Abstract
Small cell lung cancer (SCLC) is a disease characterized by aggressive clinical behavior and lack of effective therapy. Due to its tendency for early dissemination, only a third of patients have limited-stage disease at the time of diagnosis. SCLC is thought to derive from pulmonary neuroendocrine cells. Although several molecular abnormalities in SCLC have been described, there are relatively few studies on epigenetic alterations in this type of tumor. Here, we have used methylation profiling with the methylated CpG island recovery assay (MIRA) in combination with microarrays and conducted the first genome-scale analysis of methylation changes that occur in primary SCLC and SCLC cell lines. Among the hundreds of tumor-specifically methylated genes discovered, we identified 73 gene targets that are methylated in more than 77% of primary SCLC tumors, most of which have never been linked to aberrant methylation in tumors. These methylated targets have potential for biomarker development for early detection and therapeutic management of SCLC. SCLC cell lines had a ~3-fold greater number of hypermethylated genes than primary tumors. Gene ontology analysis indicated a significant enrichment of methylated genes functioning as transcription factors and in processes of neuronal differentiation. Motif analysis of tumor-specific methylated regions identified enrichment of binding sites for several neural cell fate-specifying transcription factors including NEUROD1, HAND1, ZNF423 and REST. We hypothesize that two potential mechanisms, loss of cell fate-determining transcription factors by methylation of their promoters and functional inactivation of their corresponding genomic binding sites by DNA methylation, can promote a differentiation defect of neuroendocrine cells thus enhancing the ability of tumor progenitor cells to transition towards SCLC.
Introduction
Lung cancer is divided by histology into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). SCLC represents about 15% of all lung cancer cases and is one of the most lethal forms of cancer with properties of high mitotic rate and early metastasis.1 It is distinctly characterized by small cells with poorly defined cell borders and minimal cytoplasm, rare nucleoli and finely granular chromatin. Although SCLC patients initially respond to chemotherapy and radiation therapy, the disease recurs in the majority of patients. Because of the aggressiveness of SCLC and the lack of effective therapy and early diagnosis, without treatment the median survival time for SCLC is only two to four months. With current treatment modalities, the median survival times for limited stage disease, less than 5% of the total, is 16–24 months, and for extensive disease, 7–12 months, in spite of the fact that 60–80% of patients respond to therapy. It is essential to gain a better understanding of the molecular pathogenesis of the disease and to identify molecular alterations, which could lead to improved results in early detection and a means of assessing response to therapy.
Several studies have identified abnormalities within tumor suppressor genes, oncogenes, signaling pathways, receptor kinases and growth factors that have a proven role in the pathogenesis of various other human cancers. About 90% of SCLC patients’ DNA samples have mutations in the TP53 gene.2, 3 Similarly, another tumor suppressor gene, retinoblastoma (RB), is either deleted or mutated in the majority (about 90%) of SCLCs.2, 4 In addition, higher expression of the MYC family of oncogenes has been found in SCLC cell lines, xenografts and fresh tumor specimens.5–7 Abnormalities in various receptor tyrosine kinase (RTK) families are commonly found in the majority of SCLC cases. These changes are associated with a more aggressive tumor growth, resistance to therapy and poor prognosis.8, 9 The phosphoinositide 3-kinase (PI3K)/AKT pathway is defective in SCLC patients’ tumors. Nearly two thirds of SCLCs have phosphorylated AKT9 and this constitutively active kinase can modulate a variety of cellular functions such as cell proliferation, survival, motility, adhesion and differentiation.8 The cellular origin of SCLC is yet to be proven definitively. Recent studies in mice indicated that neuroendocrine cells seem to be the predominant cells of origin of SCLC.10, 11
SCLC is also characterized by common deletion of the fragile histidine triad (FHIT) gene, located at 3p14.2 Similarly, chromosome 3p21 is another locus, which is frequently subjected to loss in almost all SCLCs, and this event is thought to be an early event in lung cancer pathogenesis.12 At 3p21.3, there are several candidate tumor suppresser genes, including the Ras association domain family member 1A (RASSF1A), tumor suppressor candidate 2 (TUSC2, also known as FUS1), semaphorin 3B (SEMA3B) and semaphorin 3F (SEMA3F).13, 14
In contrast to the genetic alterations discussed above, epigenetic aberrations, specifically DNA methylation changes found in SCLC tumors, have not been studied so far in a comprehensive manner. DNA methylation analysis might provide vital information that could shed light on mechanisms of disease initiation, development and progression, as well as lead to cancer biomarker discovery.15, 16 There are several gene-specific DNA methylation studies for SCLC. For example, promoter hypermethylation of the tumor suppressor gene RASSF1A and subsequent suppression of its expression is found in almost all of the SCLC tumors.17, 18 Another study found caveolin-1 (CAV1) gene methylation in over 90% the tested SCLC cell lines.19
Lack of genome-wide DNA methylation studies in SCLC prompted us to undertake this task. We applied the methylated-CpG island recovery assay (MIRA), which has shown excellent sensitivity for identification of methylated genomic regions in cancer20–23 to map DNA methylation patterns at promoters and CpG islands of primary SCLC tumors, SCLC cell lines, and normal lung control samples.
Results
Identification of methylated genes in human SCLC tissue on a genome-wide platform
The MIRA technique, used in combination with microarray analysis, is a high-resolution mapping technique and has proven successful for profiling global DNA methylation patterns in non-small cell lung cancers and other tumors.22–25 In the present study, we have applied this sensitive method to study the methylation status of CpG islands and promoters in SCLC to investigate the potential role of methylation changes in the initiation and development of SCLC, as well as to discover potential biomarkers for better management of the disease. Eighteen human primary SCLC and five SCLC cell line DNA samples were screened for methylation by MIRA-based microarrays. DNAs from five normal healthy lung tissues adjacent to the tumor and obtained at the time of surgical resection were used as controls in the MIRA analysis. DNA was subjected to MIRA enrichment as described previously26, 27 and subsequent microarray analysis was performed on 720k Nimblegen CpG island plus promoter arrays.
Microarray data analysis
In order to increase the specificity of MIRA-based enrichment signals, we chose to call peaks based on different quantiles of four neighboring probes. Peaks were then calculated using the base functions of the Bioconductor package Ringo.28 Table 1 shows the specificity and sensitivity of this approach relative to different quantile ranges using DNA from the SCLC cell line SW1271. Based on the validations conducted by COBRA single gene methylation assays, we chose an 80% cutoff for medium to strongly methylated regions and a cutoff below 56% defined as not methylated. Thus, compared to the conventional NimbleScan method using the default settings, we could increase the sensitivity of methylation peak detection to 94% without decreasing specificity. As this threshold was defined for one SCLC cell line, we tested the same settings for primary small lung cancer samples and did not observe a significant increase of false positive predicted hypermethylated regions.
Table 1.
Validation of microarray results by COBRA assays
| Top Quantile | No of targets tested a | Met | UnMet | PCR fails | %Met | %UnMet |
|---|---|---|---|---|---|---|
| 99% | 10 | 9 | - | 1 | 100 | 0 |
| 95% | 10 | 9 | - | 1 | 100 | 0 |
| 90% | 10 | 9 | 1 | - | 90 | 10 |
| 85% | 10 | 9 | 1 | - | 90 | 10 |
| 80% | 10 | 7 | 3 | - | 70 | 30 |
| 70% | 14 | 3 | 5 | 6 | 37.5 | 62.5 |
| 60% | 19 | 3 | 12 | 4 | 20 | 80 |
| 50% | 13 | 2 | 11 | - | 15 | 85 |
COBRA was performed for each quantile category with bisulfite-converted DNA from the SW1271 cell line. Results were tabulated for number of methylated (Met) and unmethylated (UnMet) genes in these various categories.
Using the peak identification algorithm described in the Materials and Methods section, we identified approximately 15, 000 methylation peaks in each sample (Supplementary Table 1). Our clustering analysis of tumor samples and controls showed that SCLC cell lines clustered together and that four of the five normal samples were close to each other, but different tumor samples occupied different spaces in the dendrogram (Supplementary Figure 1).
