Abyssomicin C, produced by the marine actinomycete Verrucosispora maris AB-18-032, is active against Gram-positive bacteria including methicillin-resistant Staphylococcus aureus (MRSA) and inhibits p-aminobenzoate formation during tetrahydrofolate synthesis; it is the first natural product active against this therapeutic target. To investigate the biosynthesis of this small but structurally complex secondary metabolite, we carried out feeding studies using 13C labelled polyketide building blocks. Formation of abyssomicin C requires two propionates, five acetates and one glucose-derived metabolite. Identification and sequencing of the abyssomicin biosynthetic gene cluster revealed a 57 kb segment of Verrucosispora maris AB-18-032 DNA that contained all of the genes necessary for abyssomicin biosynthesis. The identity of the biosynthetic gene cluster was confirmed by gene inactivation and complementation experiments (the first genetic manipulation of a member of this genus) and a model for abyssomicin C biosynthesis is proposed.
The search for novel antibiotics has led to the exploration of previously unscrutinised and extreme habitats, such as the deep oceans, caves, deserts and mountains, to isolate new microbial strains with untapped biosynthetic potential.1, 2 Concomitantly, rational approaches have identified new therapeutically useful bacterial targets.2, 3 The folate pathway is essential in bacteria for the synthesis of aromatic amino acids and purine nucleotides, and plays an essential role in pathogenesis. The pathway is also present in algae, higher plants, fungi and apicomplexan parasites, but absent from mammals, and is, thus, an excellent potential target for the development of new therapeutic compounds.4 So far, only synthetic drugs are known to inhibit proteins in this pathway; foremost are the sulfonamides (sulfa drugs) and diaminobenzylpyrimidines (e.g., trimethoprim).5 In 2004, targeted screening for inhibitors of the folate pathway led to the discovery of abyssomicins produced by the marine actinomycete Verrucosispora maris AB-18-032.6 Abyssomicin C is active against Gram-positive bacteria, including MRSA,6 and inhibits the biosynthesis of p-aminobenzoic acid (pABA), a constituent of the folate pathway.7 Recently, we found that abyssomicin C covalently binds to PabB, the aminodesoxychorismate synthase of Bacillus subtilis, and inhibits pABA formation and, consequently, folate biosynthesis.7, 8
Several strategies for the synthesis of abyssomicin C have been published and provide valuable new insights into the biosynthesis and activity of abyssomicin C.9 The biomimetic synthesis by Sorensen et al.9d provides an interesting model for the biosynthesis of abyssomicin C. In 2007, while working on another route for total synthesis of abyssomicin C, the Nicolaou group discovered atrop-abyssomicin C, the atrop-isomer of abyssomicin C. Atrop-abyssomicin C is even more active than the initially reported abyssomicin C.7, 9a, 10 Shortly thereafter, atrop-abyssomicin C (2) was also detected in fermentations of Verrucosispora maris AB-18-032.7 In fact, 2 is the main product synthesised by Verrucosispora maris AB-18-032, and abyssomicin C (1) is a minor by-product formed from 2 by rearrangement under acid conditions. To date, seven abyssomicins have been identified but only abyssomicin C and atrop-abyssomicin C have defined bioactivity.7, 8
Atrop-abyssomicin C and abyssomicin C are structurally related to tetronomycin-like antibiotics, such as chlorothricin,11 tetronomycin and tetrocarcin, which all contain a tetronic acid moiety (4-hydroxy-(5H)furan-2-one). The abyssomicins contain an oxo-bridge from the tetronate that forms a bicyclic system, which is unique among the tetronomycin-like antibiotics. Tetronic acid antibiotics are an important subclass of polyketide antibiotics with various bioactivities and molecular targets, and the biosynthetic gene clusters of several have been elucidated: chlorothricin, kijanimicin, tetronomycin and tetrocarcin.12 Comparison of these gene clusters was used to identify a unique set of genes required for the biosynthesis of tetronic acid-containing compounds.12b In this work, we propose a model for the biosynthesis of atrop-abyssomicin C based on a combination of feeding studies with 13C-labelled biosynthetic precursors and the identification of the abyssomicin biosynthetic gene cluster. We propose that abyssomicin is synthesised as a linear polyketide chain from five acetates, two propionates and a metabolite from the glycolytic pathway. Polyketide synthase (PKS) processing is followed by formation of the tetronate moiety, a Diels–Alder reaction, and oxygenation.
By analogy to other tetrocarcin-type antibiotics, it was assumed that the abyssomicins are formed by a polyketide biosynthetic pathway.3a To confirm this assumption, the 13C-labelled building blocks [1-13C]acetate, [1,2-13C]acetate and [1-13C]propionate were fed to exponentially growing shake-flask cultures of Verrucosispora maris AB-18-032. Once the cultures had reached stationary phase, atrop-abyssomicin C was isolated, as described previously, and analysed by 13C NMR spectroscopy.3a The incorporation of 13C carbons into the polyketide backbone resulted in a considerable increase in signal intensity of the labelled carbon positions. Feeding of [1-13C]acetate led to signal increase at C-1, C-7, C-9, C-11 and C-13 (Scheme 1). In a subsequent experiment, [1,2-13C]acetate was fed, and intact incorporation was found for C-1/C-2, C-7/C-8, C-9/C-10 and C-11/C-12. The resulting doublets with their 13C–13C coupling constants as well as all other 13C NMR data are given in Table S1 in the Supporting Information.
Scheme 1.
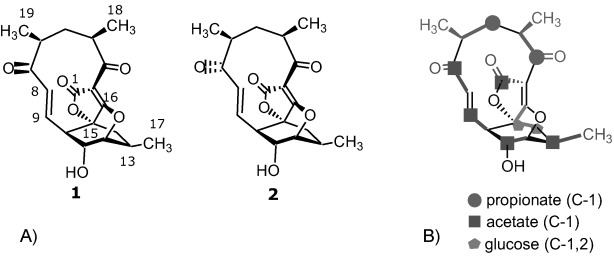
A) Structures of the antibacterial polyketides abyssomicin C (1) and atrop-abyssomicin C (2). B) Incorporation of 13C-labelled precursors into the polyketide backbone of abyssomicin C.
This pairwise 13C–13C coupling is indicative of direct assembly into the polyketide chain. When [1-13C]propionate was fed, increased signals were obtained for C-3 and C-5. The assembly of five acetate and two propionate units as biosynthetic precursors into the abyssomicin C carbon skeleton leaves a three carbon unit (C-14–C-16) unassigned. Since earlier studies on tetronic acid antibiotics had reported the incorporation of glycerol at this position,13 [1-13C]glycerol was added to the fermentation broth of Verrucosispora maris AB-18-032. Subsequently isolated atrop-abyssomicin C was analysed by 13C NMR spectroscopy, but no incorporation of glycerol was detected. Likewise, feeding of other putative 13C-labelled C3 precursors, such as serine, alanine and succinic acid, did not yield specifically labelled atrop-abyssomicin C.14 Remarkably, only [1,2-13C]glucose was successfully incorporated into the C3 unit of the abyssomicin carbon backbone; this indicates that it originated from the glycolytic pathway. The 13C–13C coupling of C-14 and C-15 (J=36.5 Hz) clearly shows that C-14 and C-15 are derived from the same glucose molecule. Furthermore, while glucose leads to doubly labelled acetate (for C-3 through C-6 and C-18 and C-19) propionate is formed from only one 13C carbon, indicated by singlet peaks (Figures S1–S6 and Table S1 in the Supporting Information). This implies that in Verrucosispora, glycolysis preferentially ends in acetate14b rather than lactate14b and leads directly to propionate. In summary, feeding experiments showed that abyssomicin C is derived from five acetates, two propionates and one metabolite from the glycolytic pathway (Scheme 1).
