Abstract
The Agrobacterium-mediated transformation system is the most commonly used method in soybean transformation. Screening of soybean genotypes favorable for Agrobacterium-infection and tissue regeneration is the most important step to establish an efficient genetic transformation system. In this study, twenty soybean genotypes that originated from different soybean production regions in China were screened for transient infection, regeneration capacity, and stable transgenic efficiency. Three genotypes, Yuechun 04-5, Yuechun 03-3, and Tianlong 1, showed comparable stable transgenic efficiencies with that of the previously reported American genotypes Williams 82 and Jack in our experimental system. For the Tianlong 1, the average stable transformation efficiency is 4.59%, higher than that of control genotypes (Jack and Williams 82), which is enough for further genomic research and genetic engineering. While polymerase chain reaction (PCR), LibertyLink strips, and β-glucuronidase (GUS) staining assays were used to detect the insertion and expression of the transgene, leaves painted with 135 mg/L Basta could efficiently identify the transformants.
Keywords: Soybean, Agrobacterium-mediated transformation, Genotypes, Stable transgenic efficiency
1. Introduction
Soybean [Glycine max (L.)] is an important food and oil crop with more than 5 000 years of cultivation history in China. China was the largest soybean exporter in the world before 1996. Since then, commercial transgenic soybean production increased in the United States of America, Brazil, and Argentina. China has gradually become the world’s largest soybean importer due to the high quality and low price of imported transgenic soybean. The soybean production has continued to decrease in China and the domestic soybean industry is facing an unprecedented crisis. Over the past decade, transgenic soybeans have become the most widely cultivated genetically modified crop in the world, accounting for 47% of the global biotech crop area (75.4 million hectares) in 2011 (James, 2012). Though transgenic technology is considered to be one of the most important ways to breed new varieties of soybean, transgenic soybean breeding is still in its infancy in China (Qiu et al., 2007).
Particle bombardment-based transformation and Agrobacterium-mediated transformation are the two predominant methods for plant transformation (Hinchee et al., 1988; McCabe et al., 1988). Particle bombardment-based transformation has some shortcomings; e.g., it requires expensive equipment and often results in transformants with complicated and multiple copies (Dai et al., 2001; Shou et al., 2004; Travella et al., 2005). Contrastingly, Agrobacterium-mediated transformation is less expense, allows for easier manipulation, requires a lower transgene copy number, and results in higher stable gene expression (Dai et al., 2001; Shou et al., 2004; Travella et al., 2005). Thus, the Agrobacterium-mediated transformation is widely used for the transformation of a large number of crops, such as soybean, maize, rice, cotton, and canola (Gelvin, 2003).
The efficiency of Agrobacterium-mediated plant transformation depends on the success of the plant-Agrobacterium interaction (Gelvin, 2000). Plant genotype, explants vigour, Agrobacterium strain, vector, selection system, and culture conditions are all important factors in Agrobacterium-mediated plant transformation (Cheng et al., 2004). The common Agrobacterium strains include LBA4404, disarmed C58, EHA101, and their derivatives (Cheng et al., 2004). The plant tissues used to produce transgenic soybean plants vary but include shoot meristems (McCabe et al., 1988), embryogenic suspension cultures (Finer and McMullen, 1991), immature cotyledons (Parrott et al., 1989), axillary meristematic tissue located in seedling cotyledonary-nodes (Hinchee et al., 1988), and embryonic plant tissues used for commercial (Monsanto Technology LLC, 2008). The reasons that genotypes influence the transformation efficiency are complex. Overexpression of AtHTA1 gene increased Agrobacterium transformation efficiency in Arabidopsis and rice, which demonstrated that plant genes could alter the transformation efficiency (Mysore et al., 2000; Yi et al., 2006; Zheng et al., 2009). It has been well documented that genetic factors determine the susceptibility of soybean genotypes to Agrobacterium-infection and regeneration capacity in the transformation process (Meurer et al., 1998; Donaldson and Simmonds, 2000; Paz et al., 2004; Liu and Wei, 2005; Ying et al., 2008). Therefore, screening soybean genotypes for the germplasm resources suitable to Agrobacterium-mediated genetic transformation has become the focus for optimizing the soybean transformation system and improving transformation efficiency.
Although great progress has been made in soybean transformation, there still remain many challenges. Our work has focused on screening the soybean genotypes that are currently grown in China for high transformation efficiency. The improved cotyledonary-node transformation method and vector pTF102 with β-glucuronidase (GUS) reporter gene were employed in this study (Paz et al., 2004). Twenty soybean varieties originating from different soybean production regions in China have been screened for Agrobacterium-mediated genetic transformation. Four genotypes showed a high transient transformation efficiency and regeneration rate. The stable transformation efficiencies of Yuechun 04-5, Yuechun 03-3, and Tianlong 1 are all greater than 2% and comparable to the American control genotypes Williams 82 and Jack in our experimental conditions.
2. Materials and methods
2.1. Plant materials
Twenty-two soybean genotypes were used in this study. Among them, fifteen soybean cultivars are spring soybean varieties commonly planted in Northeast China, including Heihe 06, Heihe 19, Heihe 25, Heihe 38, Heihe 43, Heihe 48, Hefeng 25, Hefeng 35, Heinong 35, Heinong 37, Heinong 44, Heinong 48, Heinong 51, Heinong 58, and Ha 03-3. Zhonghuang 30 and Zhonghuang 39 are summer soybean varieties that originated from the Huanghuai region of China. Tianlong 1, Yuechun 03-3, and Yuechun 04-5 are southern spring soybean varieties. The well-studied American varieties Jack and Williams 82 were used as control genotypes.