Taking into account that we had 18 tumor samples and 5 normal samples for microarray data analysis, we defined a stringent tumor-specific methylated region as the overlapping region that meets the minimum 80% quantile criterion in 14 of 18 tumors and is below the 56% quantile in 4 of 5 normal tissues. A less stringent set was defined as an overlap between at least 6 peaks from tumor samples out of 18, using the same criteria as above. Thus, we were mainly comparing strongly methylated regions versus poorly methylated regions. Although small methylation level differences could not be picked up this way, the aim of discovering uniquely strongly methylated and tumor-specific regions was well supported by this approach.
Methylated genes in primary SCLC
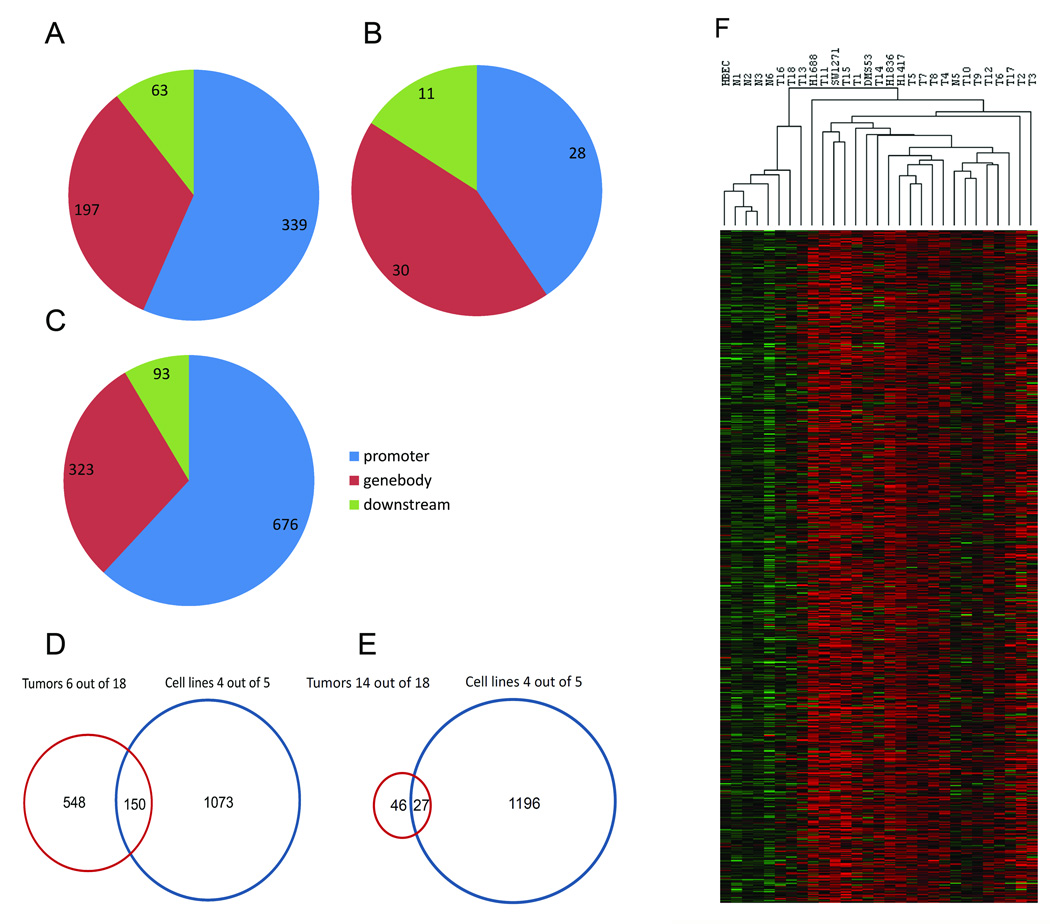
Supplementary Figure 2 shows examples of tumor-specific methylation peaks at the PROX1, CCDC140, PAX3, and SIM1 genes located on chromosomes 1, 2, and 6, respectively. Supplementary Figure 3 shows extensive tumor-specific methylation of the HOXD cluster on chromosome 2. Compilation of tumor-specific methylation peaks revealed a total of 698 regions in 6 out of 18 tumors (≥ 33% of SCLC tumors) compared to normal lung DNA, which represented 339 ensembl gene IDs for promoter-related tumor-specifically methylated regions (defined as −5000 to +1000 relative to the TSS), 197 ensembl gene IDs related to peaks mapped to the gene bodies and 63 ensembl gene IDs for peaks mapped downstream of the corresponding genes (Figure 1A; Supplementary Table 2). Individual primary SCLCs contained between 366 and almost 1, 500 tumor-specific methylation peaks (Supplementary Table 3).
Figure 1. Mapping of tumor-specific methylation peaks in primary SCLC and SCLC cell lines.
A. Localization of the methylation peaks in primary SCLC (6 or more out of 18 tumors methylated; i.e. peaks that meet the minimum 80% quantile criterion in 6 of 18 tumors) relative to gene position. B. Localization of the methylation peaks in primary SCLC (14 or more out of 18 tumors methylated) relative to gene position. C. Localization of the methylation peaks in SCLC cell lines (4 or more out of 5 cell lines methylated) relative to gene position. D. Overlap of methylation peaks between SCLC primary tumors (6 or more out of 18 tumors methylated) and SCLC cell lines (4 or more out of 5 cell lines methylated). E. Overlap of methylation peaks between SCLC primary tumors (14 or more out of 18 tumors methylated) and SCLC cell lines (4 or more out of 5 cell lines methylated). F. Cluster analysis of methylation peaks. Methylation peaks found in at least 33% of tumor samples but not in normal samples were identified. Then the data were subjected to hierarchical clustering with Euclidean distance and average linkage method using Cluster v3.0 and visualized in Java TreeView. Red, methylated state; green, unmethylated state.
There were 73 tumor-specific methylated peaks which were found in at least 14 out of 18 SCLC tumors (>77% of SCLC tumors) that corresponded to 28 ensembl gene IDs for promoters, 30 ensembl gene IDs for gene bodies and 11 for downstream regions (Figure 1B). These methylated genes from 77% or more of the SCLC tumors are presented in Table 2 and in Supplementary Table 4, for more detailed information.
Table 2.
Gene targets methylated in 77% or more of primary SCLCs.
