BAK1 interaction is consistent with our previous report (20) but contradicts a report by Lu et al. (19). Together these data indicated that BIK1 can interact with PEPR1 KD.
Fig. 1.
BIK1 interacts with the PEPR1 KD. (A) BIK1 interacts with PEPR1 KD in yeast. Serial dilution of yeast cells containing the indicated plasmids were spotted on the indicated medium for lacZ and His reporter assays (four independent experiments). PEPR1-tKD, a prey plasmid containing a truncated PEPR1 KD (amino acids 1025–1123); BIK1, a bait plasmid containing full-length BIK1; pGADT7, empty prey plasmid; pGBKT7, empty bait plasmid; SD/TL−, SD medium lacking tryptophan and leucine; SD/TLH−, SD medium lacking tryptophan, leucine, histidine, and adenine; 3-AT, 3-amino-1,2,4-triazole. The center panel contains X-gal in the medium. (B) BIK1 interacts with the intact PEPR1 KD (amino acids 827–1123) in GST pull-down assay (three independent experiments). Anti-His immunoblot (IB) shows amounts of PEPR1-KD-His bound by the indicated GST-tagged proteins. Coomassie Brilliant Blue (CBB) staining indicates amounts of GST-tagged proteins. (C) BIK1 interacts with full-length PEPR1 in Arabidopsis protoplasts. BIK1-HA was expressed in WT Arabidopsis protoplasts, along with PEPR1-FLAG or BAK1-FLAG, immunoprecipitated with anti-HA antibody (α-HA IP), and the bound protein was detected by immunoblot with the indicated antibodies (four independent experiments).
BIK1 Is Required for Pep1-Induced Defenses.
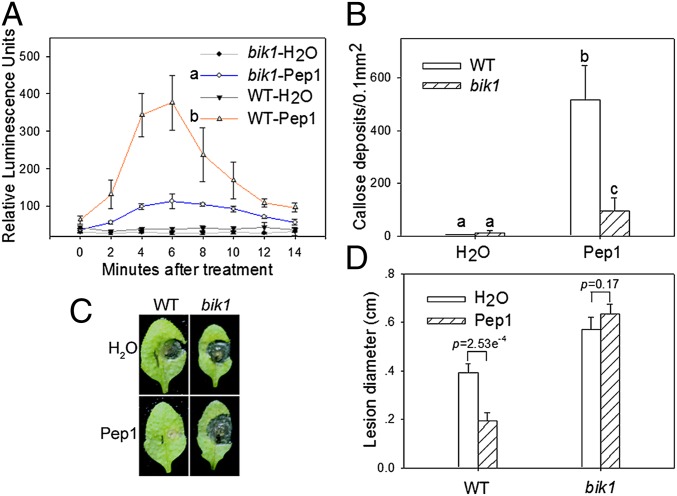
To determine the biological significance of the observed BIK1–PEPR1 interaction, we tested Pep1-indcued defenses in the bik1 mutant. As reported in previous studies (12), treatment of WT plants with Pep1 induced rapid accumulation of H2O2 and callose deposition (Fig. 2). The Pep1-induced H2O2 production in bik1 was reduced to 20–30% compared with the WT control (Fig. 2A). Similarly, the Pep1-induced callose deposition in bik1 seedlings was reduced to ∼20% of that in WT (Fig. 2B). The same treatments failed to induce H2O2 production and callose deposition in pepr1/pepr2 double mutants (Fig. S1). To determine whether BIK1 is required for Pep1-induced disease resistance, we pretreated plants before inoculation of B. cinerea. Although the Pep1 treatment reduced disease lesion size on WT plants by ∼50%, it failed to protect bik1 plants (Fig. 2 C and D). Together these results demonstrated that BIK1 is important for Pep1 signaling.
Fig. 2.
BIK1 is required for Pep1-induced defenses. (A) bik1 is compromised in Pep1-induced oxidative burst. Relative luminescence units indicate relative amounts of H2O2 production in leaf strips treated with 1 μM Pep1 at the indicated times. Different letters indicate significant difference (mean ± SD; n ≥ 4; P < 0.01, Student’s test; nine biological repeats). (B) bik1 is severely compromised to Pep1-induced callose deposition (mean + SD; n ≥ 15; P < 0.01, Student's t test; four biological repeats). (C and D) Pep1 pretreatment was unable to protect bik1 from B. cinerea infection. Leaves were infiltrated with Pep1 18 h before inoculation with B. cinerea. Disease lesion was measured 2 d later (mean + SD; n ≥ 7; P < 0.01, Student's t test; two biological repeats). P value is given above the bars.