A dual cosmid and whole genome sequencing (WGS) approach was used to identify the abyssomicin biosynthetic gene cluster (aby). A cosmid library of Verrucosispora maris AB-18-032 genomic DNA was hybridised with PKS type I-specific probes generated by PCR with primers KSIIFOR and ATIREV developed at Combinature Biopharm, AG.15 To identify which of the 49 hybridising cosmids contained genes responsible for atrop-abyssomicin C production, all were digested with two different restriction enzymes (BamHI, SmaI) and the resulting restriction profiles were used to classify the cosmids into families containing overlapping sequences. Three major families (with five to eight members each) and four smaller families (two members each) were identified, as well as 14 apparently unique cosmids. Representative cosmids from each family were subjected to restriction digestion with BamHI and the resulting fragments were cloned into the nonreplicating vector pK18mob2. After being sequenced, PKS positive clones were used for gene inactivation in Verrucosispora by selecting for single crossover integration into the host genome.
The resulting Verrucosispora maris AB-18-032 insertional mutants were screened for the absence of atrop-abyssomicin C production by analytical HPLC-ESI-MS, which revealed that cosmid family II contained genes required for abyssomicin C biosynthesis. One cosmid from this family (C17) was sequenced. Cosmids containing genomic segments overlapping that of C17 were identified by Southern analysis of restriction digests of members of cosmid family II by using abyA2 (upstream of abyB; i.e., PKS genes) and abyF1 (downstream of abyB genes) specific probes. Cosmids C49 and C45 were identified by using probes abyA2 and abyF1, respectively, and sequenced to expand the extent of the aby gene cluster. Blastx searches of the WGS contig library were then carried out by using sequences from abyX (upstream) and abyT (downstream) to identify flanking regions. WGS contigs containing these genes and additional sequences were included in the assembly with data from cosmids C17, C45 and C49. This resulted in about 57 kb of contiguous genomic DNA sequence that appeared to contain the entire aby biosynthetic gene cluster (Figure 1).
Figure 1.
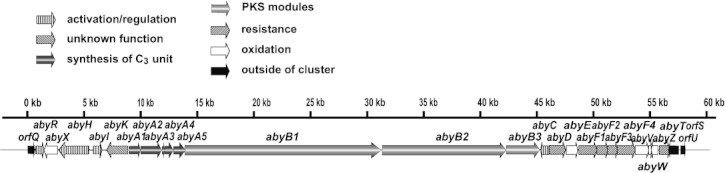
Biosynthetic gene cluster of atrop-abyssomicin C and assignment of the main gene functions.
Bioinformatics analysis of the 57 kb sequence with Glimmer and blastx revealed 24 putative protein coding sequences (pcs) that appeared to be involved in abyssomicin biosynthesis (Table 1 and Figure 1). These pcs comprise the anticipated PKS I genes (abyB1, abyB2 and abyB3) and five genes (abyA1–A5) homologous to chlM and chlD1–4 of the chlorothricin biosynthetic gene cluster involved in the assembly of the tetronic acid moiety.12c Genes encoding oxygenases, as well as genes with putative regulatory and export functions, were also identified.
Table 1.
The abyssomicin biosynthetic gene cluster with functions assigned by xblast and InterProScan searches.
| Name | Gene start [bp] | Length [aa] | Proposed function | Similarity with | Identity/ similarity [%] |
|---|---|---|---|---|---|
| orfQ | 1 | 208 | NUDIX hydrolase | Frankia sp. EAN1pec | 53/66 |
| putative MutT family protein | Nocardia farcinica IFM 10152 | 43/52 | |||
| abyR | 680 | 248 | transcriptional regulator, SARP family | [Frankia sp. EUN1f] | 54/71 |
| Frankia symbiont of Datisca glomerata | 58/69 | ||||
| abyX | 2629 | 396 | cytochrome P450 | Frankia sp. EAN1pec Streptomyces griseolus | 66/77 56/70 |
| abyH | 5389 | 889 | LuxR family transcriptional regulator | Frankia alni ACN14a | 33/46 |
| putative activator | S. carzinostaticus | 35/45 | |||
| abyI | 5745 | 252 | transcriptional regulator, SARP family | S. longisporoflavus | 50/64 |
| activator protein | S. hygroscopicus ATCC 53653 | 48/63 | |||
| abyK | 8827 | 579 | YD repeat | Frankia sp. EAN1pec | 46/57 |
| RHS repeat-containing protein | Amycolatopsis mediterranei U32 | 36/50 | |||
| abyA1 | 8911 | 341 | β-ketoacyl-acyl-carrier protein synthase I | Frankia sp. EAN1pec | 68/81 |
| 3-oxoacyl-(acyl-carrier protein) synthase III | Streptomyces sp. NRRL 11266 | 64/75 | |||
| abyA2 | 9933 | 622 | phosphatase and glyceryl transferase | Streptomyces sp. NRRL 11266 | 57/68 |
| ChlD1 | S. antibioticus | 57/68 | |||
| abyA3 | 11 798 | 78 | discrete ACP | Streptomyces sp. NRRL 11266 | 58/75 |
| ChlD2 | S. antibioticus | 53/72 | |||
| abyA4 | 12 031 | 251 | dehydrogenase catalytic domain-containing protein | Frankia sp. EAN1pec | 64/78 |
| pyruvate/2-oxogluatarate dehydrogenase | Streptomyces sp. NRRL 11266 | 60/73 | |||
| abyA5 | 12 783 | 355 | hydrolase superfamily dihydrolipoamide acyltransferase-like protein | Actinomadura kijanata | 54/68 |
| ChlD4 | S. antibtioticus | 51/65 | |||
| abyB1 | 13 847 | 5781 | PKS I (module 1: KS, ATa, ACP; module 2: KSQ, ATa, DH, KR, ACP; module 3: KS, ATa, DH, KR, ACP; module 4: KS, ATa, DH, KR, ACP) | S. avermitilis MA-4680 | 48/59 |
| abyB2 | 31 269 | 3645 | PKS I (module 5: KS, ATp, DH, KR, ACP; module 6: KS, ATp, DH, ER, KR, ACP) | Streptomyces sp. DSM 21069 | 50/61 |
| abyB3 | 42 203 | 992 | PKS I (module 7: KS, ATa, ACP) | S. antibioticus | 54/65 |