| chr | start.peak | end.peak | hgnc_symbol | description |
|---|---|---|---|---|
| 6 | 27647872 | 27648246 | ||
| 1 | 91189238 | 91189687 | BARHL2 | BarH-like homeobox 2 [Source:HGNC Symbol;Acc:954] |
| 10 | 124901911 | 124902685 | HMX2 | H6 family homeobox 2 [Source:HGNC Symbol;Acc:5018] |
| 15 | 53087134 | 53087683 | ONECUT1 | one cut homeobox 1 [Source:HGNC Symbol;Acc:8138] |
| 9 | 100611180 | 100611554 | FOXE1 | forkhead box E1 (thyroid transcription factor 2) [Source:HGNC Symbol;Acc:3806] |
| 17 | 59529794 | 59530268 | TBX4 | T-box 4 [Source:HGNC Symbol;Acc:11603] |
| 1 | 214153078 | 214153777 | PROX1 | prospero homeobox 1 [Source:HGNC Symbol;Acc:9459] |
| 14 | 95239173 | 95240547 | GSC | goosecoid homeobox [Source:HGNC Symbol;Acc:4612] |
| 21 | 38068981 | 38069055 | SIM2 | single-minded homolog 2 (Drosophila) [Source:HGNC Symbol;Acc:10883] |
| 6 | 117584283 | 117584857 | VGLL2 | vestigial like 2 (Drosophila) [Source:HGNC Symbol;Acc:20232] |
| 14 | 37124350 | 37124799 | PAX9 | paired box 9 [Source:HGNC Symbol;Acc:8623] |
| 2 | 177004205 | 177004604 | ||
| 14 | 36991675 | 36992549 | NKX2-1 | NK2 homeobox 1 [Source:HGNC Symbol;Acc:11825] |
| 1 | 197879403 | 197880252 | LHX9 | LIM homeobox 9 [Source:HGNC Symbol;Acc:14222] |
| 11 | 32455050 | 32455624 | WT1-AS | WT1 antisense RNA (non-protein coding) [Source:HGNC Symbol;Acc:18135] |
| 13 | 112719925 | 112720174 | SOX1 | SRY (sex determining region Y)-box 1 [Source:HGNC Symbol;Acc:11189] |
| 21 | 38069706 | 38069780 | SIM2 | single-minded homolog 2 (Drosophila) [Source:HGNC Symbol;Acc:10883] |
| 2 | 176956605 | 176956754 | HOXD13 | homeobox D13 [Source:HGNC Symbol;Acc:5136] |
| 9 | 129566330 | 129566704 | ZBTB43 | zinc finger and BTB domain containing 43 [Source:HGNC Symbol;Acc:17908] |
| 3 | 172167182 | 172167256 | GHSR | growth hormone secretagogue receptor [Source:HGNC Symbol;Acc:4267] |
| 1 | 230777303 | 230777452 | COG2 | component of oligomeric golgi complex 2 [Source:HGNC Symbol;Acc:6546] |
| 3 | 27765097 | 27765996 | EOMES | eomesodermin [Source:HGNC Symbol;Acc:3372] |
| 20 | 30639265 | 30639939 | HCK | hemopoietic cell kinase [Source:HGNC Symbol;Acc:4840] |
| 3 | 183274057 | 183274331 | KLHL6 | kelch-like 6 (Drosophila) [Source:HGNC Symbol;Acc:18653] |
| 12 | 114846668 | 114847217 | TBX5 | T-box 5 [Source:HGNC Symbol;Acc:11604] |
| 4 | 122685401 | 122685475 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7ENT1] | |
| 2 | 182547581 | 182547655 | ||
| 20 | 44880344 | 44880693 | CDH22 | cadherin 22, type 2 [Source:HGNC Symbol;Acc:13251] |
| 9 | 21402751 | 21403100 | IFNA12P | interferon, alpha 12, pseudogene [Source:HGNC Symbol;Acc:5443] |
| 7 | 97360940 | 97362189 | TAC1 | tachykinin, precursor 1 [Source:HGNC Symbol;Acc:11517] |
| 2 | 223162732 | 223163206 | CCDC140 | coiled-coil domain containing 140 [Source:HGNC Symbol;Acc:26514] |
| 7 | 129422815 | 129423514 | ||
| 2 | 192711381 | 192711755 | ||
| 6 | 27107272 | 27107346 | HIST1H4I | histone cluster 1, H4i [Source:HGNC Symbol;Acc:4793] |
| 2 | 176969205 | 176970504 | HOXD11 | homeobox D11 [Source:HGNC Symbol;Acc:5134] |
| 19 | 9608951 | 9609250 | ZNF560 | zinc finger protein 560 [Source:HGNC Symbol;Acc:26484] |
| 7 | 27282651 | 27282900 | EVX1 | even-skipped homeobox 1 [Source:HGNC Symbol;Acc:3506] |
| 2 | 223163332 | 223163406 | PAX3 | paired box 3 [Source:HGNC Symbol;Acc:8617] |
| 7 | 27282951 | 27283025 | EVX1 | even-skipped homeobox 1 [Source:HGNC Symbol;Acc:3506] |
| 7 | 8474326 | 8475225 | NXPH1 | neurexophilin 1 [Source:HGNC Symbol;Acc:20693] |
| 4 | 174452351 | 174452925 | Nbla00301 (NBLA00301), non-coding RNA [Source:RefSeq DNA;Acc:NR_003679] | |
| 4 | 13545178 | 13545427 | NKX3-2 | NK3 homeobox 2 [Source:HGNC Symbol;Acc:951] |
| X | 111325120 | 111325194 | TRPC5 | transient receptor potential cation channel, subfamily C, member 5 [Source:HGNC Symbol;Acc:12337] |
| 6 | 100911555 | 100911904 | SIM1 | single-minded homolog 1 (Drosophila) [Source:HGNC Symbol;Acc:10882] |
| 14 | 29243250 | 29243899 | C14orf23 | chromosome 14 open reading frame 23 [Source:HGNC Symbol;Acc:19828] |
| 5 | 172660770 | 172660844 | NKX2-5 | NK2 transcription factor related, locus 5 (Drosophila) [Source:HGNC Symbol;Acc:2488] |
| 2 | 220196257 | 220197006 | RESP18 | regulated endocrine-specific protein 18 homolog (rat) [Source:HGNC Symbol;Acc:33762] |
| 9 | 126776030 | 126776479 | LHX2 | LIM homeobox 2 [Source:HGNC Symbol;Acc:6594] |
| 1 | 165323302 | 165323951 | LMX1A | LIM homeobox transcription factor 1, alpha [Source:HGNC Symbol;Acc:6653] |
| 2 | 119603031 | 119603180 | EN1 | engrailed homeobox 1 [Source:HGNC Symbol;Acc:3342] |
| 12 | 63543634 | 63544008 | AVPR1A | arginine vasopressin receptor 1A [Source:HGNC Symbol;Acc:895] |
| 8 | 97170050 | 97170499 | GDF6 | growth differentiation factor 6 [Source:HGNC Symbol;Acc:4221] |
| 1 | 47694839 | 47695213 | TAL1 | T-cell acute lymphocytic leukemia 1 [Source:HGNC Symbol;Acc:11556] |
| 13 | 84453425 | 84453824 | SLITRK1 | SLIT and NTRK-like family, member 1 [Source:HGNC Symbol;Acc:20297] |
| 4 | 174448251 | 174448725 | HAND2 | heart and neural crest derivatives expressed 2 [Source:HGNC Symbol;Acc:4808] |
| 2 | 176977280 | 176977729 | HOXD10 | homeobox D10 [Source:HGNC Symbol;Acc:5133] |
| 5 | 37835994 | 37836168 | GDNF | glial cell derived neurotrophic factor [Source:HGNC Symbol;Acc:4232] |
| 9 | 37029751 | 37030525 | PAX5 | paired box 5 [Source:HGNC Symbol;Acc:8619] |
| 14 | 29247325 | 29247499 | C14orf23 | chromosome 14 open reading frame 23 [Source:HGNC Symbol;Acc:19828] |
| 7 | 8483051 | 8483825 | NXPH1 | neurexophilin 1 [Source:HGNC Symbol;Acc:20693] |
| 6 | 154360508 | 154360857 | OPRM1 | opioid receptor, mu 1 [Source:HGNC Symbol;Acc:8156] |
| 20 | 58569381 | 58569455 | CDH26 | cadherin 26 [Source:HGNC Symbol;Acc:15902] |
| 9 | 21968201 | 21968875 | C9orf53 | chromosome 9 open reading frame 53 [Source:HGNC Symbol;Acc:23831] |
| 16 | 49311725 | 49312274 | CBLN1 | cerebellin 1 precursor [Source:HGNC Symbol;Acc:1543] |
| 8 | 9756191 | 9756540 | MIR124-1 | microRNA 124-1 [Source:HGNC Symbol;Acc:31502] |
| 5 | 170741921 | 170741995 | TLX3 | T-cell leukemia homeobox 3 [Source:HGNC Symbol;Acc:13532] |
| 20 | 21488326 | 21488925 | NKX2-2 | NK2 homeobox 2 [Source:HGNC Symbol;Acc:7835] |
| 5 | 170743496 | 170744170 | TLX3 | T-cell leukemia homeobox 3 [Source:HGNC Symbol;Acc:13532] |
| 5 | 172672295 | 172672844 | Y RNA [Source:RFAM;Acc:RF00019] | |
| 2 | 177027180 | 177027529 | HOXD4 | homeobox D4 [Source:HGNC Symbol;Acc:5138] |
| 4 | 85402627 | 85403376 | NKX6-1 | NK6 homeobox 1 [Source:HGNC Symbol;Acc:7839] |
| 15 | 96911497 | 96912071 | MIR1469 | microRNA 1469 [Source:HGNC Symbol;Acc:35378] |
| 15 | 89949372 | 89949871 | MIR9-3 | microRNA 9-3 [Source:HGNC Symbol;Acc:31646] |
| 3 | 50377447 | 50378846 | RASSF1A (*) | Ras association (RalGDS/AF-6) domain family member 1 [Source:HGNC Symbol;Acc:9882] |
indicates a previously validated gene with a lower threshold for normal tissues than used for the other regions.
