Abstract
The methyl–methyl NOESY experiment plays an important role in determining the global folds of large proteins. Despite the high sensitivity of this experiment, the analysis of methyl–methyl NOEs is frequently hindered by the limited chemical shift dispersion of methyl groups, particularly methyl protons. This makes it difficult to unambiguously assign all of the methyl–methyl NOE crosspeaks using 3-D spectroscopy. The recent development of sparse sampling methods enables highly efficient acquisition of high-resolution 4-D spectra, which provides an excellent solution to resolving the degeneracy of methyl signals. However, many reconstruction algorithms for processing sparsely-sampled NMR data do not provide adequate suppression of aliasing artifacts in the presence of strong NOE diagonal signals. In order to overcome this limitation, we present a 4-D diagonal-suppressed methyl–methyl NOESY experiment specifically optimized for ultrasparse sampling and evaluate it using a deuterated, ILV methyl-protonated sample of the 42 kDa Escherichia coli maltose binding protein (MBP). Suppression of diagonal signals removes the dynamic range barrier of the methyl–methyl NOESY experiment such that residual aliasing artifacts in the CLEAN-reconstructed high-resolution 4-D spectrum can be further reduced. At an ultrasparse sampling rate of less than 1%, we were able to identify and unambiguously assign the vast majority of expected NOE crosspeaks between methyl groups separated by less than 5 Å and to detect very weak NOE crosspeaks from methyl groups that are over 7 Å apart.
Keywords: Diagonal suppression, Fast NMR, Sparse sampling, Methyl–methyl NOE, FFT-CLEAN, Large proteins
1. Introduction
NMR spectroscopy has become an increasingly important tool for exploring the structure, function and dynamics of biological macromolecules. The optimization of NMR pulse sequences and development of new isotope labeling schemes [1–6] have allowed NMR spectroscopists to work with ever larger systems. Among the many new developments, the methyl-specific labeling strategy [7–9], which produces samples with a high level of deuteration except for selected methyl groups, is particularly attractive for determining the global folds of large proteins, as most methyl groups are located in the hydrophobic cores of proteins. Unfortunately, methyl groups have limited chemical shift dispersion, which makes it challenging to unambiguously assign observed NOE crosspeaks to specific pairs of methyl groups using conventional 3-D experiments. High-resolution 4-D spectroscopy provides a natural solution to resolve the overlapping signals, but it typically takes a very long time (many weeks) to acquire data due to the restriction of Nyquist sampling in each of the individual dimensions. In order to increase the efficiency of NMR data acquisition, several groups have developed sparse sampling techniques and reconstruction methods that have enabled the collection of high-resolution 4-D spectra using only a fraction of the measurement time required conventionally (reviewed in [10,11]).
Although sparse sampling improves the efficiency of NMR data acquisition, due to violation of the Nyquist sampling theorem, it inevitably introduces aliasing artifacts that are often significantly larger than the true thermal noise, even when the aliasing artifacts are dispersed uniformly by randomized sampling patterns. The form and location of the aliasing artifacts are not random, however, and they can be determined through knowledge of the underlying sampling pattern and the location of real signals. A number of reconstruction algorithms have been proposed to suppress aliasing artifacts for high-quality reconstruction of 4-D spectra, each having its own strengths and limitations [12–16]. While all of these reconstruction algorithms can remove the bulk of the aliasing artifacts, they typically are unable to eliminate the aliasing artifacts completely. Therefore, their performance is less than optimal for spectra containing signals with a very large dynamic range. For example, NOESY spectra are particularly challenging because they contain very strong diagonal signals that can be over 100-fold stronger than the weakest crosspeak signals. For these diagonal signals, any residual artifacts left over by reconstruction algorithms may still be significantly higher than the very weak NOE crosspeaks. This highlights the need for further development and optimization of sparse sampling technology, either by improving the performance of reconstruction algorithms or by employing spectroscopic methods to reduce the dynamic range of observable NMR signals.
Here, we demonstrate the application of a diagonal-suppressed 4-D methyl–methyl NOESY experiment for use with ultrasparse sampling and FFT-CLEAN processing. The benefit of diagonal suppression and signal separation in the high-resolution 4-D dataset on the completeness of observed NOE crosspeaks is evaluated using a deuterated, ILV methyl-protonated sample of the 42 kDa maltose binding protein (MBP).