| abyC | 45 942 | 230 | regulatory protein, TetR | Frankia sp. EAN1pec | 60/75 |
| Brucella abortus biovar 1 str. 9-941 | 40/56 | ||||
| abyD | 46 026 | 475 | drug resistance transporter EmrB/QacA subfamily | Frankia sp. EAN1pec | 66/79 |
| export protein | S. antibioticus | 44/60 | |||
| abyE | 47 524 | 335 | luciferase; alkanal monooxygenase α-chain | Frankia sp. EAN1pec | 60/74 |
| flavin-utilizing monooxygenases | Brucella melitensis 16M | 45/64 | |||
| abyF1 | 48 619 | 538 | ABC transporter oligopeptide binding protein | Frankia alni ACN14a | 45/59 |
| peptide ABC transporter, periplasmic peptide-binding protein | Klebsiella pneumoniae 342 | 40/55 | |||
| abyF2 | 50 232 | 311 | ABC transporter oligopeptide permease | Frankia alni ACN14a | 53/74 |
| binding protein-dependent transport systems inner membrane component | Frankia sp. CcI3 | 51/69 | |||
| abyF3 | 51 164 | 283 | binding protein-dependent transport systems inner membrane component | Frankia sp. EAN1pec | 54/70 |
| oligopeptide ABC transporter permease protein | Symbiobacterium thermophilum IAM 14863 | 45/62 | |||
| abyF4 | 52 005 | 539 | peptide ABC transporter ATP-binding protein | Frankia alni ACN14a | 59/71 |
| ABC transporter ATP-binding protein | Janthinobacterium sp. Marseille | 52/63 | |||
| abyV | 53 621 | 395 | cytochrome P450 | Frankia sp. EAN1pec | 63/72 |
| cytochrome P450 hydroxylase | S. avermitilis MA-4680 | 51/63 | |||
| abyW | 54 771 | 302 | alcohol dehydrogenase zinc-binding domain protein | S. bingchenggensis BCW-1 | 64/76 |
| oxidoreductase | Streptomyces hygroscopicus ATCC 53653 | 60/74 | |||
| abyZ | 55 601 | 165 | NAD(P)H-dependent FMN reductase | S. clavuligerus ATCC 27064 | 73/81 |
| S. viridochromogenes DSM 40736 | 75/86 | ||||
| abyT | 55 726 | 298 | thioesterase | Nostoc punctiforme PCC 73102 | 35/52 |
| oleoyl-(acyl-carrier protein) hydrolase | Haliangium ochraceum DSM 14365 | 42/55 | |||
| orfU | 57 429 | 302 | alcohol dehydrogenase zinc-binding domain protein | S. bingchenggensis BCW-1 | 64/76 |
| oxidoreductase | S. hygroscopicus ATCC 53653 | 60/74 | |||
| orfS | 57 520 | 197 | transcriptional regulator, TetR family protein | S. bingchenggensis BCW-1 | 59/70 |
| putative transcriptional regulator | S. hygroscopicus ATCC 53653 | 57/72 |
Upstream of abyA1–A5, five genes were considered to be part of the aby cluster: abyH, a LuxR transcriptional regulator homologue; abyI and abyR, genes encoding putative pathway specific activator proteins (SARP family); abyX, a cytochrome P450 gene and abyK encoding a protein with YD repeats. Gene orfQ, encoding a putative NUDIX hydrolase, is not considered to be necessary for abyssomicin C biosynthesis. Downstream of the PKS I genes, those encoding a TetR-like regulatory protein (abyC), a drug resistance transporter belonging to the EmrB/QacA subfamily (abyD), a monooxygenase (abyE), an ABC-transporter system (abyF1–F4), a cytochrome P450 system (abyV, abyW and abyZ) and a type II thioesterase (abyT) form part of the putative gene cluster, whereas orfU, encoding an alcohol dehydrogenase-like protein, and orfS, a TetR-like regulatory protein, were not considered to be involved in abyssomicin formation. Finally, the identity of the gene cluster was confirmed by inactivation of PKS I in abyB1 by single crossover integration by using a fragment internal to the gene cloned into the nonreplicating vector pK18mob2; this abolished atrop-abyssomicin C production as judged by LC-ESI-MS analysis.
Three genes in the aby cluster code for a seven-module type I PKS assembly line for the polyketide backbone of abyssomicin C (Scheme 2). The first gene, abyB1, consists of four modules with the minimal set of ketosynthase (KS), acyltransferase (AT) and acyl carrier protein (ACP) for the first module. The subsequent three modules consist of the same basic set, each extended by dehydratase (DH) and ketoreductase (KR) domains arranged collinearly with their functions in the biosynthetic assembly line. Module 5 of abyB2 has the same arrangement as module 4; the presence of a DH and KR would appear to contradict the collinearity rule.16 However, consistent with the proposed model of biosynthesis (Scheme 2) the DH and KR domains of module 5 (AbyB2–DH1 and AbyB2–KR1, respectively) are likely to be inactive. The active site for DH domains is a His–Asp catalytic dyad,17 while an active site triad of Lys, Ser and Tyr residues is required for KR activity.18 Using AbyB2 numbering, AbyB2–DH1 has substitutions His→Leu953 and Asp→Thr1184, while AbyB2–KR1 has Lys→Ile1402 and Tyr→Val1439. Module 6 contains an additional enoylreductase (ER) domain while module 7 of AbyB3 adds an additional acetate unit. The specificity of AbyB2–AT1 and AbyB2–AT2 for propionate incorporation is determined by the amino acids following Arg56: RVDVV-7M-1-S-1-AXhW (the motif described by Haydock et al.18). The published consensus sequence for acetate specificity of the rest of the AT domains is less well conserved in the abyC cluster (corresponding conserved residues are shown in bold): instead of ETGYA-7-Q-1A-1-FGLL,18 RTE-1-AQPAlFa-1-E-1-AL-2-LL is found in a sequence alignment of all acetate-carrying AT domains.
Scheme 2.
