Abstract
The lateral nucleus of the amygdala (LA) has been implicated in the formation of long-term associative memory (LTM) of stimuli associated with danger through fear conditioning. The current study aims to detect genes that are expressed in LA following associative fear conditioning. Using oligonucleotide microarrays, we monitored gene expression in rats subjected to paired training where a tone co-terminates with a footshock, or unpaired training where the tone and footshock are presented in a non-overlapping manner. The paired protocol consistently leads to auditory fear conditioning memory formation whereas the unpaired do not. When the paired group was compared to the unpaired group 5 hrs after training, the expression of genes coding for the limbic system-associated membrane protein (Lsamp), kinesin heavy chain member 2 (Kif2), N-ethylmaleimide sensitive fusion protein (NSF) and Hippocalcin-like 4 protein (Hpcal4) was higher in the paired group. These genes encode proteins that regulate neuronal axonal morphology (Lsamp, Kif2), presynaptic vesicle cycling and release (Hpcal4 and NSF), and AMPA receptor maintenance in synapses (NSF). Quantitative real-time PCR (qPCR) revealed that Kif2 and Lsamp are expressed hours following fear conditioning but minutes after unpaired training. Hpcal4 is induced by paired stimulation only 5 hours after training. These results show that fear conditioning induces a unique temporal activation of molecular pathways involved in regulating synaptic transmission and axonal morphology in LA, which is different from non-associative stimulation.
Keywords: Fear conditioning, lateral amygdala, gene expression, memory, microarray
Introduction
Long-term memory (LTM) formation is believed to involve alterations of synaptic efficacy produced by modifications in neural transmission due to physiochemical and/or structural modifications of synaptic communication within neuronal networks (Konorski, 1948; Hebb, 1949; Bliss & Collingridge, 1993; Martin et al., 2000; Tsien, 2000; Lamprecht & LeDoux, 2004). These modifications in turn are made possible by gene expression, which leads to synthesis of RNA and proteins (Davis and Squire 1984; Goelet et al., 1986; Dudai, 1989; Bailey et al, 1996; Kandel 2001). The aim of the current study was to identify genes that are expressed during LTM formation after Pavlovian fear conditioning, a leading model for the study of cellular and molecular mechanisms of memory storage (Fanselow & LeDoux, 1999; LeDoux, 2000; Davis & Whalen, 2001; Blair at el., 2001; Sah et al., 2003; Maren, 2005; Dityatev & Bolshakov, 2005; Fanselow & Poulos, 2005). In fear conditioning, an association is formed between a neutral conditioned stimulus (CS), such as a tone, and an aversive unconditioned stimulus (US), typically a mild footshock. This behavioral paradigm is especially useful as a tool for studying the molecular basis of long-term memory because the putative site of memory, the lateral nucleus of the amygdala (LA), has been identified (Fanselow and LeDoux, 1999; Schafe et al., 2001; Rodrigues et al., 2004; Maren, 2005; Fanselow & Poulos, 2005). Fear conditioning LTM formation requires gene expression and protein synthesis. Indeed, blockade of RNA or protein synthesis in the LA and nearby areas prevents the formation of fear conditioning LTM without disrupting short-term memory (STM) (Bailey et al, 1999; Schafe & LeDoux, 2000; Maren et al., 2003; Duvarci et al., 2008). This observation indicates that RNA and protein syntheses are essential for the formation of fear conditioning memory in the amygdala.
To detect gene expression in the LA following fear conditioning, we used the Affymetrix microarray technology, which allows the simultaneous detection of changes in the expression of thousands of genes. While other studies have investigated gene expression in the amygdala after fear conditioning (e.g. Stork et al., 2001; Mei et al., 2005; Keeley et al., 2006; Pollak et al., 2008,) those have not focused specifically on the LA, a key site of synaptic plasticity in fear conditioning, but instead on the broader region known as the basolateral amygdala (BLA; which includes LA and other areas) (Stork et al., 2001; Keeley et al., 2006; Pollak et al., 2008). To dissect the LA from adjacent amygdala areas we used the laser microdissection technique. To specifically test memory that reflects the CS-US association, as opposed to non associative factors, such as generalized stress that can also induce gene expression, we compared the expression of genes in LA in animals that received associative training (paired CS and US presentations where the US occurs during the CS) vs. animals received the same number of CS and US presentations but in a non-overlapping manner. We initially focused on gene expression in animals sacrificed 5 hrs after training, since genes involved in key neuronal functions and synaptic plasticity are expressed around this time after synaptic stimulation (e.g. Hong et al., 2004). The differential expression between the paired and unpaired groups at 5 hours was verified using qPCR. In addition, qPCR was performed on the same genes but in a different group of animals sacrificed 30 min after training to identify possible temporal changes in the expression of these genes following the training procedure.