Specificity of BIK1 Family Members in Pep1 Signaling.
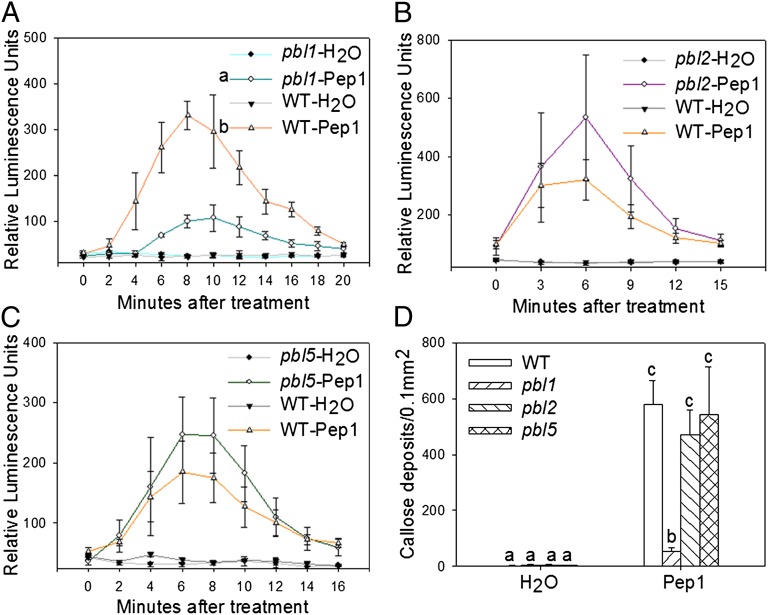
In Arabidopsis, the BIK1 family contains more than 40 members. Among these, BIK1 and PBL1 belong to the same clade, whereas PBL2 and PBL5 are more distantly related to BIK1. We therefore sought to determine the specificity of PEPR1 interaction with PBL1, PBL2, and PBL5. Yeast two-hybrid assay showed that only PBL1, but not PBL2 and PBL5, interacted with the PEPR1 KD (Fig. S2), suggesting that PEPR1 specifically interacts with the BIK1-PBL1 clade. We then tested Pep1-induced H2O2 accumulation and callose deposition in the pbl1 and pbl2 mutants (20) and the newly isolated pbl5 mutant (Fig. S3A). Consistent with the yeast two-hybrid data, pbl1, but not pbl2 and pbl5, displayed marked reduction in Pep1-induced H2O2 accumulation and callose deposition compared with WT (Fig. 3). We previously showed that BIK1, PBL1, and PBL2 play an additive role in defenses triggered by different PAMPs (20). Similarly, the pbl5 mutant was also compromised in flg22-induced H2O2 accumulation (Fig. S3A). Together these results indicate that BIK1 and PBL1 are specifically required for Pep1 signaling, whereas multiple BIK1 family members are involved in PAMP-triggered defenses.
Fig. 3.
Specificity of PBLs in Pep1 responses. (A–C) pbl1, but not pbl2 and pbl5, is compromised in Pep1-induced H2O2 production. Different letters denote significant difference between WT and pbl1 (mean ± SD; n ≥ 4; P < 0.01, Student’s test; four biological repeats). (D) pbl1, but not pbl2 and pbl5, is compromised in Pep1-induced callose deposition (mean + SD; n ≥ 14; P < 0.01, Student’s test; two biological repeats).
PEPRs Are Required for Seedling Growth Inhibition by ET.