Identification of methylated genes in human SCLC lines
Due to the limited availability of primary SCLC tissue, we added several SCLC cell lines originally derived from primary tumor sites. Due to the unavailability of neuroendocrine cells, which are believed to be the cell of origin of SCLC, 10 we chose normal bronchial epithelial cells as a control for these studies. Clustering analysis based on the total methylation peaks of SCLC cell lines showed that all cell lines cluster tightly together (Supplementary Figure 1). Further analysis of these methylated peaks for tumor cell line-specific peaks revealed 1223 unique tumor-specific peaks found in 4 out of 5 SCLC cell lines (≥80% of SCLC cell lines) compared to methylated peaks form normal bronchial epithelial cells (Supplementary Table 5). These peaks represented 676 ensembl gene IDs mapped to promoter regions, 323 ensembl gene IDs corresponding to methylated regions in the gene body and 93 ensembl gene IDs where the hypermethylated regions could be located downstream of genes (Figure 1C). Individual cell lines contained between 2779 and 4485 cell line-specific methylation peaks (Supplementary Table 3), numbers that are greater than the tumor-specific methylation peaks found in primary SCLCs. To further determine if the SCLC cell line model is appropriate to study the role of SCLC tumor methylation, we compared SCLC tumor-specific methylated regions with SCLC cell line-specific methylated regions. Strikingly, there was only a relatively small group of (<20%) of SCLC cell line-specific genes found to be commonly (>6 of 18) methylated in primary SCLC tumors and vice versa (i.e., only ~21% of SCLC primary tumor peaks matched with those of frequent SCLC cell line methylation) (Figure 1D). When we determined the overlap between peaks methylated in 14/18 tumors and 4 of 5 cell lines, the number of overlapped genes dropped to 27 (Figure 1E). This small overlap confirms that methylation patterns in SCLC cell lines do not well recapitulate primary SCLC. We mapped the location of tumor-specific methylation peaks relative to promoters, gene bodies and locations downstream of genes (Figure 1A–C). The distribution patterns were similar for peaks found in ≥6/18 tumors and in cell lines but for the most frequently methylated genes (≥14/18) the peaks tended to be more commonly localized in gene bodies and downstream (Figure 1B). Cluster analysis of methylation peaks in normal and tumor samples is shown in Figure 1F.
Validation of gene-specific methylation in SCLC samples
We further validated tumor-specific methylation peaks discovered by microarray analysis for several of the targets by combined bisulfite restriction analysis (COBRA) assay. In this assay, bisulfite-converted DNA is PCR-amplified using gene-specific primers and is then digested with a restriction endonuclease, either BstUI or TaqI, which recognize the sequences 5'-CGCG- 3’ or 5’TCGA-3’, respectively. The cytosines in unmethylated restriction sites are converted by sodium bisulfite and PCR and resist digestion, whereas methylated sites remain unchanged and are cleaved by the enzymes. The digested fragments visualized on agarose gels are thus indicative of methylated restriction sites in the region analyzed. We performed extensive validation analysis by COBRA to confirm the tumor-specific methylated regions (Supplementary Figure 4). Representative examples of COBRA results are shown for the genes DMRTA2, MIR-129-2 and GALNTL1. In total, we inspected the methylation status of 11 genes (GALNTL1, MIR-10A, MIR-129-2, MIR-196A2, MIR-615, MIR-9-3, AMBRA1, HOXD10, PROX1, ZNF672 and DMRTA2) based on the various degrees of methylation obtained from the list of differentially methylated targets. Results for all the targets are presented in Supplementary Table 6. COBRA analysis revealed that our microarray analysis is highly reliable with over 93% accuracy and only ~4% false negative and ~3% false positive hits.
To further confirm the COBRA results of the methylated genes GALNTL1 and DMRAT2, we sequenced bisulfite-converted DNA from SCLC tumor and matched normal lung samples (Supplementary Figure 4). Normal control lung DNA samples showed either no or very low levels of methylation across the CpG dinucleotides tested in contrast to SCLC tumor DNA samples, which were heavily methylated.
Gene expression and methylation status
For the SCLC cell lines SW1271, H1836 and H1688, and HBECs, Affymetrix gene expression analysis was performed and hypermethylated regions in the SCLC cell lines were compared with their associated probe expression changes. On a global level, we could not detect a correlation between the tumor-specific hypermethylated regions and downregulation of associated genes. This phenomenon has been observed in other tumor methylation studies. Some of the reasons for this lack of correlation are that (1) genes that become methylated in tumors frequently are already expressed at very low levels in corresponding normal tissues, 29–32 (2) methylation-independent mechanisms (such as chromatin modifications) are responsible for expression changes, 33 and (3) methylation of alternative promoters obscures such correlations.27, 34 Unlike the methylation patterns, the expression signals of the individual tumor cell lines were not highly correlated to each other when compared to the control cell line (as seen by principal component analysis; data not shown).
Functional pathway analysis of methylated genes
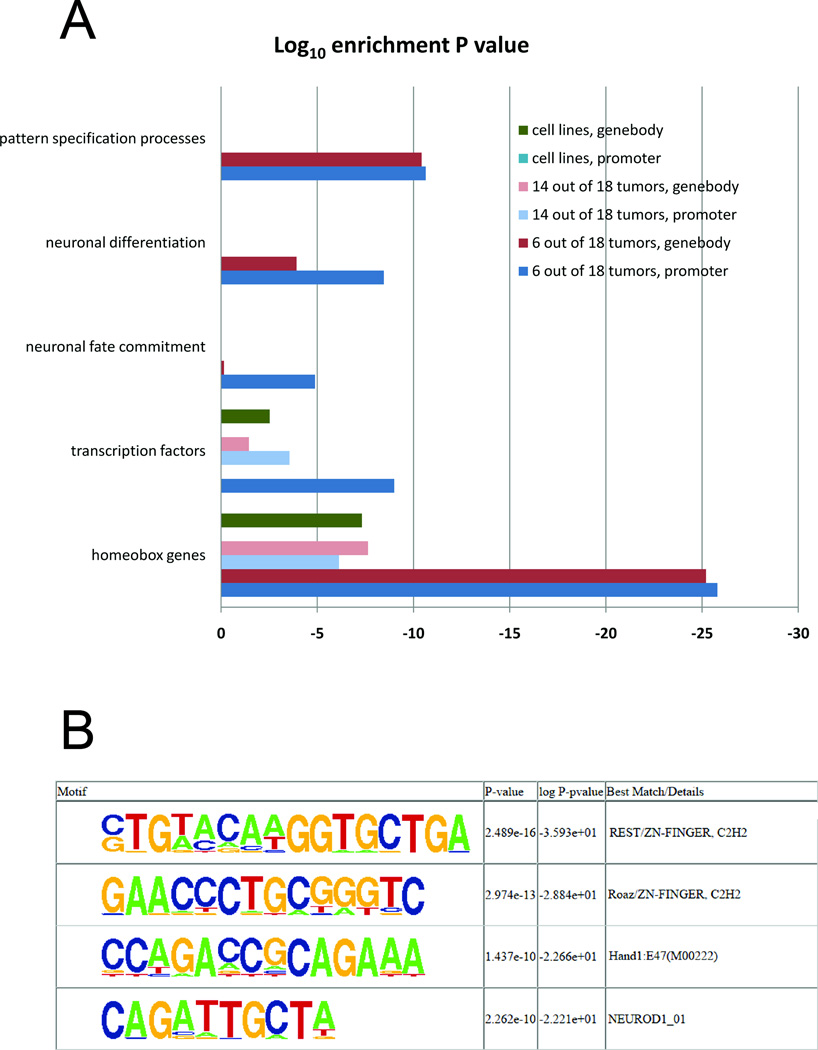
For the two stringencies that were defined (≥6 out of 18 tumors specifically hypermethylated and ≥14 out of 18 tumors specifically hypermethylated), we performed a functional annotation clustering, for promoter proximal tumor-specifically methylated regions and gene body-associated tumor-specifically methylated regions. For ≥6 out of 18 tumor-specific promoter proximal methylated regions, two main annotation clusters could be identified, one for homeobox genes (P-value 1.6E-26, Bonferroni corrected) and one for transcription factors in general (1.0E-09) (Figure 2A; Supplementary Table 7). More specifically, clusters for neuronal fate commitment (1.3E-5), neuronal differentiation (3.5E-9) and pattern specification processes (2.3E-11) showed the strongest enrichment. In comparison, hypermethylated regions in gene bodies showed similar functional enrichment clusters for homeobox genes (6.2E-26) and pattern specification processes (3.8E-11), but significantly less enrichment for neuronal fate commitment (7.0E-1) and for neuronal differentiation (1.2E-4) (Supplementary Table 8), suggesting that the latter functional categories are more related to promoter-specific methylation (Figure 2A).