Methods
Animals
All studies involved male Sprague Dawley rats (Hilltop Labs, Scottdale, Pennsylvania) weighing 250–300 g. The animals were housed separately in plastic Nalgene cages and placed on a 12 hr light/dark cycle with ad libitum food and water. All procedures were in accordance with the National Institutes of Health guide and were approved by the New York University Animal Care and Use Committee.
Fear conditioning
Fear conditioning took place in a Plexiglas rodent conditioning chamber with a metal grid floor dimly illuminated by a single house light and enclosed within a sound-attenuating chamber (Coulbourn Instruments, Lehigh Valley, Pennsylvania). Rats were habituated to the training chamber for 15 min for 4 consecutive days. On the fifth day the rats were divided into three groups: (1) naïve group exposed to the chamber for 15 minutes with no CS or US; (2) paired group was acclimated for 90 seconds in the conditioning chamber and presented with five pairings of tone for 20 sec (CS: conditioned stimulus; 5 kHz, 75 dB) that co-terminated with a foot shock (US: unconditioned stimulus; 0.5 sec, 1.3 mA). The inter trial interval (ITI) was of 120 seconds in average; and (3) unpaired group was acclimated for 90 seconds in the conditioning chamber and presented with five unpaired stimulation of shock and tone where a 0.5 sec footshock (1.3 mA) was followed by ITI of 60 seconds in average, then a 20 sec tone (5 kHz, 75 dB), and another ITI of 120 seconds in average.
Testing of conditioned fear memory
Rats were tested 24 hrs (LTM) after conditioning for tone memory in a different chamber with black walls and a plastic floor in order to minimize the effect of the context. Animals were subjected to 5 tones (20 sec 5 kHz, 75 dB) with an average ITI of 120 sec. Animal behavior was recorded and the video images were transferred to a computer equipped with an analysis program. The percentage of changed pixels between two adjacent 1 sec images was used as a measure of activity. Time period spent freezing was measured, results of individual 5 test tones were summed and percentage freezing from the total (100 seconds) was calculated for each animal.
Laser Microdissection (LMD)
Rats were sacrificed at 30 minutes or 5 hrs after paired or unpaired stimulation or 30 minutes after non-stimulation training (naïve group) and their brains were rapidly removed, frozen on dry ice and kept in −80°C until further use. The brains were subsequently cut at 20 μm serial frozen sections on a standard cryostat, and sections at the level of the LA were mounted on special polyethylene naphthalate (PEN) slides (Leica, Bannockburn, IL) (sections were collected from the anterior part of LA through the posterior end of LA). Slides were immediately frozen on dry ice and kept at −80°C. At the day of LMD, sections were removed from the freezer, immediately fixed in 100% ethanol for 1 min, and then rehydrated in a graded series of ethanol/water (95%, 75%, 50%; each one minute). Sections were then stained for 20 sec in 1% Thionin in 0.05M NaCH3COO buffer, pH 4.5, dehydrated (50%, 75%, 95%, 100% ethanol, 1 min each), dried for 2 min, and immediately used for microdissection. All solutions were prepared with diethylpyrocarbonate (DEPC)-treated water. The LA was identified on a computer screen and the entire region of interest was laser-microdissected from sections throughout the LA (AS LMD Leica; Figure 1) into a microcentrifuge tube containing RNA extraction solution (PicoPure™ RNA Isolation Kit, Molecular Devices, Sunnyvale, CA).