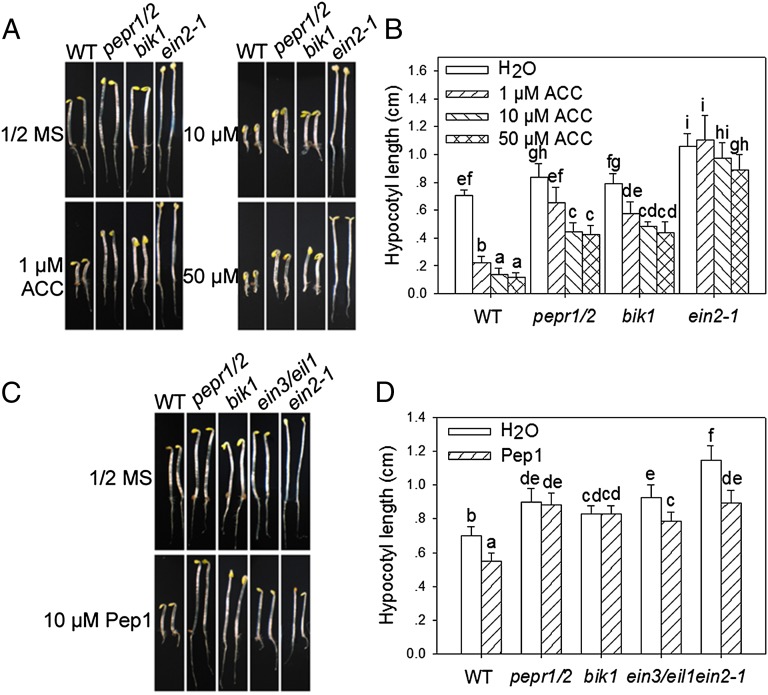
The facts that BIK1 is partially required for ET-induced growth inhibition (28) and that members of the PROPEP family are transcriptionally induced by ET (9) prompted us to test whether PEPR1 and PEPR2 are required for ET responses. In etiolated seedlings, ET is known to induce a triple response, namely exaggerated apical hook and shortened hypocotyl and root. Consistent with previous studies, WT seedlings germinated in the presence of ACC, the precursor of ET, displayed typical triple response, whereas the ein2-1 mutant is completely insensitive to ACC (Fig. 4 A and B). The bik1 mutant and pepr1/pepr2 double mutant, however, showed reduced responsiveness to ACC treatment. At 1 μM ACC, both pepr1/pepr2 and bik1 seedlings exhibited elongated hypocotyl compared with WT seedlings, although the root growth and apical hook were not significantly affected in these mutants. Although 1 μM ACC reduced hypocotyl length to ∼25% of the control in WT seedlings, it had only a minor effect (70–75% of the control) in pepr1/pepr2 and bik1 seedlings (Fig. 4B). At 10 and 50 μM ACC, the hypocotyl of pepr1/pepr2 and bik1 seedlings was reduced to ∼50% of the control, compared with ∼15% in WT (Fig. 4 A and B). These results support that the ET-induced growth inhibition is controlled, in part, by PEPR1, PEPR2, and BIK1. We also tested ET-induced seedling growth inhibition in pbl1, pbl2, and pbl5 mutants. All three mutants displayed WT response to 10 μM ACC (Fig. S3 B and C). Thus, among the four BIK1 family members tested, only BIK1 is required for ET-induced seedling growth responses, a result consistent with a previous study (28). These observations indicate a specific involvement of BIK1, but not other members of the BIK1 family, in ET-induced growth inhibition in seedlings.
Fig. 4.
PEPRs are required for ET- and Pep1-induced seedling growth inhibition. (A and B) pepr1/pepr2 (pepr1/2) double-mutant plants are compromised in ACC-induced growth inhibition on hypocotyl. (A) Photograph of seedlings; (B) hypocotyl length. Different letters denote significant difference (mean + SD; n ≥ 20; P < 0.01, Student’s test; five biological repeats). (C) Photograph of 5-d-old etiolated seedlings grown in the presence of 10 μM Pep1. (D) Hypocotyl length. Different letters denote significant difference (mean + SD; n ≥ 27; P < 0.01, Student’s test; three biological repeats).
Peps Inhibit Seedling Growth Independent of EIN3/EIL1.
To further investigate the role of PEPRs and BIK1 in ET-induced growth inhibition, we germinated seedlings in darkness in the presence of Pep1. The WT seedlings exhibited a significant inhibition of hypocotyl and root elongation, whereas bik1 and pepr1/pepr2 seedlings were completely insensitive to the Pep1 treatment (Fig. 4 C and D). Pep1 did not seem to induce apical hook, an observation consistent with the PEPR-independent induction of apical hook by ET. Similar growth inhibition was observed with when seedlings were germinated in the presence of Pep2 and, to a lesser extent, Pep3 (Fig. S4). We reasoned that Peps, PEPRs, and BIK1 form a unique pathway inhibiting hypocotyl growth downstream of the canonical ET signaling pathway. Indeed, Pep1 and Pep2 were fully capable of inhibiting the hypocotyl elongation in ein2-1 and/or ein3/eil1 seedlings (Fig. 4 C and D and Fig. S4). Together these results confirm that Peps act downstream of the canonical ET signaling pathway to inhibit hypocotyl growth. It should be noted, however, that the pepr1/pepr2 seedlings retained root growth inhibition by ET (Fig. 4 A and B), suggesting that some of the seedling growth inhibition occurs through a pathway independent of PEPRs.
PEPRs and BIK1 Mediate ET-Induced Defenses.