Figure 2. Functional annotation and motif finding analysis.
A. Shown are DAVID functional analysis clusters which contained the highest enrichment scores in all three categories: 33% or more of tumors, 77% or more of tumors and cell lines. For more details see Materials and Methods. B. Motif finding analysis. Significantly enriched consensus motifs for REST, Roaz/ZNF423, Hand1, and NEUROD1 are shown.
Concerning functional enrichment for tumor-specifically hypermethylated regions for the majority of tumors (≥14 out of 18 tumors), clusters with significantly less enrichment compared to their less significant counterpart (≥6 out of 18) could only be obtained for homeobox genes (7.5E-7 for promoter regions and 2.3E-8 for gene bodies) and transcription factors (2.8E-4 for promoter regions and 3.6E-2 for gene bodies), which can be partly explained by the lower number of genes in this category (Supplementary Tables 9 and 10). Lung development was another significantly enriched category for promoter methylation (Supplementary Table 9).
With regards to the cell lines, genes associated with hypermethylated regions in the five SCLC cell lines compared to the control cell line, homeobox-related functional terms and transcription factor-related terms were significantly enriched only for gene body associated tumor peaks (4.8E-8 for homeobox genes and 3.0E-3 for transcription factors, Bonferroni corrected) but the strong enrichment for these categories observed for promoter regions in the tumor tissues was not present at all for the cell line models (Supplementary Tables 11 and 12). This probably reflects a much greater number and higher diversity of methylation events observed in the cell lines.
For targets methylated in ≥14 out of 18 tumors and in ≥4 out of 5 cell lines (Supplementary Table 13), we again observed an enrichment in the same functional categories. This group of genes contained a number of genes involved in neuronal or neuroendocrine differentiation, such as EOMES/TBR2, the gene TAC1, which encodes the neuropeptide substance P, and RESP18, encoding a neuroendocrine-specific protein.
Motif discovery
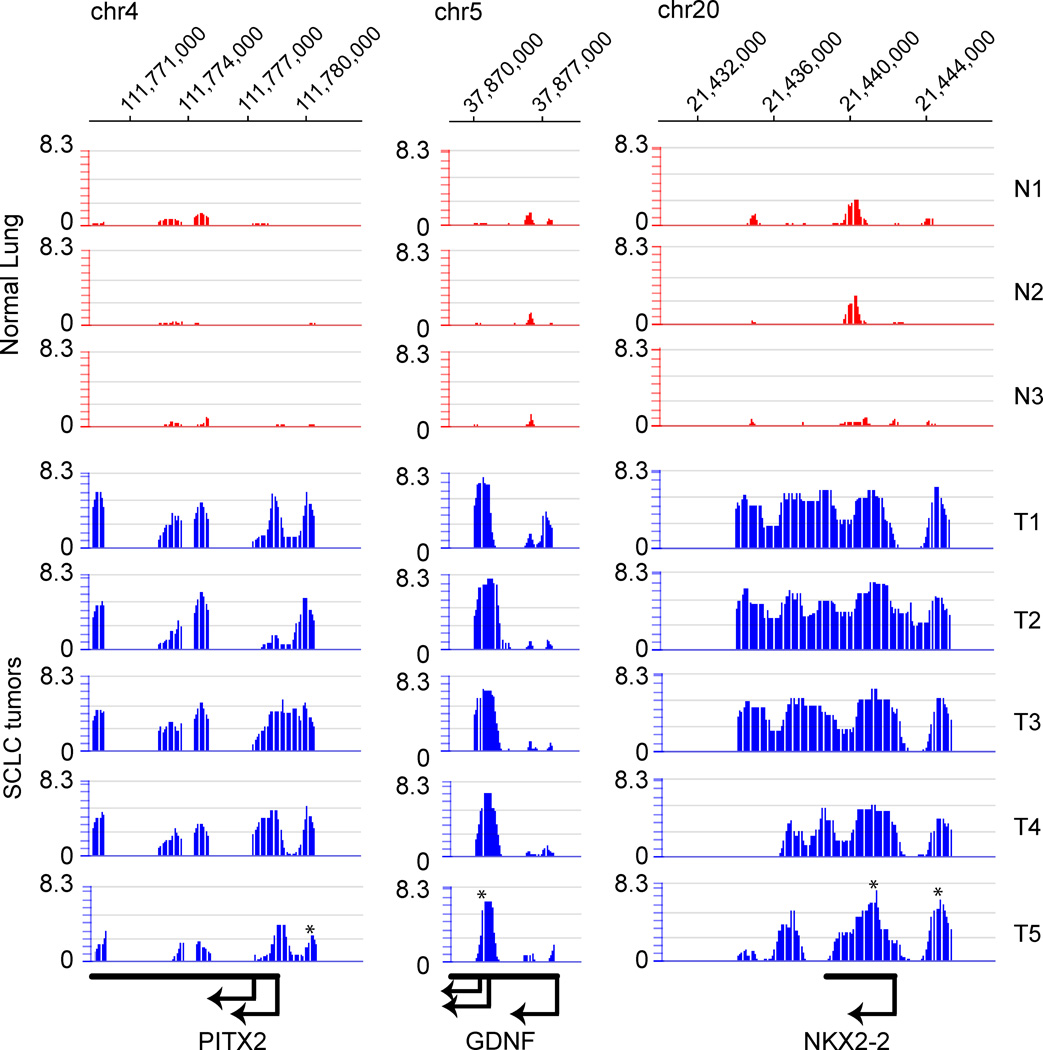
We next used the de novo motif discovery algorithm HOMER35 to search for sequence patterns that are associated with regions that are specifically methylated in SCLC tumor samples for at least 33% of the tumors and were able to identify a set of non-redundant sequence motifs that were highly enriched in comparison with all non-tumor-specifically methylated regions on the array. Transcription factors, which were falling into this category, were REST/NRSF (2.5e-16), ZNF423 (3.0e-13), HAND1 (1.44e-10) and NEUROD1 (2.3e-10) (Figure 2B). Examples of methylated NEUROD1 targets are shown in Figure 3. The majority of the sequence motifs identified in methylated regions were enriched within the proximal promoter regions of known genes. The highest enrichment was based on redundant sequence structures and for those that were not, we demanded a stringent alignment with matching transcription factor binding sites and a low number of occurrences in the background set, which contained all possible methylation sites. REST, ZNF423, HAND1 and NEUROD1 contained non-redundant sequences, a maximal mismatch of 2 bp to the identified de novo motif and were selectively enriched in the target sequence set. As such, the identified motifs might not be representative for the whole tumor-specific target set but shed light on sub-regulatory networks with a possibly major impact on the phenotype of SCLC. For example, NEUROD1 and HAND1 binding sites were found in methylated targets representing genes involved in neuronal cell fate commitment such as GDNF, NKX2-2, NKX6-1, EVX1, and SIM2 (Suppl. Tables 2 and 14). Methylation of these binding sites suggests a model in which these transacting factors were lost during tumorigenesis rendering their target sites susceptible to methylation. To analyze this scenario further, we focused on the NEUROD1 transcription factor. Indeed, expression of NEUROD1 proved to be undetectable by a sensitive RT-PCR assay (Supplementary Figure 5) in the four SCLC cell lines tested and it was expressed at very low levels in human bronchial epithelial cells. In SCLC cell lines and, importantly, also in primary SCLC tumors, the promoter of NEUROD1 was heavily methylated (Supplementary Figure 6A, B) consistent with a lack of expression. In addition, we found increased methylation at the promoters of HAND1 and REST in SCLC cell lines and in primary tumors (Supplementary Figure 6).