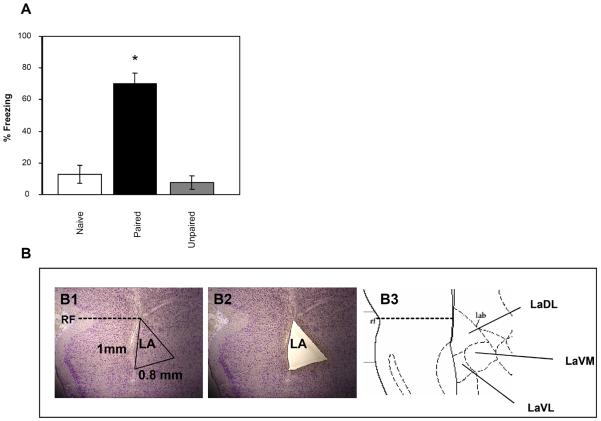
Figure 1. Behavioral outcome of training and representative photograph depicting the laser microdissection of the lateral amygdala.
A- Animals were subjected to the naïve, paired or unpaired protocols (n=4 each) and tested for freezing responses when subjected again to the CS tones 24 hrs later. *=p<0.001. Values are mean ± SEM. B- A thionine stained brain before (B1) and after (B2) laser microdissection. Only the dorsal and ventral lateral amygdala were dissected (B3).
RNA isolation, probe preparation, microarray hybridization and image analysis
Total RNA was extracted from the LMD samples using PicoPure™ RNA Isolation Kit (Molecular Devices, Sunnyvale, CA) according to manufacturer protocol. Quality of RNA was ensured before labeling by analyzing it using the RNA 6000 NanoAssay and a Bioanalyzer 2100 (Agilent, Santa Clara CA). Samples with a 28S/18S ribosomal peak ratio of 1.8–2.0 were considered suitable for labeling. Total RNA was used for cDNA synthesis utilizing an oligo(dT)-T7 primer and the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA). Synthesis, linear amplification, and labeling of cRNA were accomplished by in vitro transcription using the MessageAmp aRNA Kit (Ambion, Austin, TX) and biotinylated nucleotides (Enzo Diagnostics, Farmingdale, NY). Ten micrograms of labeled and fragmented cRNA were then hybridized to the rat array 230A (Affymetrix, Santa Clara, CA), at 45°C for 16 h. Post hybridization staining and washing were processed according to manufacturer (Affymetrix). Finally, chips were scanned with a high-numerical Aperture and flying objective (FOL) lens in the GS3000 scanner (Affymetrix). The image was quantified using MAS 5.1 (MicroArray Suite, Affymetrix) with the default parameters for the statistical algorithm and all probe set scaling with a target intensity of 500. Quality control of hybridization was assessed. No significant differences (p>0.05; exact Mann-Whitney test) in 3' and 5' ratios of GAPDH, Hexokinase and beta-actin RNAs in microarrays were found between the paired and unpaired groups. Furthermore, no significant changes in level of housekeeping genes beta-2-microglobulin (B2M) and cyclophilin A (PPIA) between paired and unpaired groups were detected.
Data analysis
The abundance of each gene transcript was scored as an average difference value by comparing the intensity of hybridization to 20 sets of perfect match 25-mer oligonucleotides relative to 20 sets of mismatched oligonucleotides using Affymetrix Genechip® Operating software (1.4). Results of individual genechips from paired animals were compared with those from unpaired animals using all possible pair wise comparisons. Genes were not included in the analysis unless they were given a score of “I” (increased) or “D” (decreased) and “P” (present) by the Affymetrix Genechip® software in all comparisons. Genes that were considered to be differentially expressed were those that met criteria for fold change (average >1.5 fold increase) and signal strength (average >100-twice the background).