2.2. Vector and Agrobacterium strain
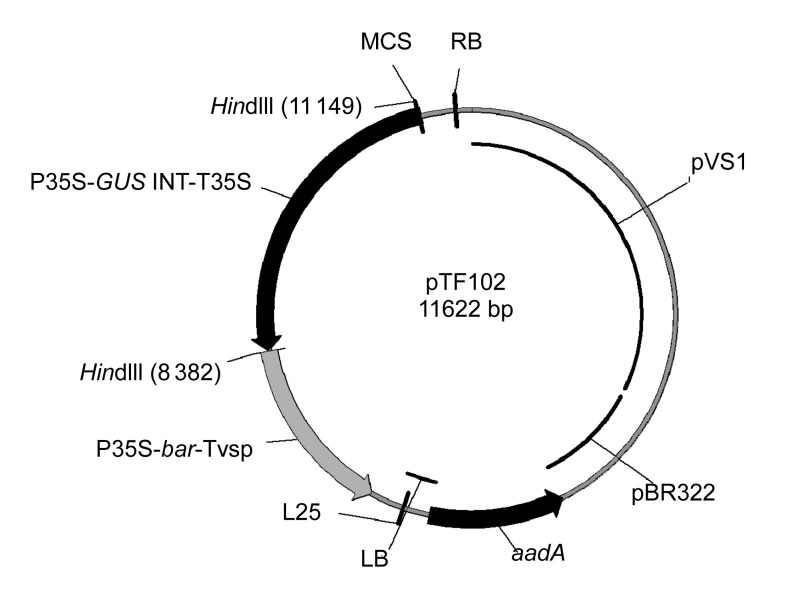
The Agrobacterium tumefaciens strain EHA101 (Hood et al., 1986) and the binary plasmid pTF102 (Fig. 1) (Frame et al., 2002) were used in this study. The vector includes a phosphinothricin acetyl transferase (bar) gene from Streptomyces hygroscopicus that confers resistance to herbicide phosphinothricin, an intron-containing GUS gene, and a spectinomycin-resistant marker (aadA) gene for bacterial selection.
Fig. 1.
Map of vector pTF102
P35S/T35S, CaMV 35S promoter/terminator; GUS INT, an intron-containing β-glucuronidase gene; bar, encoding phosphinothricin acetyl transferase; Tvsp, soybean vegetative storage protein terminator; aadA gene, a spectinomycin-resistant marker gene; pBR322, origin of replication for Escherichia coli; pVS1, origin of replication for Agrobacterium; LB, left border; RB, right border; MCS, multiple cloning site. HindIII is one of the unique restriction enzyme sites
Liquid YEP medium (10 g/L peptone, 5 g/L NaCl, 5 g/L yeast extract, pH 7.0) containing 50 mg/L kanamycin, 20 mg/L chloramphenicol, and 100 mg/L spectinomycin was inoculated with the A. tumefaciens EHA101/pTF102 and shaken at 28 °C (250 r/min) until the optical density at 650 nm (OD650) reached 0.8–1.0. The A. tumefaciens culture was centrifuged (20 °C, 4 000 r/min, 10 min) and the cell pellet was subsequently re-suspended in liquid co-cultivation medium (CCM) comprising 1/10 B5 salts and vitamins (Gamborg et al., 1968), 3.9 g/L 2-[N-morpholino] ethanesulfonic acid (MES), 3% sucrose (30 g/L) (pH 5.4), filter-sterilized 0.25 mg/L gibberellic acid (GA3), 400 mg/L L-cysteine, 1.67 mg/L N-6-benzylaminopurine (BAP), 154.2 mg/L dithiothreitol (DTT), and 200 μmol/L acetosyringone (As).
2.3. Explant preparation and inoculation
The cotyledonary-node method used in this study followed the procedure described by Paz et al. (2004). Briefly, soybean seeds were surface-sterilized by placing seeds into a tightly sealed chamber containing chlorine gas for 16 h. The sterilized seeds were soaked with sterile water at 28 °C under an 18-h photoperiod for 12–15 h. The imbibed soybean seeds were placed on sterile filter paper and cut longitudinally along the hilum. The cotyledons and hypocotyls (~5 mm) from a single seed were split evenly into two explants. Approximately fifty coat-removed explants were then inoculated in liquid CCM for 30 min. After inoculation, seven explants (adaxial side up) were randomly placed in Petri dishes (90 mm in diameter×15 mm deep) on sterile filter paper placed on solid CCM containing 0.5% purified agar (Difco Agar, Noble), and incubated in the dark at 25 °C for 3–5 d.
2.4. Selection and plant regeneration
After co-cultivation, explants were then transferred into solidified shoot induction (SI) medium containing B5 salts and vitamins, 3% sucrose, 0.8% agar (8 g/L; Sigma, USA), 0.58 mg/L MES (pH 5.8), filter-sterilized 1.67 mg/L BAP, 250 mg/L ticarcillin (Tic), 100 mg/L cefotaxime (Cef), and 5 mg/L glufosinate, and incubated in a growth room at 28 °C under an 18-h photoperiod for two weeks. Then, the hypocotyl and shoots were trimmed from the explants and the remaining cotyledons with developing nodules were sub-cultured to fresh SI medium for another two weeks.