Figure 3. Examples of tumor-specific methylation of NEUROD1 target genes in SCLC.
The top of the Figure indicates the chromosomal coordinates according to the UCSC Genome browser hg19. Gene names and direction of transcription are shown at the bottom of the Figure. The Nimblegen array data (methylated fraction versus input) are shown for three normal lung tissues (red) and five primary SCLC tumors (blue). The methylation signal is shown plotted along the chromosome as a P value score. Therefore, the minimum number on the y-axis is 0 (when P = 1). The P value score was obtained by NimbleScan software and is derived from the Kolmogorov-Smirnov test comparing the log2 ratios (MIRA versus input) within a 750 bp window centered at each probe and the rest of the data on the array. The asterisks indicate the location of the NEUROD1 target sites.
Discussion
To identify frequently methylated genes in SCLC tumor patients and SCLC cell lines, we have combined the use of a sensitive method for identifying methylation in CpG rich regions, the methylated-CpG island recovery (MIRA) assay26, 27 with genome-wide CpG island and promoter array analysis. Global profiling of 18 SCLC tumor samples compared to normal lung samples resulted in 698 and 73 tumor-specifically methylated and ensembl-annotated gene targets for 33% or more (≥6 of 18) of tumors, representing a substantial subgroup of patients, and in 77% or more of SCLC tumors (methylation in at least 14 of 18 samples), representing the majority of all patients, respectively. The 73 gene targets methylated in such a large fraction of the patient population may be of particular value for designing DNA methylation-based biomarkers for early detection of SCLC, for example in serum or sputum, and for disease management.
We randomly selected and validated 11 targets, which were predicted by the array analysis, by using bisulfite-based COBRA assays. The validated targets fell into various major functional categories, including transcription factors and noncoding RNAs like GALNTL1, MIR-10A, MIR-129-2, MIR-196A2, MIR-615, MIR-9-3, AMBRA1, HOXD10, PROX1, ZNF672 and DMRTA2. Validation of this set of samples revealed the specificity of the analysis. Some of the validated genes are epigenetically altered in various other cancers (MIR-10A, MIR-129-2, MIR-196A2, HOXD10 and PROX1) but other genes have not yet been identified as methylated in any cancer type (GALNTL1, MIR-615, AMBRA1, ZNF672 and DMRTA2). DMRTA2 methylation was found in 94% of the SCLC tumor patients. The only fact that is known about DMRTA2 is that there is crosstalk of expression with the transcription factor NFIA.36 Interestingly, there is evidence that NFIA is a key factor for the differentiation of neuronal progenitor cells by downregulating the activity of the Notch signaling pathway via repression of the key Notch effector Hes1.37 Given the strong enrichment for neuronal differentiation pathways in tumor-specific methylated regions in SCLC (Figure 2) it is tempting to speculate that there is a contribution of DMRTA2 methylation to impaired homeostasis between DMRTA2 and NFIA. There is no functional evidence yet for GALNTL1. These two targets, as well as the many other very frequently methylated genes (Table 2) have the potential to be used as biomarkers for this cancer type.
Gene annotation analysis of tumor-specific promoter methylated targets revealed a substantial subgroup of genes that are specific for neuronal fate commitment, neuronal differentiation and pattern specification processes, along with homeobox and other transcription factors. In comparison, hypermethylated regions in gene bodies showed similar functional enrichment clusters for homeobox genes and pattern specification processes, but significant less enrichment for neuronal fate commitment and for neuronal differentiation suggesting that the latter functional categories are more specific for promoter specific methylation. This striking tendency for methylation of neuronal specific genes may suggest an essential part in SCLC tumor initiation played by these genes.
Methylation of surrounding proximal promoters is often tightly associated with transcriptional silencing, whereas gene body methylation seems to be associated with transcriptional activation.27, 38 Loss of expression of genes, which are methylated in their proximal promoters, could lead to SCLC tumor initiation. Further studies in this direction will be required to establish experimental evidence. What we do not know at present is whether these genes are unmethylated and expressed in pulmonary neuroendocrine cells and their precursors, the likely cells of origin for SCLC. This specific cell type is currently not available for analysis. This issue does indeed apply to virtually all DNA methylation studies done in human cancer to date. The exact cell of origin, the cell from which the tumor initiates, is often not known, or these cells are not available in sufficient quantities. Therefore – at least theoretically – all DNA methylation ‘changes’ found in tumor DNA may already preexist in the cell of origin. However, we argue that methylation of genes that promote the differentiation of neuroendocrine cells would be unlikely to occur in such cells since that would interfere with their normal differentiated state.
The small cell lung cancer patients investigated in this study showed a strong enrichment of tumor-specific methylation at homeobox genes (Supplementary Tables 15 and 16). Homeobox genes and other transcriptional regulators are important for developmental processes, playing important roles in cellular identity, growth, differentiation and cellular interactions within the tissue environment. Given the results of our study, we developed a theory that disruptions in the early phase of these processes would increase the probability of the cell to become malignant, as this would lead to a pool of cells, which are aberrantly kept in a proliferation loop without a decision towards a specific cell fate. As already mentioned, it is thought that the cells of origin for SCLC are neuroendocrine cells, as shown in mice.10, 11 Given the fact that many of the tumor-specifically methylated targets we identified are important for cell fate decisions towards the neuronal lineage, it is intriguing to speculate that one way of shifting the balance towards the emergence of SCLC would be through the repression of key factors critical for differentiation of neuroendocrine cells. One potential way of aberrant shutdown of these critical factors would be by promoter-targeted methylation. Being freed of their normal developmental program by the absence or reduction of cell fate specification factors, some of these cells could acquire additional malignant traits, according to the “hallmark” model defined by Hahnan and Weinberg.39 This means that the observed hypermethylated regions are more probable to arise at an early stage of perturbed differentiation rather than during the later stages of tumorigenesis. Concerning other tumor-driving aberrant methylation events, which might increase the tumorigenic potential, it is interesting to note that we could rarely detect any promoter-specific methylation close to known tumor suppressor genes. Exceptions were tumor-specific methylation of TCF21, 40 which was detected downstream of the gene in the tumors but overlapping with the TSS in the cell lines and methylation of the promoter of the RASSF1A gene confirming earlier gene-specific studies.17, 18
Another potential way of disrupting cell fate decisions is not by merely reducing the responsible factors but by altering the selectivity towards their genomic recognition sites by aberrant methylation at these regulatory regions, leading to the prevention of binding. Indeed, it has long been known that DNA methylation can prevent transcription factor binding leading to the inhibition of active transcription or the recruitment of methyl binding proteins, causing gene suppression.38 When looking for binding sites of important cell fate specificators in our tumor-specific methylated regions, we could indeed identify such a correlation, especially concerning the transcription factors NEUROD1, ZNF423, HAND1 and REST (Figure 2B).
ZNF423 (also known as Ebfaz, Roaz, or Zfp423), a gene required for brain development, 41 may also have a role in neuroblastoma. ZNF423 is a transcription factor critically required for cerebellar development and retinoic acid-induced differentiation.42 Downregulation of ZNF423 expression by RNA interference in neuroblastoma cells results in a growth advantage and resistance to retinoic acid-induced differentiation. Loss of the NF1 tumor suppressor activates RAS-MEK signaling, which in turn represses ZNF423, a critical transcriptional coactivator of the retinoic acid receptors. Neuroblastomas with low levels of both NF1 and ZNF423 have poor clinical outcome.43
REST/NRSF is a transcription factor involved in complex regulatory pathways controlling neuronal differentiation, 44 having both oncogenic and tumor-suppressive roles.45 As shown in several other cancer types, there seems to be a correlation between the level of active REST and the tendency to initiate cancer.46 Inactivation of the REST/NRSF network may have a role in derepression of some neuroendocrine genes in SCLC.47 Interestingly Kreisler et al.48 found that three CpG islands associated with the REST gene were methylated in SCLC lines and we also found increased methylation near the REST promoter (Supplementary Figure 6). The loss of REST was linked to the malignant progression of SCLC.48 We present evidence that methylation of REST binding sites might also contribute to the SCLC phenotype.