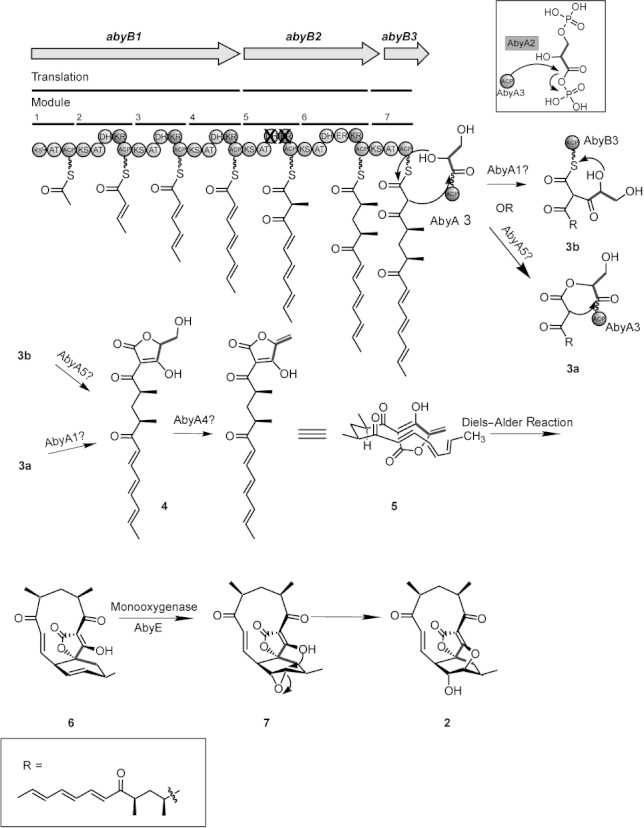
Model of the atrop-abyssomicin C biosynthesis shows the formation of a linear precursor from acetate and propionate precursors on AbyB1, AbyB2 and AbyB3. Bisphosphoglycerate from primary metabolism is attached to the discrete ACP, AbyA3, by the acyl transferase, AbyA2. Subsequently, transfer of the linear polyketide on AbyA3 carrying glyceraldehyde is followed by the formation of the tetronate unit 4. The order of reactions for the C–C and C–O bonds remains undefined. AbyA1 is similar to a β-ketoacyl-ACP-synthase III, which is known to catalyse the first condensation step in fatty acid biosynthesis, and could catalyse formation of the C–C bond to yield derivative 3 b and finally 4.12a AbyA5 shows homology to the α/β-hydrolase protein family and could catalyse the C–O bond and lead to derivative 3 a and finally 4. In other tetronic acid antibiotic biosynthetic gene clusters, an FAD-dependent oxidoreductase has been proposed to catalyse the dehydration of 4 to yield 5.12a, d However, in the atrop-abyssomicin C biosynthetic gene cluster, this gene is not present. Instead, AbyA4, a dehydrogenase catalytic domain-containing protein could catalyse this reaction. Following the formation of the tetronic acid moiety, a Diels–Alder reaction is proposed to form product 6. After suggested epoxide formation by the oxygenase AbyE, ring opening of 7 leads to formation of atrop-abyssomicin C (2).
All four previously sequenced tetronate biosynthetic gene clusters (chlorothricin: chl;12c kijanimicin: kij;12d tetronomycin: tmn;12a tetrocarcin: tca12b) contain a unique set of four or five genes. In the case of tetrocarcin and kijanimicin, two genes (homologues of chlD3 and clhD4) are fused together to form a single bifunctional enzyme (TcaD3 and KijE, respectively).12b Originally, it was proposed that only four of these genes are involved in tetronic acid biosynthesis since inactivation of chlM of the chlorothricin gene cluster did not abolish chlorothricin production. Recent elucidation of the biosynthetic gene clusters of three more tetronic acid-containing antibiotics, and the identification of additional copies of chlM in the genome of the chlorothricin producer (which would explain why inactivation of chlM in the chl cluster did not interrupt chlorothricin biosynthesis) suggests that all five conserved genes play a role.12a, b, d The presence of homologues of these five genes (abyA1–A5) in the abyssomicin gene cluster strengthens this hypothesis (see Scheme 2 for a proposed biosynthetic pathway).
In this study, in-frame deletions of abyA1, abyA2, abyA3 and abyA4 completely abolished abyssomicin production; this confirms their involvement in atrop-abyssomicin C biosynthesis (Scheme 2). However, biochemical evidence for the individual steps in tetronic acid biosynthesis remains scarce and only the first two steps have been examined in vitro.12a Firstly, a glyceryl moiety from d-1,3-bisphosphoglycerate is transferred to Tmn7a and ChlD2 (discrete ACPs homologous to AbyA3), by Tmn16 (a homologue of FkbH and AbyA2), leading to glyceryl-S-ACP (Scheme 2). The second step, that is, the binding of glyceryl-S-ACP to the nascent polyketide chain and detachment of the polyketide from the polyketide synthase, remains elusive together with the subsequent dehydration. Sun et al.19 used a “minimal set” of only six genes and suggested that rkD, a 3oxoacyl-acyl carrier protein synthase III (homologue of abyA1), is capable of catalysing both C–C and C–O bond formation during biosynthesis of the tetronate moiety.12a
However, it remains to be seen whether these results can be directly transferred to the more complex biosynthetic pathways for tetronates, such as abyssomicin, tetronomycin or tetrocarcin. Further experiments will be needed to determine the precise mechanisms involved. For initiation of tetronate ring formation of abyssomicin, we propose the C–O bond as the first step (Scheme 2). This is performed by attack of the primary hydroxyl group of the glyceryl-ACP (AbyA3) on the polyketide thioester on AbyB3. This reaction might be catalysed by an acyltransferase/thioesterase (AbyA5). Formation of the C–O bond would release the polyketide from the polyketide synthase enzyme, AbyB3. Subsequent formation of the tetronate ring could occur by nucleophilic attack of the C-2 carbon. This step could be catalysed by AbyA1 but also an uncatalysed reaction from the enol-form is conceivable. Alternatively, AbyA5, ChlD4 and Tmn17, as predicted acyltransferases, could catalyse this C–O bond formation step.12a
Interestingly, a gene encoding a FAD-dependent oxidoreductase like KijA or Tmn9, which has been suggested to be involved in the formation of the tetronic acid moiety by catalysing dehydration and subsequent Diels–Alder reaction, was not found in the abyssomicin gene cluster. For the chlorothricin and tetrocarcin gene clusters, cross complementation experiments, in vivo, have shown that the homologues of AbyA3 and AbyA4 (encoding an ACP and dehydrogenase, respectively) are interchangeable, which is not surprising given the strong structural similarity between chlorothricin and tetronomycin.12b Sequence comparison of the five tetronic acid biosynthetic gene clusters at the protein level revealed that kijanimicin and tetrocarcin are very closely related (78 % overall identity), followed by chlorothricin (63 % identity with kijanimicin). Atrop-abyssomicin and tetronomycin show 61 % amino acid sequence identity (see the Supporting Information, phylogenetic analysis and percentage identity table).
After formation of the tetronic acid moiety to give linear intermediate 5 (Scheme 2), we propose the occurrence of an intramolecular Diels–Alder reaction to yield intermediate 6. Many natural products are postulated to be Diels–Alder products.20 However, so far only two natural enzymes have been purified to homogeneity and their role as catalysts for Diels–Alder reactions confirmed.21 Analysis of the aby gene cluster did not identify a candidate for such an enzyme, neither did any of the gene-inactivation experiments yield a linear precursor. From work on the total synthesis of abyssomicin, it is known that the methoxy-protected analogue of the linear tetronate is converted to the corresponding Diels–Alder product under very mild conditions:9c after one week at 25 °C in CDCl3, 40 % had reacted to form the methoxy-protected analogue of 6. Hence, it is reasonable to assume that atrop-abyssomicin C can be formed under physiological conditions in the absence of a putative Diels–Alderase. Likewise, the gene clusters for other members of the tetronate family, kijanimicin, chlorothricin and tetrocarcin, for which Diels–Alder reactions have also been proposed,12c, d do not suggest enzyme candidates for a Diels–Alderase. The only candidate proteins for supporting or execution of Diels–Alder reaction are the polyketide synthases (AbyB1–3) and proteins involved in post-PKS modifications.