2. Materials and methods
2.1. NMR sample
We tested the 4-D methyl–methyl diagonal-suppressed NOESY experiment on the 42 kDa MBP in complex with β-cyclodextrin. The U-[15N, 2H], Ile δ1-[13CH3], Leu δ-[13CH3, 12CD3], Val γ-[13CH3, 12CD3] (ILV)-labeled MBP sample was prepared as described previously [17]. In this labeling scheme, only one of two methyl groups (either pro-R or pro-S) of each Val and Leu residues is protonated, and the other one remains deuterated. U-[2H]-glucose and 15NH4Cl were used as the main carbon and nitrogen sources, respectively. 100 mg of 2-keto-3-d2-4-13C-butyrate and 100 mg of 2-keto-3-methyl-d3-3-d1-4-13C-butyrate were added into 1 L of D2O M9 medium 1 h before induction in order to achieve selective protonation of ILV methyl groups. The final NMR sample contained 1 mM MBP, 2 mM β-cyclodextrin, 20 mM sodium phosphate, 100 μM EDTA, 1 mM DTT and 99.9% D2O.
2.2. Pulse sequence
Various strategies for suppressing diagonal signals in NOESY experiments have been proposed [18–26]. Initially, we focused on strategies using ZQ-TROSY [21] or spin-state-filter elements [23–26] in the methyl NOESY experiment. However, the level of methyl diagonal suppression was unsatisfactory due to the fast relaxation of HzCα/β spin states during the NOE mixing period. Therefore, we turned to a more general method for diagonal suppression [22] that involves collecting two separate experiments. The first experiment uses scheme A in Fig. 1, a modified version of the conventional 4-D methyl–methyl 13C-HMQC-NOESY-13C-HMQC experiment that records diagonal signals and off-diagonal NOE crosspeaks. The methyl-TROSY effect [27] is preserved through a modified HMQC element for enhanced sensitivity. The second experiment uses scheme B in Fig. 1 in order to collect only diagonal signals. Scheme B works in the following way. For two neighboring methyl groups 1 and 2, the magnetization from methyl group 1 has the form at point a. During the NOE mixing period, part of this magnetization is transferred to methyl group 2, which gives magnetization that is unobservable during acquisition. However, the untransferred magnetization can be refocused to by inserting a period of 2τ before acquisition for the selective observation of diagonal signals. Finally, subtracting the spectra collected with schemes A and B gives a single spectrum containing only the off-diagonal NOE crosspeaks. In practice, the unequal pulse sequence lengths of schemes A and B and the different relaxation properties of and magnetization during the NOE mixing period lead to discrepancies in the diagonal signal intensities for the two experiments. Therefore, a scaling factor was introduced for the second experiment before subtraction in order to achieve optimal diagonal suppression.
Fig. 1.
Pulse sequence for the 4-D diagonal-suppressed methyl–methyl NOESY experiment. Narrow and wide bars represent 90° and 180° pulses, respectively. All pulses are along the x-axis unless indicated otherwise. The delay τ is 1.8 ms, and the NOE mixing times MIX1 and MIX2 are 200 ms. Two datasets were collected to achieve diagonal suppression as described in the text. For the second dataset, scheme A is replaced by scheme B. The phase cycling is ϕ1 = [x, –x], ϕ2 = [x, x, x, x, –x, –x, –x, –x], ϕ3 = [x, x, –x, –x], ϕ4 = [y], ϕ5 = [–y], ϕrec = [x, –x, –x, x, –x, x, x, –x]. Quadrature detection in F1, F2 and F3 is achieved using the States-TPPI method by changing ϕ1, ϕ2 and ϕ3, respectively.
2.3. Experimental setup and CLEAN algorithm
A randomized concentric shell sampling scheme [14] was used for acquisition of the high-resolution 4-D diagonal-suppressed methyl–methyl NOESY spectrum. 2400 sampling points in the time domain were used for the three indirect dimensions and were adapted to a regular grid of 64 × 64 × 64, corresponding to only 0.9% of the sampling points used conventionally. For each time point, 8 FIDs were recorded to achieve quadrature detection. The carrier frequencies for the 1H and 13C dimensions were set to 0.1 ppm and 16.4 ppm, respectively. The spectral widths for the indirect 1H and 13C dimensions were set to 1502 Hz and 2641 Hz, with maximum evolution times of 42.6 ms and 24.2 ms, respectively. All experiments were recorded at 37 °C on a Bruker AVANCE 500 MHz spectrometer equipped with a cryoprobe. The NOE mixing time in both schemes was set to 200 ms. With 8 scans per FID and a recycle delay of 0.9 s, the acquisition time was ~3 days for each spectrum, for a total of 6.25 days overall. For optimal performance, these two experiments (scheme A and scheme B in Fig. 1) were collected in an interleaved manner, using an identical sampling pattern. The time domain data of the two schemes were subtracted to generate a difference dataset, which was used for spectral reconstruction. The 4-D dataset in this work was calculated at 128-point resolution in all three indirect dimensions and processed with the FFT-CLEAN algorithm [14] with a gain of 30% and a stopping threshold of 4σnoise. The processing time is ~70 min on a dual-core 2.6 GHz Linux computer.