Four weeks after SI, the cotyledons were removed from the explants and the remaining tissues were transferred into shoot elongation (SE) medium. SE medium was composed of MS salts and vitamins (Murashige and Skoog, 1962), 3% sucrose, 0.8% agar (Sigma, USA), 0.58 mg/L MES, filter-sterilized 50 mg/L L-asparagine, 50 mg/L glutamine, 0.1 mg/L indole acetic acid (IAA), 0.5 mg/L GA3, 1 mg/L zeatin riboside (ZR), 250 mg/L Tic, 100 mg/L Cef, and 5 mg/L glufosinate (pH 5.8). Explants were transferred to fresh SE medium every two weeks until the regenerated shoots were suitable for rooting. The incubation conditions were the same as the conditions for SI.
Elongated shoots (3–4 cm in length) were excised and placed into rooting medium (RM) containing MS salts and vitamins, 3% sucrose, 0.8% agar (Sigma, USA), 0.58 mg/L MES (pH 5.8), filter-sterilized 50 mg/L L-asparagine, 50 mg/L glutamine, 0.1 mg/L indole butyric acid (IBA), 250 mg/L Tic, and 100 mg/L Cef. After 1–2 weeks, the roots were fully developed to 2–3 cm in length, and eventually transplanted in the pots grown in the greenhouse.
2.5. Determination of the transient expression and regeneration rate
After 3 d of co-cultivation, the explants were first washed using sterile distilled water, and subsequently cut off at the upper side of the cotyledon (away from hypocotyl) and 3–5 mm below the cotyledonary node on the hypocotyl. The explants were harvested for GUS staining. According to the staining results, the explants were divided into three categories (strong, weak, and none). The transient rate of each category is calculated as: n e/n t×100%, where n e is the number of explants in each category and n t is the number of total stained explants. The regeneration rate is defined as: n ea/n eb×100%, where n ea is the number of explants after four weeks screening and n eb is the number of explants before screening.
2.6. Detection of transgenic soybean plants
Transgenic soybean plants were verified by leaf painting, polymerase chain reaction (PCR) analysis, GUS staining, and LibertyLink strip analysis. Leaf painting was performed following the procedure described by Ying et al. (2008) and Ma et al. (2012). Stock of LibertyLink® Basta (135 g/L) was diluted 1000-fold and painted on half the upper surface of tested soybean leaves. After 3 to 5 d, the painted leaves of the negative and positive plants died and remained healthy, respectively. For PCR analysis, genomic DNA of transgenic soybean was extracted using a simple and quick DNA extracting method (Edwards et al., 1991). The 459-bp bar gene coding region was amplified using the pairs of primers: 5′-GTACCGGCAGGCTGAAGTCC-3′ and 5′-CGGTCTGCACCATCGTCAAC-3′. The amplified products were separated by electrophoresis on a 1% agarose gel (10 g/L) for about 20 min and photographed with a gel imaging system. For GUS staining, the leaves of transgenic soybeans were harvested, immediately submerged in GUS staining solution, and placed under a vacuum for 10 min (Jefferson et al., 1987). The samples were incubated overnight in darkness at 37 °C. Chlorophyll was removed by submerging the tissue in 70% ethanol. LibertyLink strips (Envirologix, USA) were used to determinate genetically modified plants containing the phosphinothricin N-acetyltransferase (PAT) protein following the manufacturer’s instruction. Briefly, a circular leaf tissue was isolated by closing the cap of the Eppendorf tubes. The tissue was ground with a pestle for 20 to 30 s and 0.25 ml of protein extraction buffer was added before re-grinding. Development of the control line indicated that the strip had functioned properly. The second line (test line) would show up when the tested sample was positive.
2.7. Determination of the efficiency of soybean transformation
Positive transgenic seedlings were verified by PCR, leaf painting, GUS staining, and LibertyLink strip analysis. Transformation efficiency is defined as: n p/n i×100%, where n p is the number of positive transgenic seedlings and n i is the number of explants initially infected.
2.8. Molecular analysis
Southern (1975) blot analysis was conducted on T1 generations. Soybean genomic DNA was extracted from their leaf tissue using the cetyltrimethylammonium bromide (CTAB) method described by Doyle and Doyle (1987). Approximately 30 μg of genomic DNA was completely digested with HindIII (Fermentas, Canada). The restriction fragments were separated on 0.8% agarose gel by electrophoresis and transferred onto a Hybond-N+ nylon membrane (Amersham, UK). The PCR-fragments from the bar gene were labeled with Digoxigenin (DIG)-high prime according to the DIG High Prime DNA Labeling and Detection Starter Kit II instructions (Roche Applied Science, Germany). Prehybridization, hybridization, membrane washing, and detection were also conducted following the kit’s instruction manual (Roche Applied Science, Germany).