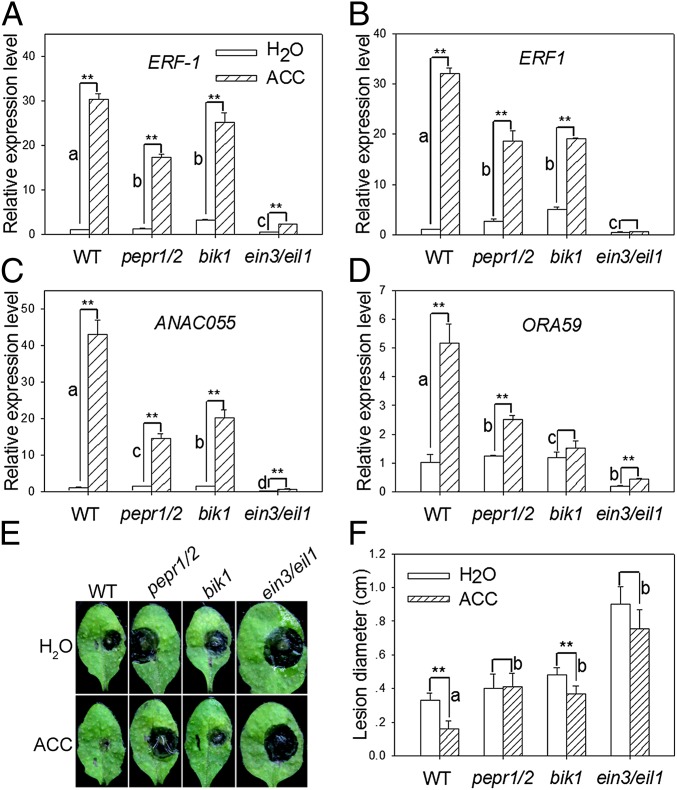
To further assess the role of PEPRs in the ET signaling network, we examined ET response gene expression in the pepr1/pepr2 double mutant. A preliminary RNA-Seq analysis suggested that 164 ET response genes were down-regulated in pepr1/pepr2. In particular, a number of genes encoding transcription factors, including ANAC055, ORA59, ERF1, and ERF-1 (29–31), that are implicated in defenses were down-regulated in pepr1/pepr2 seedlings. We confirmed the expression of ANAC055, ORA59, ERF1, and ERF-1 by real-time RT-PCR (Fig. 5 A–D). In ACC-treated pepr1/pepr2 and bik1 seedlings, the levels of ANAC055 and ORA59 transcripts were reduced to less than half compared with WT seedlings. The levels of ERF-1 and ERF1 transcripts were also reduced, albeit to a lesser extent. The effect of pepr1/pepr2 and bik1 mutations on the ET inducibility was more pronounced when fold change of ACC vs. water treatment was compared. The ET-induced expression of all four genes was nearly abolished in ein3/eil1 seedlings, indicating that these genes are controlled by the canonical ET pathway. All four genes were strongly induced by both Pep1 and Pep2 in a PEPR-dependent manner (Fig. S5), indicating that these genes are controlled by both ET- and PEPR-mediated signaling.
Fig. 5.
PEPRs and BIK1 are required for ET-induced defenses. (A–D) pepr1/pepr2 and bik1 are compromised in ET-induced defense gene expression. Real-time RT-PCR analyses of the indicated genes in plants treated with ACC and H2O. **Significant difference between H2O and ACC treatments. Different letters denote significant difference of difference among different lines (mean + SD; n = 3; P < 0.01; ANOVA; three biological repeats). (E and F) pepr1/pepr2 and bik1 are compromised in ET-induced resistance to B. cinerea. Leaves were pretreated with ACC 13 h before inoculation with B. cinerea. (E) Photograph of disease symptoms taken 2 d after inoculation. (F) Lesion size of B. cinerea-infected leaves. **Significant difference between H2O and ACC treatments. Different letters denote significant difference of difference among different lines (mean + SD; n ≥ 9; P < 0.01; ANOVA; four biological repeats).
To further investigate the role of PEPRs and BIK1 in ET-induced defenses, we examined ET-induced resistance to B. cinerea. A pretreatment of plants with ACC reduced the lesion size to 49% in WT plants but failed to protect pepr1/pepr2 and ein3/eil1 plants (Fig. 5 E and F). In bik1, the ACC treatment reduced the lesion size to ∼70% of the H2O control, and the protection was significantly smaller than in WT. The fls2 mutant showed WT protection by the ACC treatment (Fig. S6), suggesting that PEPRs are specifically required for the ET-induced resistance B. cinerea. Together these results further support an important role of PEPRs and BIK1 in ET-mediated immune regulation.