The biosynthetic logic of abyssomicin assembly further requires the formation of epoxide 7 (Scheme 2), which can then be opened by the tetronate hydroxyl group. AbyE is a putative monooxygenase, which is highly homologous to a large family of bacterial luciferase-like proteins. Little is known about this family and most members are not biochemically characterised. InterProScan22 analysis shows that bacterial luciferases are flavin monooxygenases that catalyse the oxidation of long-chain aldehydes and release energy in the form of visible light by using flavin as a substrate rather than a cofactor. In-frame deletion of abyE led to a significant reduction of atrop-abyssomicin C production (less than 10 % of wild-type levels). Detailed analysis of the culture filtrate and extract by using HPLC-ESI-MS showed no candidate masses for possible intermediary structures.
AbyX is a cytochrome P450, and thus, also a candidate for oxygenation of Diels–Alder product 6 (Scheme 2). In-frame deletion of abyX led to reduction of abyssomicin production to about 3 % of wild-type levels. One further candidate for oxygenation of 6 is cytochrome P450, AbyV. Interruption of abyV with single crossover recombination led to loss of abyssomicin production; however, polar effects on the expression of downstream genes cannot be ruled out. Detailed analysis of the culture filtrates of both mutants by using HPLC-MS did not show any candidate masses for possible intermediary structures. Failure to detect any biosynthetic intermediate by LC-ESI-MS prior to oxidative epoxide formation was unexpected, since precursors 5 and 6 were expected to be released by the polyketide assembly line, and mass spectrometry is the method of choice even if the compounds are unstable. Ultimately, in vitro experiments will be needed to determine the exact mechanism for oxygenation of the proposed Diels–Alder product 6.
Upstream of the PKS I genes, there are three genes encoding homologues of known regulators of secondary metabolism: abyH, encoding a LuxR family transcriptional regulator, and abyI and abyR, both encoding putative pathway specific activator proteins belonging to the SARP (Streptomyces antibiotic regulatory protein) family of transcriptional activators. As expected, in-frame deletion of abyI abolished abyssomicin production. Complementation of the mutant with a copy of the gene by using vectors pSETermEΔHindIII and pUWLoriT restored production to wild-type levels in both cases (for details see the Supporting Information). In-frame deletion of abyR led to a reduction in abyssomicin production; this suggests some degree of involvement in regulation of abyssomicin biosynthesis.
LuxR regulators, like AbyH can be activators or repressors, which, acting through a quorum sensing mechanism, induce transcription when a certain cell density is reached.23 The role of AbyH in abyssomicin production remains to be determined.
According to its sequence data AbyD belongs to the major facilitator superfamily (MFS) of bacterial transporters. MFS transporters are capable of transporting small solutes in response to chemiosmotic ion gradients.24 AbyD belongs to the EmrB/QacA subfamily of drug efflux pumps, related to the tetracycline resistance protein, TetB. Many multidrug transporters in this superfamily are negatively regulated by the TetR family of transcriptional regulators.25 Consistent with this is the location of abyC (predicted to encode a TetR-like transcriptional regulator), which is upstream of, and divergently transcribed from, abyD. Examination of the intergenic region between abyC and abyD revealed a 16 bp perfect palindromic repeat sequence, TGAACTGATCAGTTCA, that is likely to be the operator binding sequence for AbyC. In-frame deletions of abyC and abyD both resulted in a reduction of atrop-abyssomicin C production to less than 10 % of the wild-type level. Self-resistance might, therefore, stem from the efficient export of abyssomicin from the cell; the low level of abyssomicin production observed in the ΔabyC and ΔabyD strains can be explained by a degree of passive diffusion from the cell or by partial suppression of ΔabyD by another MFS transporter with broader substrate tolerance. The former possibility has been suggested to explain the excretion of heterologously produced polyfunctional aromatic compounds by mutants of Streptomyces coelicolor that lack the actinorhodin transporters.26
Downstream of abyD are abyF1–4, which are predicted to encode an ABC transporter system. Mutations in these genes have yet to be made. In all cases studied so far, ABC transporters that contain an extracellular solute binding protein, such as that encoded by AbyF1, are importers—although sometimes efflux is also facilitated, it always leads to a net uptake of solutes.27 This suggests that these genes might be involved in the biosynthesis, rather than export of atrop-abyssomicin C, and possibly in the import of a compound required for atrop-abyssomicin C production. While the precursors for biosynthesis should be present in the intracellular metabolite pool, Verrucosispora maris AB-18-032 requires the presence of glycerol in the fermentation medium for atrop-abyssomicin C production. Glycerol is not the origin of the three-carbon unit incorporated into the C-14–C-16 position of atrop-abyssomicin C, but it functions as an autoinducer of pimaricin biosynthesis in Streptomyces natalensis and stimulates the production of six different macrolide antibiotics.28 Glycerol is imported by an ABC transporter complex in Mycobacterium species,27 and thus one possible role for AbyF1–4 is the uptake of glycerol or another solute involved in stimulating atrop-abyssomicin C production.
For many of the aby genes, the highest similarity is to a counterpart in Frankia alni ACN14a and/or Frankia sp. EAN1pec, both found in whole genome sequencing projects.29 Hence, an interesting profile of secondary metabolite production can be expected from these organisms. Examination of the genome of Frankia alni ACN14a (GenBank: CT573213.2) revealed a putative spirotetronate biosynthetic gene cluster that contains homologues of most of the genes in the aby cluster, including abyK (Figure 2). This region (FRAAL4069–4097) contains abyF1–4 homologues in proximity to those for a MFS protein, a TetR regulator, a monooxygenase and a cytochrome P450. Elsewhere in the genome of Frankia alni (ddpD-FRAAL0342) this gene organisation is repeated with greater synteny to the aby cluster, but the product of the biosynthetic gene cluster in this region is not predicted to be a tetronate as it lacks homologues of the genes required for the biosynthesis of the tetronic acid moiety (see above).
Figure 2.
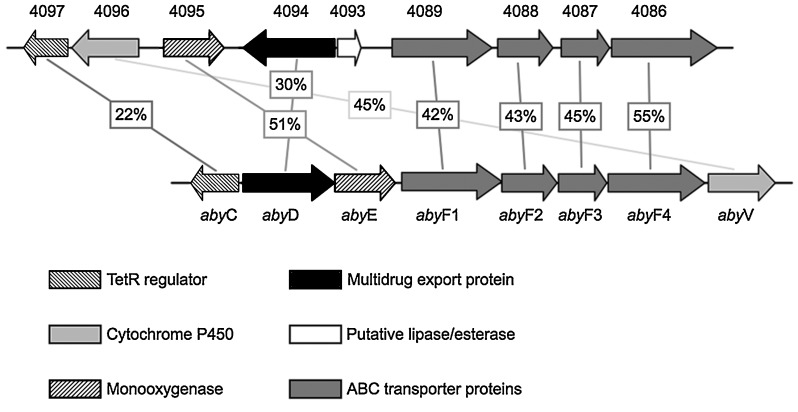
Comparison of the genes predicted to be involved in the export of abyssomicin and a putative spirotetronate biosynthetic gene cluster present in the genome of Frankia alni ACN14a (GenBank accession: CT573213.2). The numbers in boxes represent the percentage identify (amino acid) between the proteins (joined by lines). The putative lipase/esterase might function as an editing thioesterase as predicted for abyT (located downstream of abyV).