2.4. 3-D control experiments
The 3-D 13C-edited NOESY-HMQC and 13C-HMQC-NOESY-13C-HMQC experiments were recorded using conventional sampling with carrier frequencies and spectral widths identical to those of the 4-D experiment. Both control experiments also used a NOE mixing period of 200 ms and 8 scans per FID. For the 3-D 13C-edited NOESY-HMQC experiment, the maximum evolution times were 42.7 ms (64 complex points) and 18.9 ms (50 complex points) for the F1 (H1) and F2 (C2) dimensions, respectively, and the total experimental time was 45 h. For the 3-D 13C-edited HMQC-NOESY-HMQC experiment, the maximum evolution times of both the F1(C1) and F2 (C2) dimension were 24.2 ms (64 complex points), and the total experimental time was 2.5 days.
3. Results and discussion
Despite the clear advantages of the selective methyl labeling approach, the limited chemical shift distribution of methyl groups can cause severe signal degeneracy for large proteins, requiring high-resolution 4-D data for optimal signal separation and assignment. The benefit of high-dimensionality data for methyl–methyl NOESY is clearly demonstrated in the example planes of Fig. 2. In both cases, every NOE crosspeak can be unambiguously identified in the high-resolution 4-D spectrum, but not in the 3-D control spectra due to peak overlap.
Fig. 2.
Selected 2-D F1/F2 planes at the F3/F4 positions of I108 δ1 (A) and L290 δ2 (B). Red peaks in both panels are the residual diagonal signals. Strips along each dimension are from the corresponding 3-D control experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
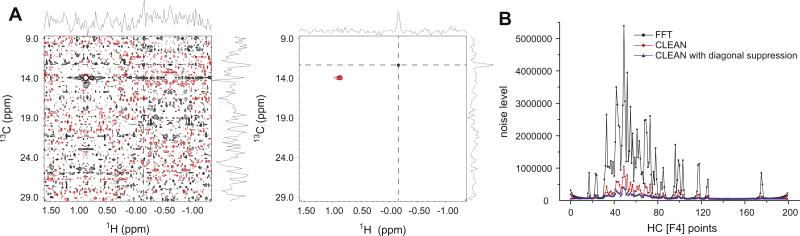
Due to the restriction of independent Nyquist sampling of each indirect dimension, conventionally-sampled high-resolution 4-D NOESY spectra require prohibitively long measurement times. For example, achieving the same digital resolution of the 4-D methyl–methyl NOESY experiment described here with conventional sampling would take over 340 days of spectrometer time (or 42 days with 2-fold linear prediction in each of the individual dimensions). These unfeasible acquisition times have prevented the routine application of high-resolution 4-D spectroscopy. The recent development of sparse sampling and associated reconstruction techniques has provided an elegant solution to this dilemma and has made it possible to acquire high-resolution 4-D NOESY spectra within a couple of days. However, sparse sampling under the Nyquist sampling rate inevitably introduces aliasing artifacts. With optimal sampling patterns, these aliasing artifacts take the form of random noise, and they are typically much weaker in intensity than the real signals. The CLEAN algorithm iteratively identifies the strongest peak that is well above the noise in a spectrum, generates its aliasing artifacts based on the underlying sampling pattern, and substrates a fraction of the signal and its associated aliasing artifacts, leaving behind a spectrum containing reduced peak intensity and a reduced artifact level. This process is repeated until no signals can be reliably identified above the noise level, at which point the clean signals without aliasing artifacts were added back to generate the final reconstruction spectrum. In this study, we show that the residual artifacts left over by the FFT-CLEAN algorithms in a 4-D methyl–methyl NOESY spectrum containing strong diagonals can overshadow very weak NOE crosspeaks and result