PEPR1 Kinase Domain Directly Phosphorylates BIK1.
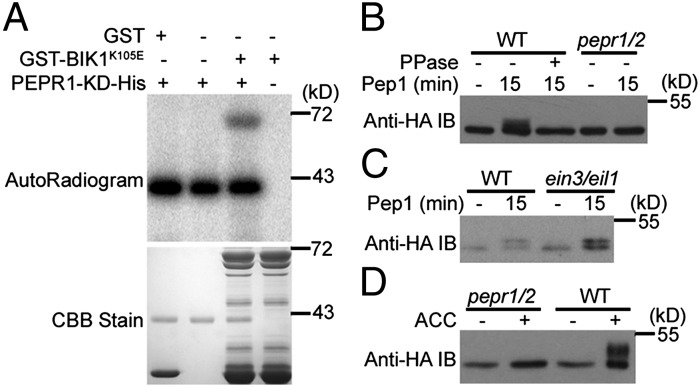
The direct interaction between PEPR1 KD and BIK1 raises the question of whether PEPR1 phosphorylates BIK1. The GST-BIK1K105E recombinant protein, which carries a mutation in the ATP-binding site rendering BIK1 unable to autophosphorylate, was incubated with PEPR1-KD-His in an in vitro kinase assay. GST-BIK1K105E, but not GST, was phosphorylated by the PEPR1 KD (Fig. 6A). GST-BIK1 was also able to phosphorylate the inactive PEPR1-KDK855E-His (Fig. S7), suggesting a reciprocal phosphorylation between PEPR1 and BIK1. Mass spectrometry identified phosphorylation on serine 233, serine 236, and threonine 237 in BIK1K105E when it was coexpressed with PEPR1-KD-His in Escherichia coli, indicating that these residues are major sites phosphorylated by PEPR1 KD (Fig. S8). Serine 236 and threonine 237 are known to be required for BIK1 function in PAMP signaling and ET responses (19, 20, 28, 32). As expected, overexpression of the BIK1S236A/T237A mutant in protoplasts completely blocked Pep1-induced FRK1 reporter expression (Fig. S9A), indicating that these phosphorylation sites are necessary for Pep1 signaling. To determine whether Pep1 induces BIK1 phosphorylation in vivo, we generated transgenic plants carrying a BIK1-HA transgene under the control of the BIK1 native promoter (BIK1::BIK1-HA) in various genetic backgrounds and examined BIK1-HA phosphorylation after Pep1 treatment. As expected, the Pep1 treatment of BIK1::BIK1-HA transgenic plants generated in the WT or ein3/eil1 background induced a protein phosphatase-reversible BIK1-HA mobility shift indicative of phosphorylation (Fig. 6 B and C), indicating that Pep1 induces BIK1 phosphorylation independent of EIN3/EIL1. Likewise, Pep1 also induced BIK1-HA phosphorylation when the latter was transiently expressed in WT protoplasts (Fig. S9B). However, Pep1 failed to induce BIK1-HA phosphorylation in BIK1::BIK1-HA transgenic plants generated in the pepr1/pepr2 background (Fig. 6B), indicating that PEPRs are essential for Pep1-induced BIK1 phosphorylation. These data further support that BIK1 is a direct substrate of PEPR1, and likely PEPR2.
Fig. 6.
PEPRs mediate BIK1 phosphorylation in response to Pep1 and ET. (A) The PEPR1 KD phosphorylates BIK1 in vitro (four independent experiments). GST-BIK1K105E was incubated with PEPR1-KD-His in a kinase buffer containing 32P-γ-ATP, and protein phosphorylation was detected by autoradiography. CBB indicates loading of the protein. (B) Pep1-induces BIK1 phosphorylation in plants (three independent experiments). BIK1::BIK1-HA transgenic plants (WT or pepr1/pepr2 background) were sprayed with H2O (-) or 10 μM Pep1, and tissues were harvested 15 min later. BIK1-HA phosphorylation was detected as a band-shift in an anti-HA immunoblot. PPase, protein phosphatase. (C) Pep1 induces BIK1 phosphorylation in ein3/eil1 plants carrying the BIK1::BIK1-HA transgene (two independent experiments). (D) ACC induces BIK1 phosphorylation in a PEPR1/PEPR2-dependent manner. BIK1::BIK1-HA transgenic plants of pepr1/pepr2 and WT background were treated with ACC for 2 h, and BIK1 phosphorylation was detected as by anti-HA immunoblot (three independent experiments).