3. Results
3.1. Transient infection efficiency and regeneration rate of soybean genotypes
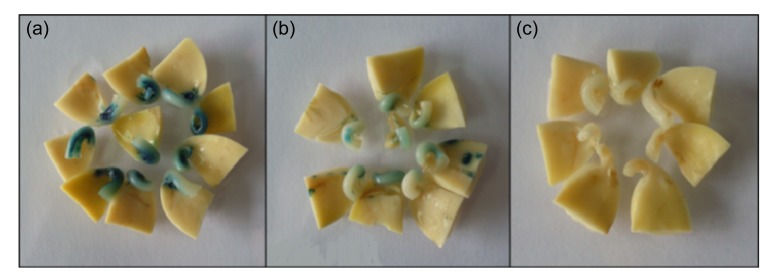
Twenty soybean genotypes cultivated in different regions of China were collected for screening. First, we tested the susceptibility of these soybean genotypes to Agrobacterium infection by transient GUS staining. The major GUS staining tissues are cotyledonary nodes and hypocotyls, which are tissues easily accessible to the Agrobacterium. According to the intensity of GUS staining, we divided the explants into three categories: strong, weak, and none (Fig. 2). The transient infection efficiencies were calculated and listed in Table 1. The control genotypes, Jack and Williams 82, showed 58.5% and 56.1%, respectively, of individuals expressing strong staining and less than 5% with no staining, indicating the effectiveness of Agrobacterium transient infection in our system. For all the tested genotypes, the average percentage of individuals demonstrating strong staining was 19.5%, indicating low efficiencies of Agrobacterium infection for most genotypes. However, Tianlong 1, Yuechun 03-3, Yuechun 04-5, Hefeng 35, Hefeng 25, and Heinong 37 exhibited above-average percentages of strong staining. Moreover, these six genotypes all exhibited “no staining” percentages of less than 50%.
Fig. 2.
Classification standards of GUS staining
(a) Strong, with deep dyeing in the cotyledon node; (b) Weak, with light dyeing in the cotyledon node or cotyledon; (c) None, with no dyeing
Table 1.
Regeneration rate and transient GUS staining of explants infected by Agrobacterium
| Genotype | Shoot regeneration |
GUS staining |
|||||||
| n 1 | Regeneration rate* (%) | n 2 | Strong |
Weak |
None |
||||
| n 3 | Ratio (%) | n 3 | Ratio (%) | n 3 | Ratio (%) | ||||
| Heihe 06 | 539 | 13.4±3.4jk | 40 | 1 | 2.5 | 17 | 42.5 | 22 | 55.0 |
| Heihe 19 | 528 | 13.6±1.9jk | 50 | 1 | 2.0 | 20 | 40.0 | 29 | 58.0 |
| Heihe 25 | 613 | 23.8±4.3hi | 55 | 0 | 0.0 | 19 | 34.5 | 36 | 65.5 |
| Heihe 38 | 608 | 27.3±2.1gh | 52 | 4 | 7.7 | 18 | 34.6 | 30 | 57.7 |
| Heihe 43 | 553 | 25.7±3.8gh | 40 | 4 | 10.0 | 25 | 62.5 | 11 | 27.5 |
| Heihe 48 | 452 | 39.8±6.5f | 59 | 0 | 0.0 | 24 | 40.7 | 35 | 59.3 |
| Hefeng 25 | 289 | 21.1±7.1hi | 37 | 9 | 24.3 | 16 | 43.2 | 12 | 32.4 |
| Hefeng 35 | 370 | 13.5±2.6jk | 40 | 11 | 27.5 | 13 | 32.5 | 16 | 40.0 |
| Heinong 35 | 604 | 17.1±3.6ij | 40 | 6 | 15.0 | 17 | 42.5 | 17 | 42.5 |
| Heinong 37 | 12 102 | 52.2±1.1de | 44 | 9 | 20.5 | 16 | 36.4 | 19 | 43.2 |
| Heinong 44 | 884 | 31.8±7.2g | 40 | 4 | 10.0 | 15 | 37.5 | 21 | 52.5 |
| Heinong 48 | 374 | 10.2±3.6k | 40 | 4 | 10.0 | 21 | 52.5 | 15 | 37.5 |
| Heinong 51 | 557 | 8.4±3.2k | 40 | 7 | 17.5 | 20 | 50.0 | 13 | 32.5 |
| Heinong 58 | 2 027 | 58.7±1.9d | 41 | 4 | 9.8 | 19 | 46.3 | 18 | 43.9 |
| Ha 03-3 | 190 | 9.0±2.7k | 40 | 0 | 0.0 | 6 | 15.0 | 34 | 85.0 |
| Zhonghuang 30 | 400 | 20.8±5.1hi | 39 | 0 | 0.0 | 22 | 56.4 | 17 | 43.6 |
| Zhonghuang 39 | 340 | 24.1±4.4h | 40 | 4 | 10.0 | 15 | 37.5 | 21 | 52.5 |
| Tianlong 1 | 437 | 80.7±4.3ab | 42 | 26 | 61.9 | 11 | 26.2 | 5 | 11.9 |
| Yuechun 04-5 | 898 | 75.4±3.2b | 40 | 16 | 40.0 | 18 | 45.0 | 6 | 15.0 |
| Yuechun 03-3 | 355 | 51.5±4.7e | 40 | 18 | 45.0 | 16 | 40.0 | 6 | 15.0 |
| Jack | 337 | 82.8±3.3a | 41 | 24 | 58.5 | 15 | 36.6 | 2 | 4.9 |
| Williams 82 | 356 | 67.7±4.7c | 41 | 23 | 56.1 | 18 | 43.9 | 0 | 0.0 |
|
| |||||||||
| Average | 34.9 | 19.5 | 40.7 | 39.8 | |||||
Regeneration rate was expressed as mean±standard deviation; means were compared using Fisher’s least significant difference test, and different small letters showed significant difference (P<0.05)