According to InterProScan analysis, abyK contains several tandem copies of a 21 residue extracellular repeat that is found in Gram-negative, Gram-positive, and animal proteins. The repeat is named for a YD dipeptide, the most strongly conserved motif of the repeat. These repeats appear to be involved in binding carbohydrates; for example, the chicken teneurin-1 YD-repeat region binds heparin.30 In-frame deletion of abyK led to a decrease of abyssomicin C production to roughly 30 % of wild-type levels. Complementation of the knockout mutant with a copy of the gene on a plasmid transcribed from the ermE* promoter restored production to wild-type levels. Interestingly, homologues of this gene are contained in the gene clusters for tetrocarcin, chlorothricin and tetronomycin, albeit as shorter versions (164, 191 and 190 aa, respectively). In the gene clusters for tetrocarcin and tetronomycin, the YD-repeat proteins are located directly upstream of the genes responsible for tetronic acid biosynthesis. Therefore, it is likely that this gene has some relevance for the biosynthesis of the aforementioned tetronates, however, its exact function remains elusive.
Discrete type II thioesterase domains (TEs) have been predicted to play a role in editing stalled polyketide chains.26 The TEs are thought to patrol the PKS assembly line in trans, and hydrolytically remove any inappropriate monomers misincorporated by the AT domains.31 The chlorothricin, kijanimicin, tetrocarcin and tetronomycin biosynthetic gene clusters all have discrete TEs (ChlK, KijB2, TcaF and Tmn6, respectively). In-frame deletion of abyT resulted in approximately 50 % lower levels of production than that of the wild type, which is consistent with an editing function in abyssomicin biosynthesis.
Spirotetronates form a highly complex, and very often, extremely functionalised class of polyketides. Among these, atrop-abyssomicin C is the smallest and simplest example of a spirotetronate, the biosynthetic gene cluster of which has been determined. Apart from the comparatively short polyketide chain (seven modules) we found five genes (abyA1–abyA5) assigned to enzymes catalysing formation of the tetronic acid moiety and three genes coding for oxidising enzymes (abypE, abyX and abyV). Apart from one gene, the function of which remains obscure (abyK), the remaining genes of the cluster are assigned to export, possibly import, and regulatory functions (abyI, abyH, abyC, abyD, abyR, abyI, abyT and abyF1–abyF4). The work reported here describes the first example of the genetic manipulation of a member of the genus Verrucosispora, and paves the way for future detailed studies of secondary metabolism in this group of scarcely studied microorganisms.
Experimental Section
Fermentation and isolation: For growth of Verrucosispora maris AB-18-032 strains, the production medium SGG (1 % dextrin, 1 % glucose, 1 % glycerol, 0.25 % corn steep powder from Marcor, Carlstadt, NJ, USA, 0.5 % peptone, 0.2 % yeast extract, 0.1 % NaCl, 0.3 % CaCO3 in 1 L tap water, adjusted to pH 7.3 prior to sterilisation) was dispersed in Erlenmeyer flasks (500 mL; 10× for each cultivation) with three baffles. The flasks were inoculated with 3 v % of a 48 h old preculture and incubated at 27 °C and 120 rpm on a rotary shaker. After 72 h the cultures were harvested by centrifugation. The supernatants were used for isolation of abyssomicins, as described previously.3a
13C-Isotope labelling studies: In parallel experiments 500 mg of 13C-labelled precursors [1-13C]acetate, [1,2-13C]acetate and [1-13C]propionate, and [1,2-13C]glucose (250 mg; Eurisotop, Saint Aubin, France) were fed to producing cultures (1 L) of Verrucosispora maris AB-18-032 in SGG liquid medium in five portions: at 48, 51, 54, 57 and 60 h after inoculation. After 72 h of cultivation, the bacteria were harvested and 13C-labelled atrop-abyssomicins C was isolated.3a
HPLC-ESI-mass spectrometry: The supernatants of 72 h cultures (5 mL) of Verrucosispora maris AB-18-032 were adjusted to pH 4 (aq. HCl) and subsequently extracted with ethyl acetate (5 mL). The organic phase was separated and the solvent evaporated, in vacuo. The dry residue was dissolved in methanol (500 μL), centrifuged and analysed on an Agilent 1100 analytical HPLC system (Waldbronn, Germany) equipped with a Luna RP-C18 column (4.6 mm i.d., Phenomenex, Aschaffenburg, Germany) and a diode array detector (5 % B (acetonitrile) to 100 % B in 10 min, flow rate: 1.5 mL min−1, tR=5.6 min). Relative production was estimated by peak areas compared to wild-type fermentations. ESI-MS measurements: single-ion monitoring set to m/z 374.1, polarity: positive, fragmentor: 135 V (ESI-Triple-Quadrupol-MS, 6460 Series, Agilent Technologies).
NMR spectroscopy: 1H- and 13C NMR spectroscopy experiments were measured on a DRX500 NMR spectrometer (Bruker) equipped with a 5 mm diameter broad band inverse probe head with z gradients. Spectra were recorded in and referenced to [D4]methanol (3.30 ppm; 49.0 ppm). Additional spectra are shown in the Supporting Information.
Bacterial strains, plasmids, and reagents: E. coli strains and plasmids used in this work were obtained from Combinature Biopharm, AG (Berlin, Germany). Biochemicals, chemicals and media were obtained from Roth unless indicated otherwise. Restriction enzymes and molecular biological reagents were obtained from standard commercial sources, such as Qiagen, New England Biolabs and Fermentas. Primers were ordered from biomers.net (Ulm, Germany).
DNA isolation, genomic library construction and screening: A cosmid library of Verrucosispora maris AB-18-032 genomic DNA (2304 clones) was constructed, as described previously.32 Homogenised bacterial cultures were immersed in low-melting agarose (0.5 %; SeaPlaque GTG, Biozym, Hessisch Oldendorf, Germany) and incubated with HEW lysozyme (2 mg mL−1; Roth) for 14 h at room temperature and with proteinase K (1 mg mL−1; Merck) for 24 h at 50 °C. The embedded DNA was partially digested with Sau3AI and extracted with Gelase (Epicentre, Madison, WI, USA) and dephosphorylated (Antarctic Phosphatase, New England Biolabs). The fragments of genomic DNA (∼50 kb long) were ligated with BamHI digested vector pOJ436 (750 ng), desalted and packaged by using Gigapack III Gold packaging extract (Stratagene).33 The cosmid clones were screened by hybridisation with a DIG-labelled PKS probe (Roche DIG-labelling kit), which was amplified from Verrucosispora maris AB-18-032 genomic DNA by using primers KSIIFOR (5′-CTSGGSGACCCSATCGAG-3′) and ATIREV (5′-GCSGCSGCGATCTCSCCCTGSSWGTGSCC-3′); 49 PKS I positive cosmid clones were identified.
Whole genome sequencing (WGS) of Verrucosispora maris AB-18-032: Genomic DNA from Verrucosispora maris AB-18-032 was prepared by using a standard CTAB procedure.33 Genomic DNA was further purified by passaging twice through a 20/G genomic-tip according to the manufacturers instructions (Qiagen). Purified DNA was used for whole-genome shotgun pyrosequencing by using a Genome Sequencer 20 instrument (Roche) and GS emPCR Kit I, according to the manufacturers instructions (Roche). Pryosequencing reads representing 130 Mb of data (ca. 20.4× coverage) were assembled by using the Newbler assembler version 1.1.03.24 (Roche) into 1276 contigs with an N50 contig size of 8455 bp.