in the loss of critical distance constraint information. This problem is clearly illustrated in Fig. 3A, which contains 2-D planes from the 4-D methyl–methyl NOESY spectrum at the F3/F4 position of I178 δ1 without (left panel) and with (right panel) diagonal suppression. In the presence of diagonal signals, the residual artifact level after FFT-CLEAN processing is still unsatisfactorily high, and the crosspeak from I333 δ1 (at the cross of the dashed lines) is buried in the aliasing noise and cannot be readily identified. With diagonal suppression, however, the same crosspeak can be observed clearly. The improvement in apparent noise is highlighted by the 1-D traces along each dimension. Overall, CLEAN processing significantly reduces the aliasing artifacts level (Fig. 3B). Excluding the contribution from the thermal noise (i.e., background noise in the cubes without signals), CLEAN reduces the aliasing artifacts from 4- to 10-folds compared to that of FFT processed spectrum (Fig. 3B). However, the residual aliasing artifact level after CLEAN is still noticeably higher than the background thermal noise. Using a more aggressive termination criterion does not result in significant further reduction of aliasing artifacts, suggesting that the incomplete elimination of artifacts is not due to early termination of CLEAN processing. The application of the diagonal suppression technique remedy this by reducing the aliasing artifacts even further by as much as 3- to 6-fold, generating a much more uniform distribution of aliasing artifact levels for the 3-D cubes in the reconstructed 4-D spectrum and enabling detection of weak NOE signals among methyl groups that are over 7 Å apart. Some cubes in the reconstructed 4-D spectrum after diagonal suppression still have an elevated noise level that can be as high as 3- to 4-fold of the thermal noise, suggesting that the sensitivity of this experiment can be further improved by development and optimization of the reconstruction software.
Fig. 3.
Reduction of aliasing noise by diagonal suppression. (A) Selected 2-D F1/F2 planes at the F3/F4 position of I178 δ1 from the 4-D methyl–methyl NOESY experiment without (left panel) and with (right panel) diagonal suppression. A crosspeak from I333 δ1 is denoted by the cross of the dashed lines with 1-D traces shown beside the 2-D planes. Both spectra are processed with the CLEAN algorithm for artifact suppression. (B) Aliasing noise levels of individual cubes in the directly detected HC [F4] dimension of 4-D spectra generated by FFT, CLEAN and CLEAN with diagonal suppression spectra.
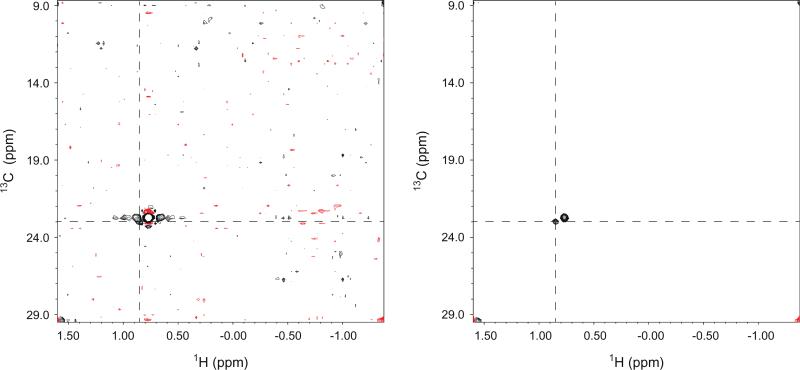
In addition to the impressive reduction of artifacts, suppression of diagonal signals has added benefits when crosspeaks are located very close to the diagonal signals. In Fig. 4, we show 2-D planes from the 4-D methyl–methyl NOESY experiment at the F3/F4 position of L121 δ2 without (left panel) and with (right panel) diagonal suppression. In this example, diagonal suppression permits the identification of a crosspeak from L139 δ2 (at the cross of the dashed lines), which would otherwise be overshadowed by truncation artifacts from the diagonal signal.
Fig. 4.
Selected 2-D F1/F2 planes at the F3/F4 position of L121 δ2 from the 4-D methyl–methyl NOESY experiment without (left panel) and with (right panel) diagonal suppression. A crosspeak from L139 δ2 is denoted by the cross of the dashed lines.