n 1: number of explants infected; n 2: total number of explants for GUS staining; n 3: number of explants
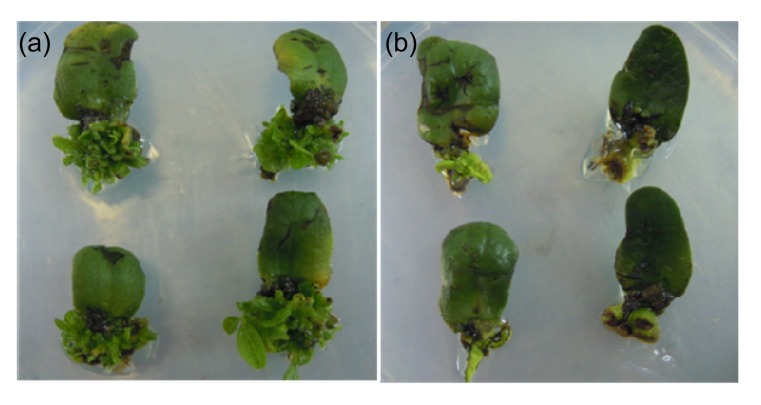
After selection with 5 mg/L glufosinate on SI medium for four weeks, healthy explants produced multiple observable buds (Fig. 3a). Some explants were dead and had no buds (Fig. 3b). Regeneration rate was calculated as the proportion of healthy explants for all the tested genotypes, with an average regeneration rate of 34.9%. As expected, Jack and Williams 82 varieties grew well and had 82.8% and 67.7% regeneration rates, respectively. Among the twenty genotypes originating from China, Tianlong 1, Yuechun 04-5, Heinong 37, Heinong 58, Yuechun 03-3, and Heihe 48 showed above-average regeneration rates.
Fig. 3.
Phenotype of explants after shoot induction (SI) for four weeks
(a) Subculture explants with a great of herbicide-resistant multiple buds; (b) Discarded explants with no or very little shoots
Based on transient infection and regeneration rates, Tianlong 1, Yuechun 04-5, Heinong 37, and Yuechun 03-3 performed better than other Chinese genotypes, and were used in the following stable transformation experiments.
3.2. Quick detection of T0 transgenic plants
Due to the large number of T0 transgenic plants that were generated in the lab, a rapid method for determining positive transgenic status was pivotal. PCR detection of a selectable marker gene is a traditional and effective way of doing this, though false positive results are sometimes observed. Therefore, we employed a two-step method for the identification of T0 transgenic plants. First, the bar marker gene was detected by PCR in the T0 transgenic plants, and the results indicated some might be positive events (Fig. 4a). Part of the PCR positive lines was randomly selected and tested using LibertyLink strips, which determined the PAT protein of transgenic plants (Fig. 4b). We also used GUS staining to determine the positive transgenic plants (Fig. 4c). If 95% of the randomly selected plants were proven to be positive transgenic plants by test strips or GUS staining, we concluded that the PCR results were believable. When less than 95% of the randomly selected plants showed positive results by test strips or GUS staining, the two-step detection was repeated.
Fig. 4.
Procedure for confirmation of T0 transgenic plants
(a) PCR analysis of genome DNA of putative transgenic soybean plants using bar gene primers. The length of PCR production was 459 bp. M: DL2000 marker DNA; Ctr−: non-transformed soybean; Ctr+: plasmid DNA; (b, c) Events of the PCR positive lines were randomly selected and tested using LibertyLink strips (b) or GUS staining (c). +: positive transgenic soybean plant; −: negative transgenic soybean plant
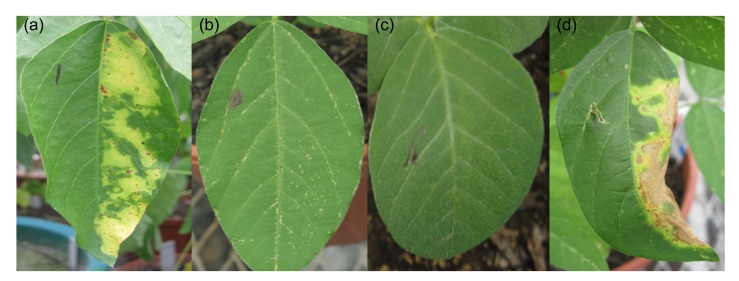
To avoid the cumbersome procedure of PCR analysis and the expensive costs of the test strips and GUS staining, we employed leaf painting to detect transgenic soybean seedlings. Halves of the leaves were painted with 135 mg/L Basta. Five days later, painted half leaves of non-transgenic control seedlings were lesioned while the unpainted halves remained green (Fig. 5a). However, the whole leaves of the positive control plants with bar gene remained green (Fig. 5b). The T0 seedlings selected by leaf painting were consistent with those of PCR and testing strips (data not shown). Thus, this method provides a low-cost, rapid, and accurate way to identify a large number of T0 transgenic plants.
Fig. 5.