Cosmid analysis and shotgun cloning: To identify cosmids carrying PKS I genes for abyssomicin C biosynthesis, all 49 cosmids containing PKS I genes were digested with BamHI and SmaI and separated on an agarose gel. After hybridisation with DIG-labelled PKS I probes, cosmids were placed into families containing overlapping genomic sequences. Three major families (five to eight members each) and four smaller families (two members each) as well as 14 apparently unique cosmids were identified. One cosmid from each family was chosen and subjected to restriction digestion with BamHI. The fragments were cloned in pBKS and sequenced. PKS I positive inserts were then cloned into pK18mob234 and used for mutational inactivation in Verrucosispora maris AB-18-032 by single crossover recombination. The resulting mutants were screened for the absence of abyssomicin C production. Cosmid family II was found to carry genes required for abyssomicin C biosynthesis and one cosmid from this family (C17) was chosen for sequencing.
Cosmid sequencing and sequence assembly: DNA sequencing was carried out by AGOWA GmbH (Berlin, Germany). Cosmid C17 (insert size 35 567 bp) was sequenced by shotgun sequencing (4× coverage); overlapping regions on cosmids C45 and C49 were identified and sequenced by chromosome walking.
Sequence analysis: Open reading frame (ORF) prediction was carried out by using Glimmer (http://cbcb.umd.edu/software/glimmer/) and Frameplot (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl). Protein functions were assigned with an xblast (http://www.ncbi.nlm.nih.gov/blast/) search as well as an InterProScan (http://www.ebi.ac.uk/InterProScan/) domain analysis. WGS contigs containing DNA from the aby gene cluster were identified by BLAST searching, and were manually annotated by using Artemis sequence visualisation and annotation software35 and BLAST search tools. The annotated sequence is available at http://www.ncbi.nlm.nih.gov/Genbank/index.html under GenBank accession number JF752342.
Gene inactivation: Two different methods of gene inactivation were applied. Function assignment and cluster boundary assignment was achieved with single crossover homologous recombination by using plasmid pK18mob2.34 In-frame deletion of genes was achieved following the ReDirect protocol by using λ-Red-mediated recombination, and classical gene deletion methods.36 Procedures, primers and constructs are described in the Supporting Information.
Acknowledgments
The authors wish to thank Dr. Bertolt Gust and Dr. Tilmann Weber for advice and helpful discussions and Dipl.-Ing. Nicole Sattler for technical assistance. This work was funded by the German Research Foundation (DFG), project SU239/8-1, the Cluster of Excellence UniCat and the Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/E017 053/1.
Supplementary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- 1a.Fiedler HP, Bruntner C, Bull AT, Ward AC, Goodfellow M, Potterat O, Puder C, Mihm G. Antonie van Leeuwenhoek. 2005;87:37–42. doi: 10.1007/s10482-004-6538-8. [DOI] [PubMed] [Google Scholar]
- 1b.Herold K, Gollmick FA, Groth I, Roth M, Menzel KD, Mollmann U, Grafe U, Hertweck C. Chem. Eur. J. 2005;11:5523–5530. doi: 10.1002/chem.200500320. [DOI] [PubMed] [Google Scholar]
- 1c.Takahashi Y, Omura S. J. Gen. Appl. Microbiol. 2003;49:141–154. doi: 10.2323/jgam.49.141. [DOI] [PubMed] [Google Scholar]
- 1d.Tsurumi Y. Kagaku (Kyoto, Japan) 2003;58:28–29. [Google Scholar]
- 2.Clardy J, Fischbach MA, Walsh CT. Nat. Biotechnol. 2006;24:1541–1550. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 3a.Bister B, Bischoff D, Ströbele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zähner H, Fiedler HP, Süssmuth RD. Angew. Chem. 2004;116:2628–2630. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2004;43:2574–2576. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 3b.von Nussbaum F, Brands M, Hinzen B, Weigand S, Häbich D. Angew. Chem. 2006;118:5194–5254. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2006;45:5072–5129. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]
- 4.Coggins CAJR, Evans LB, Frederickson M, Robinson DA, Roszak AW, Lapthorn AP. Biochem. Soc. Trans. 2003;31:548–552. doi: 10.1042/bst0310548. [DOI] [PubMed] [Google Scholar]
- 5a.Domagk G. Dtsch. Med. Wochenschr. 1935;61:250–253. [Google Scholar]
- 5b.Fuller AT. Lancet. 1937;229:194–198. [Google Scholar]
- 5c.Northey EH. Chem. Rev. 1940;27:85–197. [Google Scholar]
- 5d.Nicol CA, Welch AD. Proc. Soc. Exp. Biol. Med. 1950;74:403–411. doi: 10.3181/00379727-74-17922. [DOI] [PubMed] [Google Scholar]
- 5e.Burchall GHHJ. Mol. Pharmacol. 1965;1:126–136. [PubMed] [Google Scholar]
- 6.Riedlinger J, Reicke A, Zähner H, Krismer B, Bull AT, Maldonado LA, Ward AC, Goodfellow M, Bister B, Bischoff D, Süssmuth RD, Fiedler HP. J. Antibiot. 2004;57:271–279. doi: 10.7164/antibiotics.57.271. [DOI] [PubMed] [Google Scholar]
- 7.Keller S, Nicholson G, Drahl C, Sorensen E, Fiedler HP, Süssmuth RD. J. Antibiot. 2007;60:391–394. doi: 10.1038/ja.2007.54. [DOI] [PubMed] [Google Scholar]
- 8a.Schadt HS, Schadt S, Oldach F, Süssmuth RD. J. Am. Chem. Soc. 2009;131:3481–3483. doi: 10.1021/ja809283u. [DOI] [PubMed] [Google Scholar]
- 8b.Keller S, Schadt H, Ortel I, Süssmuth RD. Angew. Chem. 2007;119:8433–8435. doi: 10.1002/anie.200701836. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2007;46:8284–8286. doi: 10.1002/anie.200701836. [DOI] [PubMed] [Google Scholar]
- 9a.Nicolaou KC, Harrison ST. Angew. Chem. 2006;118:3334–3338. [Google Scholar]
- Angew. Chem. Int. Ed. 2006;45:3256–3260. [Google Scholar]
- 9b.Peters R, Fischer DF. Angew. Chem. 2006;118:5866–5869. [Google Scholar]
- Angew. Chem. Int. Ed. 2006;45:5736–5739. doi: 10.1002/anie.200602409. [DOI] [PubMed] [Google Scholar]
- 9c.Snider BB, Zou Y. Org. Lett. 2005;7:4939–4941. doi: 10.1021/ol0518941. [DOI] [PubMed] [Google Scholar]