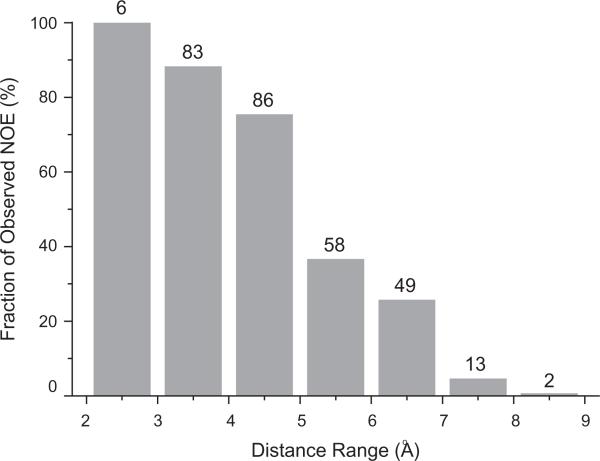
The diagonal suppression approach described here involves the subtraction of two separate spectra and doubles the measurement time. Therefore it reduces sensitivity by a factor of 2 compared to the experiment without diagonal suppresion. However, suppression of the strong diagonal signals reduces the aliasing artifact level by as much as 3- to 6-fold, thereby increasing the effective signal-to-noise ratio and allowing the detection of very weak NOE crosspeaks. In our study of MBP, we were able to identify 297 methyl–methyl NOE crosspeaks, 85% of which can be unambiguously assigned. In order to evaluate the sensitivity of our experiment, we calculated the expected NOE crosspeaks between ILV methyl groups using pseudoatom positions of methyl protons in the available X-ray structure (PDB ID: 1dmb), and compared them with the observed NOE crosspeaks (Fig. 5). For methyls less than 4 Å apart, we were able to unambiguously detect nearly all of the expected crosspeaks (>90%). The completeness level drops to ~75% for distances from 4 to 5 Å. Excitingly, we can readily identify NOE crosspeaks between methyl groups that are over 7 Å apart, highlighting the high sensitivity of our experiment.
Fig. 5.
Percentage of observed methyl–methyl NOE crosspeaks (based on the number expected from the X-ray structure [PDB ID: 1dmb]) as a function of the distance between methyl groups. The number of observed NOE crosspeaks is listed on top of the vertical bars.
Although the advantages of diagonal suppression were demonstrated here using the FFT-CLEAN processing algorithm, the ability to reduce the dynamic range of NMR signals experimentally should benefit many other reconstruction algorithms. This approach may be particularly valuable for the application of reconstruction algorithms, such as the maximum entropy reconstruction method [12], that do not maintain the linearity for signals with a large dynamic range, to high-resolution 4-D NOESY experiments for faithful extraction of distance information.
4. Conclusion
We have introduced a diagonal-suppressed 4-D methyl–methyl NOESY experiment for studies of large proteins, and tested it on the 42 kDa Escherichia coli maltose binding protein. Diagonal suppression further reduces the residual aliasing artifacts left over by the CLEAN reconstruction algorithm by as much as 3- to 6-fold and allows the identification of weak crosspeaks that would otherwise be lost to aliasing noise as well as crosspeaks that are close to diagonal signals. The excellent signal separation of high-resolution 4-D spectroscopy and the high sensitivity of our experiment should make this approach particularly valuable for protein global fold determination of large proteins by NMR.
Acknowledgments
This work was supported by the National Institutes of Health (GM079376) to P. Zhou and the National Basic Research Program of China (973 Program, Grant 2011CB966302) to J. Wu. The authors thank Dr. Weiwei Wang for assistance in sample preparation, Jiahai Zhang for assistance with instrumentation, and Dr. Ronald A. Venters (Duke University NMR Center) and Dr. Jonathan W. Werner-Allen for critical reading of the manuscript.