Identification of the herbicide resistance of the leaves of transgenic soybean plants using 135 mg/L Basta
Half of a leaf with the black mark is not painted with herbicide, and the other side is painted with herbicide. (a) Non-transgenic plant; (b, c) Positive transgenic plants; (d) Negative transgenic plant
3.3. Stable transformation efficiencies of the screened soybean genotypes
The whole procedures of Agrobacterium-mediated cotyledonary-node transformation method used in this study were illustrated in Fig. 6. Under our conditions, the four Chinese genotypes tested (Tianlong 1, Yuechun 04-5, Yuechun 03-3, and Heinong 37) and the two control genotypes from the USA (Jack and Williams 82) produced positive transgenic plants. However, the stable transformation efficiencies varied among these genotypes. The average efficiencies of Tianlong 1 and Williams 82 were 4.59% and 4.39%, respectively, which were significantly higher than those of Yuechun 04-5, Yuechun 03-3, and Heinong 37 (Table 2). Yuechun 04-5 and Yuechun 03-3 showed similar average efficiencies to that observed in the Jack variety. The average efficiency of Heinong 37 was only 0.31%, the lowest among the tested genotypes (Table 2).
Fig. 6.
Agrobacterium-mediated soybean transformation using the cotyledonary node as explants
(a) Surface sterilization of soybean seeds; (b) A longitudinal cut between the cotyledons and through the hypocotyl to generate two identical explants; (c) Inoculation of explants with Agrobacterium; (d) Two weeks after selection on shoot induction (SI) medium containing 5 mg/L glufosinate; (e) Four weeks after selection on SI medium containing 5 mg/L glufosinate; (f) Elongated shoot in shoot elongation (SE) medium; (g) Rooting of the resistant shoot; (h) A putative transgenic plantlet was transplanted to pots in the greenhouse
Table 2.
Transformation efficiencies of six soybean genotypes
| Genotype | n 1 | n 2 | Efficiency* (%) | RI |
| Heinong 37 | 5 | 1 021 | 0.31±0.28c | + |
| Yuechun 03-3 | 5 | 1 000 | 2.40±0.65b | ++ |
| Yuechun 04-5 | 5 | 1 058 | 2.42±0.70b | ++ |
| Tianlong 1 | 5 | 1 118 | 4.59±1.54a | +++ |
| Jack | 5 | 1 014 | 2.84±1.25ab | ++ |
| Williams 82 | 5 | 1 007 | 4.39±1.85a | +++ |
Data of efficiency were expressed as mean±standard deviation; means were compared using Fisher’s least significant difference test, and different small letters showed significant difference (P<0.05)
n 1: number of experiments; n 2: number of explants infected; RI: recommendation index
3.4. Analysis of T1 seedlings
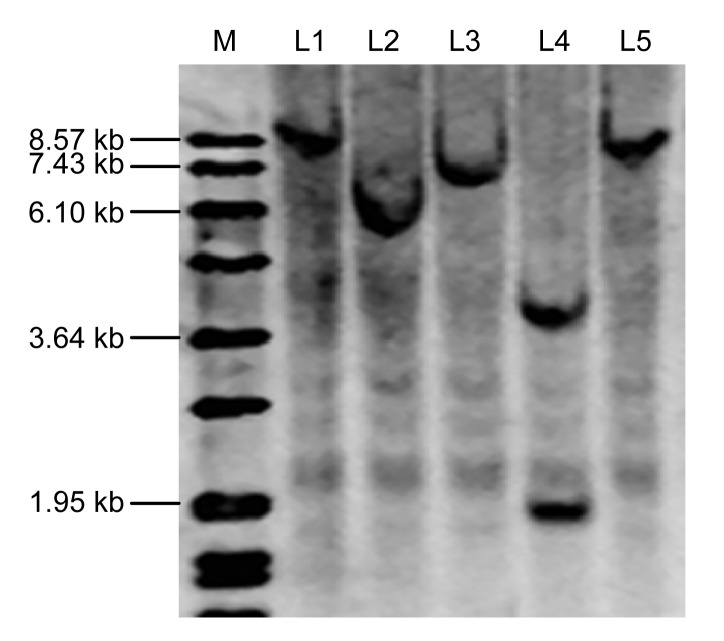
We examined T1 progenies of five independent transformed T0 lines with the integration of bar and GUS genes. All of the five independent transgenic plants produced positive T1 seedlings. Chi-square analysis was performed on the segregation ratios of the five T0 lines to determine goodness-of-fit for 3:1 and 15:1 (GUS +:GUS −). The results indicated that transgenic soybean Lines 1–4 were inherited in a Mendelian fashion and Line 5 was segregated in other ratio (Table 3). We further confirmed the copy number of inserted bar gene using Southern blots. Lines 1, 2, 3, and 5 had only one copy of the bar gene while Line 4 contained two copies (Fig. 7).
Table 3.
Segregation of GUS gene in the T1 seedlings of five transgenic soybean lines
| Transgenic line | Genotype | Number of T1 plants | Segregation assay |
||||
| GUS + | GUS − | Ratio | χ 2 | P-value | |||
| Line 1 | Yuechun 03-3 | 13 | 10 | 3 | 3:1 | 0.0256 | 0.8728 |
| Line 2 | Yuechun 03-3 | 18 | 12 | 6 | 3:1 | 0.2963 | 0.4142 |
| Line 3 | Yuechun 03-3 | 25 | 17 | 8 | 3:1 | 0.3333 | 0.3855 |
| Line 4 | Yuechun 03-3 | 10 | 10 | 0 | 15:1 | 0.0267 | 0.4142 |
| Line 5 | Yuechun 03-3 | 8 | 3 | 5 | Other | ||
Fig. 7.