- 9d.Zapf CW, Harrison BA, Drahl C, Sorensen EJ. Angew. Chem. 2005;117:6691–6695. doi: 10.1002/anie.200502119. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2005;44:6533–6537. doi: 10.1002/anie.200502119. [DOI] [PubMed] [Google Scholar]
- 9e.Zhang XJ, Parry RJ. Antimicrob. Agents Chemother. 2007;51:946–957. doi: 10.1128/AAC.01214-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9f.Rath J-P, Eipert M, Kinast S, Maier ME. Synlett. 2005:314–318. [Google Scholar]
- 9g.Rath J-P, Kinast S, Maier ME. Org. Lett. 2005;7:3089–3092. doi: 10.1021/ol0511068. [DOI] [PubMed] [Google Scholar]
- 10.Nicolaou KC, Harrison ST. J. Am. Chem. Soc. 2007;129:429–440. doi: 10.1021/ja067083p. [DOI] [PubMed] [Google Scholar]
- 11.Schindler PW, Zähner H. Eur. J. Biochem. 1973;39:591–600. doi: 10.1111/j.1432-1033.1973.tb03158.x. [DOI] [PubMed] [Google Scholar]
- 12a.Demydchuk Y, Sun YH, Hong H, Staunton J, Spencer JB, Leadlay PF. ChemBioChem. 2008;9:1136–1145. doi: 10.1002/cbic.200700715. [DOI] [PubMed] [Google Scholar]
- 12b.Fang J, Zhang Y, Huang L, Jia X, Zhang Q, Zhang X, Tang G, Liu W. J. Bacteriol. 2008;190:6014–6025. doi: 10.1128/JB.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12c.Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W. Chem. Biol. 2006;13:575–585. doi: 10.1016/j.chembiol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 12d.Zhang H, White-Phillip JA, Melancon CE, Kwon HJ, Yu WL, Liu HW. J. Am. Chem. Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Chijiwa HRPS, Furihata K, Ogata M, Endo T, Kuzuyama T, Hayakawa Y, Shin-Ya K. Tetrahedron Lett. 2003;44:5897–5900. [Google Scholar]
- 13b.Lee JPLJJ, Keller PJ, Cottrell CE, Chang CJ, Zähner H, Floss HG. J. Antibiot. 1986;39:1123–1134. doi: 10.7164/antibiotics.39.1123. [DOI] [PubMed] [Google Scholar]
- 13c.Mashimo Y, Sekiyama Y, Araya H, Fujimoto Y. Bioorg. Med. Chem. Lett. 2004;14:649–651. doi: 10.1016/j.bmcl.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 13d.Tamaoki T, Tomita F. J. Antibiot. 1983;36:595–598. doi: 10.7164/antibiotics.36.595. [DOI] [PubMed] [Google Scholar]
- 14a.Byrne KM, Arison BH, Nallinomstead M, Kaplan L. J. Org. Chem. 1993;58:1019–1024. [Google Scholar]
- 14b.Michal G. Biochemical Pathways: Biochemie-Atlas. Heidelberg: Spektrum; 1999. [Google Scholar]
- 15a.Weber T, Welzel K, Pelzer S, Vente A, Wohlleben W. J. Biotechnol. 2003;106:221–232. doi: 10.1016/j.jbiotec.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 15b.Weber KJLT, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W. Chem. Biol. 2008;15:175–188. doi: 10.1016/j.chembiol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Donadio S, Katz L. Gene. 1992;111:51–60. doi: 10.1016/0378-1119(92)90602-l. [DOI] [PubMed] [Google Scholar]
- 17.Akey JRRDL, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haydock SF, Aparicio JF, Molnar I, Schwecke T, Khaw LE, Konig A, Marsden AFA, Galloway IS, Staunton J, Leadlay PF. FEBS Lett. 1995;374:246–248. doi: 10.1016/0014-5793(95)01119-y. [DOI] [PubMed] [Google Scholar]
- 19.Sun FHY, Demydchuk Y, Chettle J, Tosin M, Osada H, Leadlay PF. Nat. Chem. Biol. 2010;6:99–101. doi: 10.1038/nchembio.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stocking EM, Williams RM. Angew. Chem. 2003;115:3186–3223. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]
- Angew. Chem. Int. Ed. 2003;42:3078–3115. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]
- 21.Kelly WL. Org. Biomol. Chem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 22.Zdobnov EM, Apweiler R. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 23.Nasser W, Reverchon S. Anal. Bioanal. Chem. 2007;387:381–390. doi: 10.1007/s00216-006-0702-0. [DOI] [PubMed] [Google Scholar]
- 24.Pao SS, Paulsen IT, Saier MH., Jr Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 27.Vilei EM, Frey J. Clin. Diagn. Lab. Immunol. 2001;8:85–92. doi: 10.1128/CDLI.8.1.85-92.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Recio E, Aparicio JF, Rumbero A, Martin JF. Microbiology. 2006;152:3147–3156. doi: 10.1099/mic.0.28953-0. [DOI] [PubMed] [Google Scholar]
- 29.Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N, Couloux A, Cournoyer B, Cruveiller S, Daubin V, Demange N, Francino MP, Goltsman E, Huang Y, Kopp OR, Labarre L, Lapidus A, Lavire C, Marechal J, Martinez M, Mastronunzio JE, Mullin BC, Niemann J, Pujic P, Rawnsley T, Rouy Z, Schenowitz C, Sellstedt A, Tavares F, Tomkins JP, Vallenet D, Valverde C, Wall LG, Wang Y, Medigue C, Benson DR. Genome Res. 2007;17:7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Hill CW, Sandt CH, Vlazny DA. Mol. Microbiol. 1994;12:865–871. doi: 10.1111/j.1365-2958.1994.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 30b.Feulner G, Gray JA, Kirschman JA, Lehner AF, Sadosky AB, Vlazny DA, Zhang J, Zhao S, Hill CW. J. Bacteriol. 1990;172:446–456. doi: 10.1128/jb.172.1.446-456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30c.Minet A, Rubin B, Tucker R, Baumgartner S, Chiquet-Ehrismann R. J. Cell Sci. 1999;112:2019–2032. doi: 10.1242/jcs.112.12.2019. [DOI] [PubMed] [Google Scholar]
- 31.Kim BS, Cropp TA, Beck BJ, Sherman DH, Reynolds KA. J. Biol. Chem. 2002;277:48028–48034. doi: 10.1074/jbc.M207770200. [DOI] [PubMed] [Google Scholar]
- 32a.Burgtorf C, Welzel K, Hasenbank R, Zehetner G, Weis S, Lehrac H. Genomics. 1998;52:230–232. doi: 10.1006/geno.1998.5444. [DOI] [PubMed] [Google Scholar]
- 32b.Beye M, Poch A, Burgtorf C, Moritz RFA, Lehrach H. Genomics. 1998;49:317–320. doi: 10.1006/geno.1998.5253. [DOI] [PubMed] [Google Scholar]
- 33.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 34. A Gene Cluster Encoding an Efomycin Polyketide Synthase from a Streptomycete, S. Pelzer, H. Dorsch, T. Spellig (Combinature Biopharm AG, Berlin), DE 102004027516, 2006.
- 35.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream M-A. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc. Natl. Acad. Sci. USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36b.Gust B, Chandra G, Jakimowicz D, Tian YQ, Bruton CJ, Chater KF. Adv. Appl. Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