We next sought to determine whether the ET-induced BIK1 phosphorylation is mediated by PEPRs. BIK1-HA was phosphorylated upon ACC treatment in the WT background but completely abolished in the pepr1/pepr2 mutant background (Fig. 6D), indicating that PEPR1, and likely PEPR2, is essential for the ET-induced BIK1 phosphorylation. Together these results support that Peps, PEPRs, and BIK1 constitute a pathway acting downstream of the canonical ET signaling cascade to regulate ET responses.
Discussion
In this study, we show that BIK1 and PBL1, but not PBL2 and PBL5, specifically interact with the DAMP receptor PEPR1. Both BIK1 and PBL1, but not PBL2 and PBL5, are required for Pep1-inudced defenses, indicating that BIK1 and PBL1 play a specific role in Pep signaling. We also show that PEPRs contribute to ET-induced seedling growth inhibition, defense gene expression, and B. cinerea resistance in plants, explaining the previous report that BIK1 is required for complete ET-induced growth inhibition (28). Although both BIK1 and PBL1 are required for Pep1-induced responses, PBL1 does not seem to be required for ET responses. The canonical ET signaling components include the ER-localized ET receptors, the CTR1 kinase, EIN2, the F-box proteins EBF1 and EBF2, and transcription factors EIN3 and EIL1 (24). The surprising results that cell surface-localized receptor kinases PEPR1 and PEPR2 play a profound role in ET responses indicate that the ET signaling network is more complex than previously thought.
Because PROPEP genes are known to be induced by ET, the PEPR-mediated ET responses are likely a result of the accumulation of Pep peptides. Indeed, Pep1 and Pep2 treatment can at least partially mimic ET responses in seedlings. Pep1 and Pep2 are fully capable of inhibiting seedling growth in ein2 and/or ein3/eil1 mutants, suggesting that Pep peptides act downstream of EIN3/EIL1 to regulate ET responses, although it is not known whether EIN3 and EIL1 directly bind to the promoter of PROPEP genes.
PEPR1, and likely PEPR2, directly phosphorylates BIK1 on serine 236 and threonine 237 in vitro, suggesting that BIK1 is a substrate of the PEPR1 kinase. In support of this, BIK1 is phosphorylated in response to ET and Pep1 treatments in a PEPR-dependent manner. Overexpression of the BIK1S236A/T237A mutant rendered protoplasts insensitive to Pep1, as indicated by a lack of FRK1 reporter expression. These two phosphorylation sites are also known to be important for ET responses, because the BIK1S236A/T237A mutant was unable to restore ET responsiveness to the bik1 mutant plants (28). Thus, the BIK1 phosphorylation by PEPRs is critical for amplifying the ET signal. BIK1 can also phosphorylate the PEPR1 KD in vitro, raising a possibility that BIK1 and PEPR1 cross-phosphorylate during the activation by Peps.
ET is known to play an important role in regulating plant defenses. ET accumulates upon PAMP-treatment through the MPK-mediated stabilization of ACS2 and ACS6 (22), and this is thought to regulate downstream defense responses. Much of the ET-induced defense responses are considered to be directly controlled by EIN3/EIL1 or other transcription factors regulated by EIN3/EIL1. For example, EIN3 is known to transcriptionally activate ERF1, an important transcription activator that integrates both JA and ET signaling to regulate defenses (29), such as the biosynthesis of indole glucosinolate (33). However, emerging evidence indicates that ET can also enhance PAMP-triggered immunity by inducing FLS2 transcription (25, 26). The companion article by Tintor et al. (34) shows that EIN2 and EIN3/EIL1 are required for elf18-induced signaling and that PEPR1/PEPR2 contributes to EFR-triggered immunity. These results are consistent with our observation that PEPRs and BIK1 are required for ET-triggered defense responses and disease resistance to B. cinerea. Together these results demonstrate that ET can amplify defense signals by activating the PRR complex defined by PEPR1/PEPR2, and BIK1, illustrating a unique mechanism by which ET and an endogenous peptide signaling pathway act in concert to amplify plant innate immunity.
Materials and Methods
DNA Constructs.
To generate the PEPR1-KD-His construct, cDNA encoding the C-terminal 297 amino acids (827–1123) of PEPR1-KD was PCR amplified and inserted between NdeI and XhoI sites of pET30a. The full-length PEPR1 coding region was PCR amplified and cloned into PUC-35S-FLAG-RBS to generate the PEPR1-FLAG construct. The previously described BIK1::BIK1-HA-RBS expression cassette (20) was PCR amplified, mobilized to PENTR/D-TOPO vector (Invitrogen), and subsequently recombined into the Gateway compatible pFAST-G01, which contains a GFP marker specifically expressed in seed coat to facilitate selection of transgenic seeds (35). The resulting plasmid pFAST-BIK1::BIK1-HA was used for plant transformation.