Southern blot analysis of transgenic soybeans
Soybean genomic DNA was sampled from five independent events in the T1 generation (L1, L2, L3, L4, and L5) and digested with the restriction enzyme HindIII, and hybridized with the bar probe labeled with DIG. A DIG-labeled DNA marker (M) was shown
4. Discussion
Soybean production in China has largely decreased in the past decade because of the high-quality and low price of imported transgenic soybean. Currently, Williams 82 and Jack soybean varieties are commonly used in international soybean transformation breeding and research. However, these two genotypes have poor agronomic traits and are only suitable to be grown in narrow regions. In order to speed up the process of transgenic soybean breeding in China, we screened soybean genotypes suitable for domestic ecological regions. In this study, twenty cultivated genotypes from different regions of China were collected and screened for their Agrobacterium-mediated cotyledonary node transformation.
Agrobacterium infection of plants is a process that resembled the pathogen-host interaction (Gelvin, 2003). Therefore, genotypes that are resistant to pathogen infection may have decreased susceptibility to Agrobacterium infection. In this research, we employed a transient transformation system to evaluate various genotypes’ susceptibility to Agrobacterium. At the same time, another key factor of evaluation is the regenerability of explants. Although transient infection efficiency and regeneration rate were significantly different among the tested genotypes, Jack and Williams 82, the control genotypes in the study, showed a relatively good performance in both transient infection and regeneration processes. Therefore, our system is vigorous to screen the soybean genotypes for transgenic breeding. According to the transient infection efficiency and regeneration rates, four Chinese genotypes (Tianlong 1, Yuechun 03-3, Yuechun 04-5, and Heinong 37) showed potential for soy bean transformation. Though all of the four genotypes could stably produce transgenic soybean plants, the efficiencies varied. The Tianlong 1 variety showed the highest stable transformation efficiency among all tested genotypes. This suggests that Tianlong 1 is suitable for further transgenic soybean breeding and functional genomics research. Yuechun 03-3 and Yuechun 04-5 also exhibited considerable transformation efficiencies and might be used in the future. Heinong 37 is a genotype with high regeneration rates and a good infection rate. However, using current protocol, the transformation efficiency was not as high as expected. Closely looking at the performance of the genotypes across the entire transformation process showed that the major obstacle to the transformation of a genotype is the elongation stage. Many putative transformed buds that initiated from the transformation process were not able to elongate. Further optimization of the hormone combination in SE medium may be necessary to improve transformation efficiency.
The method that we used for soybean transformation was a modified version of soybean cotyledonary node transformation. We observed that the cotyledonary nodes grew better when they were placed with the explants’ adaxial side up. However, we did not observe a statistically significant difference between the two ways of placements of explants (abaxial or adaxial side up) on CC medium in terms of transformation efficiency. In most published soybean cotyledonary node transformation protocols, the embryonic axis in explants are excised to promote the growth of the axillary buds. However, removing the embryonic axis increased oxidative stress due to wounding. In this study, we kept the embryonic axis to reduce the oxidative stress. The growth of the embryonic axis can be suppressed by two weeks of high levels of selection. According to our observations, retaining the embryonic axis improved the initiation of axillary buds.
As a large amount of T0 plants were generated during transgenic breeding, a quick and effective way to identify positive events was critical for subsequent growth and management of the seedlings. A two-step method, combining primary PCR testing and following marker gene detection, was employed to identify the positive T0 transgenic plants in our system. However, the primary PCR testing is cumbersome and costly. Instead of PCR procedures, a leaf painting method was chosen (Ying et al., 2008; Ma et al., 2012). As leaf painting directly detected the effect of a selectable marker gene, it showed fewer false positive results than PCR procedures. Furthermore, the leaf painting method was low-cost and easy to carry out.
Three Chinese soybean genotypes were selected for further applications in this study. The stable transformation efficiencies of these genotypes fulfill the requirements of soybean functional genomic research and transgenic breeding in China. However, these genotypes do not meet transformation efficiencies observed in commonly used North American genotypes. Further studies are needed to improve the soybean transformation efficiency of Chinese varieties and to optimize our transformation system.
Acknowledgments
We sincerely thank Dr. Kan WANG from Iowa State University (USA) for providing the control soybean genotypes, pTF102 vector and EHA101 strain. The authors also thank Dr. Chuang WANG of Zhejiang University (China) and Mr. Zachary LARSON of Iowa State University (USA) for critical reading and English editing.