Given that neuroendocrine cells are the likely cells of origin for SCLC, 10, 11 it is interesting that a significant number of NEUROD1 potential binding sites were correlated with methylation in the tumors (Figure 2B). It has been shown in mice that NeuroD deficiency resulted in both impaired alveolar septation and altered morphology of the pulmonary neuroendocrine cells, suggesting a role in the regulation of pulmonary neuroendocrine and alveolar morphogenesis.49 As such, methylation of NEUROD1 binding sites is supporting our theory of early methylation aberrations causing a defect in the developmental program of pulmonary neuroendocrine cells. Alternatively, lack of expression of transacting developmental transcription factors induced by methylation of their own promoter, which we did find for the NEUROD1 gene (Supplementary Figure 6), could lead indirectly to methylation of the transcription factor target sites. In this scenario, methylation of the binding site regions of these factors is the default state and can be prevented by in vivo binding of the factor. Although hypothetical, our model has gained support from a recent study in mouse ES cells.50 In this study, it was shown that the presence of several transcription factors, including REST, is required to create genomic regions with low DNA methylation.
In summary, we propose that probably both mechanisms, loss of key transcription factors involved in cell fate decisions or differentiation by methylation of their promoters, and functional inactivation of their corresponding binding site regions by methylation, can guide the cell of origin towards a malignant state. We note that this could be a potential explanation not only for the origin of SCLC but for tumorigenesis in general.
Materials and methods
Tissue and DNA samples
Primary SCLC tumor tissue DNAs were obtained from patients undergoing surgery at Nagoya University Hospital or Aichi Cancer Center, Nagoya, Japan. Pairs of human primary small cell lung cancer (SCLC) tumor tissue DNA and adjacent normal lung tissue DNA were obtained from Asterand (Detroit, MI), BioChain (Hayward, CA), and Cureline (South San Francisco, CA). SCLC cell lines (H1688, H1417, H1836, DMS53 and SW1271) were obtained from the ATCC. The ATCC used short tandem repeat profiling for cell line identification. Normal bronchial epithelial cells (HBECs; from Lonza; Walkersville, MD) were used as a control for the cell line analysis. All cells were cultured with DMEM/F12 with 0.5% fetal bovine serum and the bronchial epithelial growth medium (BEGM) bullet kit (Lonza). DNA and RNA from the cell lines were extracted using the DNeasy Blood & Tissue Kit and RNeasy Mini Kit (Qiagen; Valencia, CA) respectively.
MIRA and microarray hybridization
Tumor and normal tissue DNA was fragmented by sonication to ~500 bp average size as verified on agarose gels. Enrichment of the methylated double-stranded DNA fraction by MIRA was performed as described previously.26, 27 The labeling of amplicons, microarray hybridization, and scanning were performed according to the NimbleGen protocol. NimbleGen tiling arrays were used for hybridization (Human 3×720K CpG Island Plus RefSeq Promoter Arrays). These arrays cover all UCSC Genome Browser annotated CpG islands (total of 27, 728) as well as the promoters (total of 22, 532) of the well-characterized RefSeq genes derived from the UCSC RefFlat files. The promoter region covered is ~3 kb (−2440 to +610 relative to the transcription start sites). For all samples, the MIRA-enriched DNA was compared with the input DNA. All microarray data sets have been deposited into the NCBI GEO database (accession number GSE35341).
Identification and annotation of methylated regions
Analysis of the arrays was performed with R version 2.10, Perl scripts and the Bioconductor package Ringo.28 Arrays were clustered in normal tissues, cell lines and tumor tissues using hclust and Spearman correlation. Biological replicates were quantile-normalized and arrays were normalized by Nimblegen's recommended method, tuyey-biweight. Probe ratios were smoothed for three neighboring probes prior to peak calling. Instead of estimating a cutoff ratio based on a hypothetical normal distribution for non-bound probes (Ringo), a quantile-based approach was chosen to estimate methylation intensities. For this aim, peaks at different quantiles were called, where four probes were above the quantile-based threshold with a distance cutoff of 300 bp. A randomized set of peaks was validated by combined bisulfite restriction analysis (COBRA) assays51 for each quantile range. Thus a quantile range of 80% was chosen as a cutoff for methylated regions (defined as hypermethylated regions). False positives and false negatives were assessed by COBRA. To investigate if inter-sample differences had an influence on the acquired cutoff, predicted peaks were validated in different tissues by COBRA analysis.
Tumor-specific regions were defined using two different stringencies. In one case, an overlap of peaks in 6 or more out of 18 tumor samples (33%) was required above the cutoff quantile threshold of 80%; the genomic regions were defined and for those regions only one out of five normal tissues was allowed to overlap with a peak called on a 56% basis, which resulted in an at least 1.5 ratio change. Overlaps were calculated using BEDtools.52 A more stringent analysis required an overlap of peaks in at least 14 out of 18 tumors (>77%), with the same settings as above. The obtained chromosomal positions were converted to the latest hg19 genome build, using LiftOver from UCSC, requiring a minimum ratio of 0.9 of bases that must remap. The obtained positions where then annotated using the Bioconductor package ChIPpeakAnno and the latest ensembl annotation from BioMart.
Microarray expression analysis
Affymetrix human U133plus2.0 arrays for the three cell lines SW1271, H1836 and H1688 were processed by the robust multi-array average (RMA) method implemented in the Bioconductor ‘Affy’ package and the average log2 intensity of each gene across all samples was calculated. The three cell lines were clustered and compared against the control cell line, HBECs. Single expression values were obtained, using the MAS 5.0 method. Proximal promoter hypermethylated and non-hypermethylated regions, defined as −2000 bp to +1000 bp relative to transcription start sites according to the NimbleGen tilling arrays, were assigned with their respective expression probe changes of the corresponding transcript. The correlation between methylation and gene expression was based on a binary decision, linking gene promoters with differentially methylated regions with gene expression changes. A comparison with gene expression changes, where the promoter regions had a change in their methylation level (as measured by peak detected or absent) was above the significance threshold (p-value 0.05, Two sided T test).
De novo motif prediction
Motif analysis was performed by HOMER, a program developed by Chris Benner.35 More specifically, the discovery was performed using a comparative algorithm similar to those previously described by Linhart et al.53 Briefly, sequences were divided into target and background sets for each application of the algorithm (choice of target and background sequences are noted below). Background sequences were then selectively weighted to equalize the distributions of CpG content in target and background sequences to avoid comparing sequences of different general sequence content. Motifs of length 8 to 30 bp were identified separately by first exhaustively screening all possible oligos for enrichment in the target set compared to the background set by assessing the number of target and background sequences containing each oligo and then using the cumulative hypergeometric distribution to score enrichment. Up to two mismatches were allowed in each oligonucleotide sequence to increase the sensitivity of the method. The top 200 oligonucleotides of each length with the best enrichment scores were then converted into basic probability matrices for further optimization. HOMER then generates motifs comprised of a position-weight matrix and detection threshold by empirically adjusting motif parameters to maximize the enrichment of motif instances in target sequences versus background sequences using the cumulative hypergeometric distribution as a scoring function. Probability matrix optimization follows a local hill-climbing approach that weights the contributions of individual oligos recognized by the motif to improve enrichment, while optimization of motif detection thresholds were performed by exhaustively screening degeneracy levels for maximal enrichment during each iteration of the algorithm. Once a motif is optimized, the individual oligos recognized by the motif are removed from the data set to facilitate the identification of additional motifs. Sequence logos were generated using WebLOGO (http://weblogo.berkeley.edu/). Motifs, obtained from Jasper and TRANSFAC for which no high-throughput data exists were discarded for this analysis. Only those motifs with the highest alignments to known transcription factors, non-redundant matrixes and non-repetitive sequences were chosen for further analysis.