Plant Materials, Growth, and Pathogen Infection.
Arabidopsis plant materials used in this study include WT (Col-0), bik1, pbl1, pbl2 (20, 36), ein3-1/eil1-1 (37), and ein2-1 (38). Plant materials newly made include pbl5 (Fig. S3), the pepr1/pepr2 double mutant generated by crossing pepr1 [Arabidopsis Biological Resource Center (ABRC) stock #CS800015] and pepr2 (ABRC stock #CS800008), and BIK1::BIK1-HA transgenic plants generated by transforming WT, ein3/eil1, and pepr1/pepr2 plants with pFAST-BIK1::BIK1-HA.
For seedling growth inhibition assays, seeds were germinated on 1/2 MS plates with or without different concentrations of ACC or 10 μM Pep peptides and kept in darkness at 23 °C for 5 d. For other assays, plants were grown under 10 h daylight and 14 h night at 23 °C.
Oxidative Burst, Callose, and Dual-Luciferase Reporter Assays.
These assays were performed as previously reported (20). For oxidative burst assay, leaf strips of 4-wk-old plants were induced with 1 μM Pep1, and the relative luminescence units were detected by GLOMAX 96-well microplate luminometer (Promega). For callose deposition assay, 4-wk-old plants were infiltrated with 1.5 μM Pep1 for 12 h. Leaves were then harvested for callose staining and microscopy. For reporter assay, protoplasts transfected with appropriate plasmids were treated with 1 μΜ Pep1 for 3 h, and Dual-Luciferase Reporter assay was performed according to manufacturer’s instruction (Promega).
Yeast Two-Hybrid Library Construction and Screening.
To construct a yeast two-hybrid cDNA library, mRNA was isolated from tissues at different growth stages of salt-treated WT plants. cDNA was synthesized using a commercial kit (Stratagene), ligated into EcoRI/XhoI digested pGADT7 vector (Clontech), and transformed into E. coli XL1-Blue MRF’. The cDNA library contained ∼1.5 × 106 primary transformants. The BIK1 coding region was cloned into pGBKT7 (Clontech) and introduced into yeast cells carrying an Arabidopsis cDNA library via a mating procedure (Clontech). Approximately 2 × 107 yeast clones were screened with BIK1 as bait. To verify interactions between the PEPR1 C terminus (amino acids 1025–1123) and various BIK1 family members, the pGADT7 plasmid containing the PEPR1 C-terminal fragment and pGBKT7 plasmids containing BIK1, PBL1, PBL2, and PBL5 were cotransformed into yeast cells. The resulting transformants were grown on a synthetic dropout (SD) medium (Difco Yeast Nitrogen Base without amino acid; BD) lacking tryptophan and leucine but supplemented with 80 μg/mL X-gal and 25 mM phosphate buffer to detect the lacZ reporter activity. Transformants were also spotted on an SD medium containing 3 mM 3-amino-1,2,4-triazole (3-AT; Sigma) but lacking tryptophan, leucine, and histidine to detect the His reporter gene activity.
GST Pull-Down Assay, in Vitro Kinase Assay, and Coimmunoprecipitation.
GST-tagged proteins were expressed in E. coli, lysed in a buffer containing 25 mM Tris·HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT, and the soluble protein was immobilized to glutathione agarose beads (Sigma) for 1 h at 4 °C. Equal amounts of bacterial lysate containing PEPR1-KD-His recombinant protein were added to the glutathione agarose beads and incubated for 2 h at 4 °C. The beads were washed five to six times with a buffer containing 25 mM Tris·HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT, and the bound protein was eluted with 25 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, and 15 mM reduced glutathione. The amount of PEPR1-KD-His was detected by anti-His immunoblot.
For in vitro kinase assay, the protein storage buffer was changed to 25 mM Tris·HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, and 1 mM DTT using the Millipore Amicon ultra-15 centrifugal filter device. The assay was performed as previously described (20) by incubating 0.5 μg active kinase (GST-BIK1 or PEPR1-KD-His) and 5 μg substrate (PEPR1-KDK855E-His or GST-BIK1K105E) in a 20-μL reaction buffer containing 20 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 100 μM ATP, and 5 μCi 32P-γ-ATP at 30 °C for 30 min. The reaction was stopped by adding 5 μL 5× SDS loading buffer, separated by 12% (wt/vol) SDS/PAGE, and detected by autoradiography.