Footnotes
Project supported by the Ministry of Agriculture of China (Nos. 2011ZX08004 and 2009ZX08010013) and the National Natural Science Foundation of China (No. 31172024)
Compliance with ethics guidelines: Zhang-yue SONG, Jing-luan TIAN, Wei-zhe FU, Lin LI, Ling-hong LU, Lian ZHOU, Zhi-hui SHAN, Gui-xiang TANG, and Hui-xia SHOU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Cheng M, Lowe BA, Spencer TM, Ye XD, Armstrong CL. Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cell Dev Biol-Plant. 2004;40(1):31–45. doi: 10.1079/IVP2003501. [DOI] [Google Scholar]
- 2.Dai SH, Zheng P, Marmey P, Zhang SP, Tian WZ, Chen SY, Beachy RN, Fauquet C. Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breeding. 2001;7(1):25–33. doi: 10.1023/A:1009687511633. [DOI] [Google Scholar]
- 3.Donaldson PA, Simmonds DH. Susceptibility to Agrobacterium tumefaciens and cotyledonary node transformation in short-season soybean. Plant Cell Rep. 2000;19(5):478–484. doi: 10.1007/s002990050759. [DOI] [PubMed] [Google Scholar]
- 4.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 5.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19(6):1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finer J, McMullen M. Transformation of soybean via particle bombardment of embryogenic suspension culture tissue. In Vitro Cell Dev Biol-Plant. 1991;27(4):175–182. doi: 10.1007/bf02632213. [DOI] [Google Scholar]
- 7.Frame BR, Shou HX, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SEK, Li B, Nettleton DS, Pei D, et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using standard binary vector system. Plant Physiol. 2002;129(1):13–22. doi: 10.1104/pp.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamborg OL, Miller RA, Ojiama K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50(1):151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- 9.Gelvin SB. Agrobacterium and plant genes involved in T-DNA transfer and integration. Ann Rev Plant Physiol Plant Mol Biol. 2000;51(1):223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 10.Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67(1):16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinchee MAW, Conner-Ward DV, Newell CA. Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat Biotechnol. 1988;6(8):915–922. doi: 10.1038/nbt0888-915. [DOI] [Google Scholar]
- 12.Hood EE, Helmer GL, Fraley RT, Chilton MD. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol. 1986;168(3):1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James C. 2011 ISAAA report on global status of Biotech/GM crops. China Biotechnol. 2012;32(1):1–14. (in Chinese) [Google Scholar]
- 14.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu HC, Wei ZM. Recent advances in soybean genetic transformation. J Plant Physiol Mol Biol. 2005;31(2):126–134. (in Chinese) [PubMed] [Google Scholar]
- 16.Ma L, Zhou L, Hua MF, Zhou ZJ, Tang GX, Shen ZC, Shou HX. Establishment of methods to rapidly and precisely identify transgenic rice and soybean containing herbicide-resistant gene bar and EPSPS . J Zhejiang Univ (Agric Life Sci) 2012;38(6):647–654. (in Chinese) [Google Scholar]
- 17.McCabe DE, Swain WF, Martinell BJ, Christou P. Stable transformation of soybean (Glycine max) by particle acceleration. Nat Biotechnol. 1988;6(8):923–926. doi: 10.1038/nbt0888-923. [DOI] [Google Scholar]
- 18.Meurer CA, Dinkins RD, Collins GB. Factors affecting soybean cotyledonary node transformation. Plant Cell Rep. 1998;18(3-4):180–186. doi: 10.1007/s002990050553. [DOI] [PubMed] [Google Scholar]
- 19.Monsanto Technology LLC. Preparation and Use of Plants Embryo Explants for Transformation. US Patent 112628 A2. 2008
- 20.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–479. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 21.Mysore KS, Nam J, Gelvin SB. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. PNAS. 2000;97(2):948–953. doi: 10.1073/pnas.97.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrott WA, Hoffman LM, Hildebrand DF, Williams EG, Collins GB. Recovery of primary transformants of soybean. Plant Cell Rep. 1989;7(8):615–617. doi: 10.1007/BF00272042. [DOI] [PubMed] [Google Scholar]
- 23.Paz MM, Shou HX, Guo ZB, Zhang ZY, Banerjee AK, Wang K. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica. 2004;136(2):167–179. doi: 10.1023/B:EUPH.0000030669.75809.dc. [DOI] [Google Scholar]
- 24.Qiu LJ, Wang CL, Zhou GA, Chen SY, Chang RZ. Soybean molecular breeding. Sci Agric Sin. 2007;40(11):2418–2436. (in Chinese) [Google Scholar]
- 25.Shou HX, Frame BR, Whitham SA, Wang K. Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breeding. 2004;13(2):201–208. doi: 10.1023/B:MOLB.0000018767.64586.53. [DOI] [Google Scholar]
- 26.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98(3):503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 27.Travella S, Ross SM, Harden J, Everett C, Snape JW, Harwood WA. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep. 2005;23(12):780–789. doi: 10.1007/s00299-004-0892-x. [DOI] [PubMed] [Google Scholar]
- 28.Yi HC, Sardesai N, Fujinuma T, Chan CW, Veena , Gelvin SB. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell. 2006;18(7):1575–1589. doi: 10.1105/tpc.105.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ying S, He XW, Wang XR, Shou HX. Assessment of factors affecting the transformation efficiency of soybean cotyledonary-node Agrobacterium-mediated transformation system. Mol Plant Breeding. 2008;6(1):32–40. (in Chinese) [Google Scholar]
- 30.Zheng Y, He XW, Ying YH, Lu JF, Gelvin SB, Shou HX. Expression of the Arabidopsis thaliana histone gene AtHTA1 enhances rice transformation efficiency. Mol Plant. 2009;2(4):832–837. doi: 10.1093/mp/ssp038. [DOI] [PubMed] [Google Scholar]