References
- 1.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riek R, Wider G, Pervushin K, Wüthrich K. Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc. Natl. Acad. Sci. USA. 1999;96:4918–4923. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coughlin PE, Anderson FE, Oliver EJ, Brown JM, Homans SW, Pollak S, Lustbader JW. Improved resolution and sensitivity of triple-resonance NMR methods for the structural analysis of proteins by use of a backbone-labeling strategy. J. Am. Chem. Soc. 1999;121:11871–11874. [Google Scholar]
- 4.Metzler WJ, Wittekind M, Goldfarb V, Mueller L, Farmer BT. Incorporation of 1H/13C/15N-{Ile, Leu, Val} into a perdeuterated, 15N-Labeled Protein: potential in structure determination of large proteins by NMR. J. Am. Chem. Soc. 1996;118:6800–6801. [Google Scholar]
- 5.Rajesh S, Nietlispach D, Nakayama H, Takio K, Laue ED, Shibata T, Ito Y. A novel method for the biosynthesis of deuterated proteins with selective protonation at the aromatic rings of Phe, Tyr and Trp. J. Biomol. NMR. 2003;27:81–86. doi: 10.1023/a:1024710729352. [DOI] [PubMed] [Google Scholar]
- 6.Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 7.Gardner KH, Zhang X, Gehring K, Kay LE. Solution NMR studies of a 42 KDa escherichia coli maltose binding protein/β-cyclodextrin complex: chemical shift assignments and analysis. J. Am. Chem. Soc. 1998;120:11738–11748. [Google Scholar]
- 8.Tugarinov V, Kay LE. Ile, leu, and val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 9.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase G. J. Am. Chem. Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 10.Coggins BE, Venters RA, Zhou P. Radial sampling for fast NMR: concepts and practices over three decades. Prog. Nucl. Magn. Res. Spectrosc. 2010;57:381–419. doi: 10.1016/j.pnmrs.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W. Random sampling in multidimensional NMR spectroscopy. Prog. Nucl. Magn. Res. Spectrosc. 2010;57:420–434. doi: 10.1016/j.pnmrs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat. Methods. 2007;4:467–468. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- 13.Orekhov VY, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR. 2003;27:165–173. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- 14.Coggins BE, Zhou P. High resolution 4-D spectroscopy with sparse concentric shell sampling and FFT-CLEAN. J. Biomol. NMR. 2008;42:225–239. doi: 10.1007/s10858-008-9275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyberts S, Frueh D, Arthanari H, Wagner G. FM reconstruction of non-uniformly sampled protein NMR data at higher dimensions and optimization by distillation. J. Biomol. NMR. 2009;45:283–294. doi: 10.1007/s10858-009-9368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanek J, Augustyniak R, Koźmiński W. Suppression of sampling artefacts in high-resolution four-dimensional NMR spectra using signal separation algorithm. J. Magn. Reson. 2012;214:91–102. doi: 10.1016/j.jmr.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protocols. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 18.Denk W, Wagner G, Rance M, Wuthrich K. Combined Suppression of diagonal peaks and t1 ridges in two-dimensional nuclear overhauser enhancement spectra. J. Magn. Reson. 1985;62:320–355. [Google Scholar]
- 19.Harbison GS, Feigon J, Ruben DJ, Herzfeld J, Griffin RG. Diagonal peak suppression in 2D-NOE spectra. J. Am. Chem. Soc. 1985;107:5567–5569. [Google Scholar]
- 20.Zhu G, Xia Y, Sze KH, Yan X. 2D and 3D TROSY-enhanced NOESY of 15N labeled proteins. J. Biomol. NMR. 1999;14:377–381. [Google Scholar]
- 21.Pervushin KV, Wider G, Riek R, Wuthrich K. The 3D NOESY-[(1)H, (15)N, (1)H]-ZQ-TROSY NMR experiment with diagonal peak suppression. Proc. Natl. Acad. Sci. USA. 1999;96:9607–9612. doi: 10.1073/pnas.96.17.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Fan J.-s., Pascal SM, Yang D. General method for suppression of diagonal peaks in heteronuclear-edited NOESY spectroscopy. J. Am. Chem. Soc. 2004;126:15018–15019. doi: 10.1021/ja045300l. [DOI] [PubMed] [Google Scholar]
- 23.Meissner A, Sorensen OW. Suppression of diagonal peaks in TROSY-type 1H NMR NOESY spectra of 15N-labeled proteins. J. Magn. Reson. 1999;140:499–503. doi: 10.1006/jmre.1999.1860. [DOI] [PubMed] [Google Scholar]
- 24.Meissner A, Sorensen OW. Three-dimensional protein NMR TROSY-type 15N-resolved 1HN-1HN NOESY spectra with diagonal peak suppression. J. Magn. Reson. 2000;142:195–198. doi: 10.1006/jmre.1999.1961. [DOI] [PubMed] [Google Scholar]
- 25.Werner-Allen JW, Coggins BE, Zhou P. Fast acquisition of high resolution 4-D amide-amide NOESY with diagonal suppression, sparse sampling and FFT-CLEAN. J. Magn. Reson. 2010;204:173–178. doi: 10.1016/j.jmr.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diercks T, Truffault V, Coles M, Millet O. Diagonal-free 3D/4D HN, HN-TROSY-NOESY-TROSY. J. Am. Chem. Soc. 2010;132:2138–2139. doi: 10.1021/ja910523q. [DOI] [PubMed] [Google Scholar]
- 27.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]