Coimmunoprecipitation assay was performed with WT Arabidopsis protoplasts expressing BIK1-HA and BAK1-FLAG or PEPR1-FLAG. Anti-HA (Tiangen) immunoprecipitation was carried out as previously described (20), and the amount of BAK1-FLAG or PEPR1-FLAG in the immune precipitates was analyzed by anti-FLAG immunoblot.
Real-Time RT-PCR Analysis.
Eight-day-old seedlings were sprayed with H2O or 0.5 mM ACC in 0.01% silwet L-77, harvested after 10 h, and total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer’s instruction. cDNA was synthesized with the SuperScriptIII First-Strand Kit (Invitrogen). Real-time PCR was performed by using Agilent Mx3005P.
B. cinerea Inoculation.
Four- to 5-wk-old plants were infiltrated with 5 μM Pep1 or coated with 0.5 mM ACC in 0.01% silwet L-77, incubated for 13 or 18 h, and each leaf was inoculated with 5 μL B. cinerea at 5 × 105 conidia/mL. Plants were kept at high humidity for 2 d before disease lesion was measured.
BIK1 Phosphorylation in Vivo.
BIK1 phosphorylation in vivo was examined by using a BIK1 mobility shift assay (20). For Pep1-induced BIK1 phosphoryolation, 10-d-old BIK::BIK1-HA transgenic seedlings were sprayed with 10 μM Pep1 containing 0.01% silwet L-77 15 min before protein extraction. For ACC-induced BIK1 phosphorylation, 4-wk-old BIK1::BIK1-HA transgenic plants were sprayed with 20 μM ACC in 0.01% silwet L-77, and tissues were collected 2 h later.
Mass Spectrometry.
The GST-BIK1K105E protein was expressed in E. coli along with PEPR1-KD-His, purified by using glutathione agarose beads, separated by 10% NuPAGE gel (Invitrogen), and the band containing GST-BIK1K105E excised and destained for nano-LC-MS/MS analysis as previously described (20).
Supplementary Material
Acknowledgments
We thank Ikuko Hara-Nishimura for the pFAST plasmid and Hongwei Guo for advice on ET experiments. J.-M.Z. is supported by Chinese Natural Science Foundation Grant 31230007 and Chinese Ministry of Science and Technology Grants 2011CB100700 and 2010CB835301.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215543110/-/DCSupplemental.
See Commentary on page 5748.
References
- 1.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18(2):465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 3.Kaku H, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103(29):11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20(2):471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SW, et al. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 2009;326(5954):850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- 7.Willmann R, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA. 2011;108(49):19824–19829. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell. 2012;24(8):3406–3419. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA. 2006;103(26):10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi Y, Pearce G, Ryan CA. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA. 2006;103(26):10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell. 2010;22(2):508–522. doi: 10.1105/tpc.109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krol E, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2010;285(18):13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huffaker A, Dafoe NJ, Schmelz EA. ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 2011;155(3):1325–1338. doi: 10.1104/pp.110.166710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 16.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze B, et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem. 2010;285(13):9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23(6):2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Robert-Seilaniantz A, et al. The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 2011;67(2):218–231. doi: 10.1111/j.1365-313X.2011.04591.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16(12):3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo HW, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115(6):667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Q, Guo HW. Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol Plant. 2011;4(4):626–634. doi: 10.1093/mp/ssr042. [DOI] [PubMed] [Google Scholar]
- 25.Boutrot F, et al. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA. 2010;107(32):14502–14507. doi: 10.1073/pnas.1003347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mersmann S, Bourdais G, Rietz S, Robatzek S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010;154(1):391–400. doi: 10.1104/pp.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009;21(8):2527–2540. doi: 10.1105/tpc.108.065193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laluk K, et al. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell. 2011;23(8):2831–2849. doi: 10.1105/tpc.111.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15(1):165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pré M, et al. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147(3):1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng X-Y, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11(6):587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng F, et al. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485(7396):114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 33.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323(5910):95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tintor N, et al. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci USA. 2013;110:6211–6216. doi: 10.1073/pnas.1216780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada TL, Shimada T, Hara-Nishimura I. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 2010;61(3):519–528. doi: 10.1111/j.1365-313X.2009.04060.x. [DOI] [PubMed] [Google Scholar]
- 36.Veronese P, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18(1):257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso JM, et al. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA. 2003;100(5):2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139(3):1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.