Functional annotation analysis
Gene ontology analysis was performed using DAVID functional annotation tools with Biological Process FAT and Molecular Function FAT datasets.54, 55 The enriched Gene Ontology terms were reported as clusters to reduce redundancy. The P-value for each cluster is the geometric mean of the P-values for all the GO categories in the cluster. The gene list in each cluster contains the unique genes pooled from the genes in all the GO categories in the cluster. Functional terms were clustered by using a Multiple Linkage Threshold of 0.5 and Bonferroni corrected P-values.
DNA methylation analysis using sodium bisulfite–based methods
DNA was treated and purified with the EZ DNA Methylation-Gold™ Kit (Zymo Research). PCR primer sequences for amplification of specific gene targets in bisulfite-treated DNA are shown in Supplementary Table 17. The PCR products were analyzed by combined bisulfite restriction analysis (COBRA) as described previously.51 Additionally, PCR products from bisulfite-converted DNA were cloned into pCR2.1-TOPO using a TOPO TA cloning kit (Invitrogen), and individual clones were sequenced with M13 forward (−20) primer.
Transfection, reverse transcription and quantitative real-time PCR
The DMS53 SCLC line was transfected with a NEUROD1 expression plasmid (2 µg) at ~60% confluence in 35 mm dishes with FugenHD® (Roche Applied Science, Indianapolis, IN) in serum-free medium according to the manufacturer's recommendations. The cells were cultured for an additional 48 h for analysis of NEUROD1 expression. Total RNA was isolated from HBECs, all five SCLC cell lines and from DMS53 cells overexpressing NEUROD1 using the RNeasy Mini Kit (Qiagen; Valencia, CA). cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad; Hercules, CA). Quantitative PCR was performed to assess expression of NEUROD1 and 18S RNA using NEUROD1 primers (forward, 5’GTTCTCAGGACGAGGAGCAC3’and reverse 5’CTTGGGCTTTTGATCGTCAT3’) and 18S primers (forward 5’GTAACCCGTTGAACCCCATT3’ and reverse 5’CCATCCAATCGGTAGTAGCG3’). Real-time PCR was performed using iQ SYBR Green Supermix and the iCycler real-time PCR detection system (Bio-Rad). Amplicon expression in each sample was normalized to 18S RNA. The relative abundance of target mRNA in each sample was calculated as 2 raised to the negative of its threshold cycle value times 1016 after being normalized to the abundance of its corresponding 18S RNA, e.g. [2-(IL-8 threshold cycle)/2-(18 S threshold cycle)] × 106].
Supplementary Material
Acknowledgements
We thank Steven Bates for culturing SCLC cell lines. This work was supported by National Institutes of Health grant CA084469 to G.P.P and by generous funds from an anonymous donor.
Footnotes
Conflict of interest:
Under a licensing agreement between City of Hope and Active Motif (Carlsbad, CA), the methylated-CpG island recovery assay (MIRA) technique was licensed to Active Motif, and the author G.P.P. is entitled to a share of the royalties received by City of Hope from sales of the licensed technology. The other authors declare no potential conflict of interest.
References
- 1.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 2.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 3.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 4.Modi S, Kubo A, Oie H, Coxon AB, Rehmatulla A, Kaye FJ. Protein expression of the RB-related gene family and SV40 large T antigen in mesothelioma and lung cancer. Oncogene. 2000;19:4632–4639. doi: 10.1038/sj.onc.1203815. [DOI] [PubMed] [Google Scholar]
- 5.Johnson BE, Ihde DC, Makuch RW, Gazdar AF, Carney DN, Oie H, et al. myc family oncogene amplification in tumor cell lines established from small cell lung cancer patients and its relationship to clinical status and course. J Clin Invest. 1987;79:1629–1634. doi: 10.1172/JCI112999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Obata Y, Sekido Y, Hida T, Ueda R, Watanabe H, et al. Expression and amplification of myc gene family in small cell lung cancer and its relation to biological characteristics. Cancer Res. 1989;49:2683–2688. [PubMed] [Google Scholar]
- 8.Sattler M, Salgia R. Molecular and cellular biology of small cell lung cancer. Semin Oncol. 2003;30:57–71. doi: 10.1053/sonc.2003.50019. [DOI] [PubMed] [Google Scholar]
- 9.Fischer B, Marinov M, Arcaro A. Targeting receptor tyrosine kinase signalling in small cell lung cancer (SCLC): what have we learned so far? Cancer Treat Rev. 2007;33:391–406. doi: 10.1016/j.ctrv.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 11.Park KS, Liang MC, Raiser DM, Zamponi R, Roach RR, Curtis SJ, et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle. 2011;10:2806–2815. doi: 10.4161/cc.10.16.17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 13.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 14.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 15.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 16.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 17.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dammann R, Takahashi T, Pfeifer GP. The CpG island of the novel tumor suppressor gene RASSF1A is intensely methylated in primary small cell lung carcinomas. Oncogene. 2001;20:3563–3567. doi: 10.1038/sj.onc.1204469. [DOI] [PubMed] [Google Scholar]
- 19.Sunaga N, Miyajima K, Suzuki M, Sato M, White MA, Ramirez RD, et al. Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res. 2004;64:4277–4285. doi: 10.1158/0008-5472.CAN-03-3941. [DOI] [PubMed] [Google Scholar]
- 20.Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauch TA, Pfeifer GP. DNA methylation profiling using the methylated-CpG island recovery assay (MIRA) Methods. 2010;52:213–217. doi: 10.1016/j.ymeth.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, et al. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–2727. doi: 10.1158/0008-5472.CAN-09-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009;11:R14. doi: 10.1186/bcr2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauch TA, Pfeifer GP. The MIRA method for DNA methylation analysis. Methods Mol Biol. 2009;507:65–75. doi: 10.1007/978-1-59745-522-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo--an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinert T, Modin C, Castano FM, Lamy P, Wojdacz TK, Hansen LL, et al. Comprehensive genome methylation analysis in bladder cancer: identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin Cancer Res. 2011;17:5582–5592. doi: 10.1158/1078-0432.CCR-10-2659. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez J, Munoz M, Vives L, Frangou CG, Groudine M, Peinado MA. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:19809–19814. doi: 10.1073/pnas.0810133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T. The presence of RNA polymerase II active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 2009;19:1974–1982. doi: 10.1101/gr.093310.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 34.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee NH, Haas BJ, Letwin NE, Frank BC, Luu TV, Sun Q, et al. Cross-talk of expression quantitative trait loci within 2 interacting blood pressure quantitative trait loci. Hypertension. 2007;50:1126–1133. doi: 10.1161/HYPERTENSIONAHA.107.093138. [DOI] [PubMed] [Google Scholar]
- 37.Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, et al. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2010;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Smith LT, Lin M, Brena RM, Lang JC, Schuller DE, Otterson GA, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A. 2006;103:982–987. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warming S, Rachel RA, Jenkins NA, Copeland NG. Zfp423 is required for normal cerebellar development. Mol Cell Biol. 2006;26:6913–6922. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Laoukili J, Epping MT, Koster J, Holzel M, Westerman BA, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328–340. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qureshi IA, Gokhan S, Mehler MF. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–1935. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- 46.Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. 2005;15:R665–R668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 47.Coulson JM, Edgson JL, Woll PJ, Quinn JP. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 2000;60:1840–1844. [PubMed] [Google Scholar]
- 48.Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene. 2010;29:5828–5838. doi: 10.1038/onc.2010.321. [DOI] [PubMed] [Google Scholar]
- 49.Neptune ER, Podowski M, Calvi C, Cho JH, Garcia JG, Tuder R, et al. Targeted disruption of NeuroD, a proneural basic helix-loop-helix factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem. 2008;283:21160–21169. doi: 10.1074/jbc.M708692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 51.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linhart C, Halperin Y, Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–1189. doi: 10.1101/gr.076117.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 55.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.