Real-time PCR (qPCR)
The RNA of genes, which were found to be differentially expressed between paired and unpaired groups 5 hrs after training by the microarray experiments, were further subjected to qPCR performed using an ABI Prism 7900 Sequence Detector (Applied Biosystems, Foster City, CA)) and gene-specific TaqMan FAM/MGB assays (Applied Biosystems; Assays ID: Kif2–Rn01497645_m1, Lsamp–Rn00568353_m1 Hpcal4–Rn00569340_g1; Nsf–Rn01645680_g1; HSP-90β–Rn01511686_g1; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) ~Rn99999916_s1, beta-2-microglobulin (B2M) ~Rn00560865_m1, cyclophilin A (PPIA) ~Rn00690933_m1) mostly as previously described (Dracheva et al., 2008) Each 10 μl PCR reaction consisted of 2.5 μl of the relevant cDNA (diluted 25 times in H2O), a specific TaqMan assay, and 5 μl of the 2xPCR Gold Master Mix (Applied Biosystems), which contained ROX, Aplitag Gold DNA polymerase, AmpErase UNG, dATP, dCTP, dGTP, dUTP, and MgCl2. For all target and for all endogenous control genes (ECGs), the standard thermal cycling program was applied (Dracheva et al., 2008). All assays were run in triplicate for each sample, and only one cDNA was amplified in each reaction (monoplex). To account for the differences in the amount of the input material between the samples, the expression level of target genes was normalized to the expression levels of three different ECGs (GAPDH, B2M, PPIA) (Vandesompele et al., 2002). The relative expression level of the target transcripts was determined using Relative Standard Curve Method (RSCM; see Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR, Applied Biosystems). This approach provides accurate quantitative results since it accounts for differences in the efficiencies between target and control amplifications. Standard curves were generated for each target assay and for each ECG assay by the association between the threshold cycle (Ct) values and different quantities of a “calibrator” cDNA. The “calibrator” was prepared by mixing small quantities of all experimental samples. Using the linear equations of standard curves, the relative amounts of each target and each ECG mRNA were calculated in each sample. The relative expression level of the target mRNA was computed as the ratio between the target mRNA and the geometric mean of the three above-mentioned ECGs.
Statistics
Comparisons between RNA levels for main effect of groups were done using one-way ANOVA followed by Tukey's posthoc test with an α level of 0.05. Statistical significance for differences between two groups was assessed with Mann–Whitney test.
Results
Fear conditioning induces the expression of genes encoding proteins regulating neuronal morphology and transmission in LA
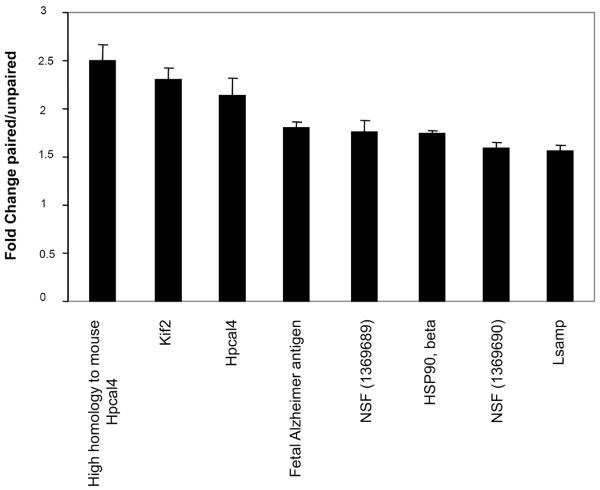
Fear learning is known to induce the expression of immediate early genes encoding transcription factors in LA (e.g. Ressler et al., 2002). We were, therefore, interested in examining the expression of late response genes in LA following fear conditioning. Thus, we sampled tissue 5 hours after completion of conditioning. Rats were trained with one of two conditions: paired training, five pairings of a tone (the conditioned stimulus, CS) that co-terminated with a footshock (the unconditioned stimulus, US) and unpaired training, in which five presentations of the CS and US occurred in a non-overlapping manner (Lamprecht et al., 2002). Two groups were then analyzed the first for behavioral outcome of training and the second for gene expression in LA. Figure 1A shows that when the CS tones were presented 24 hrs after training freezing is significantly higher in the paired group (ANOVA F(2,9)=32.26, p<0.001) when compared to unpaired (p<0.001) or naïve (no CS and US presentation; p<0.001) groups. No significant differences were detected between the naïve and unpaired animals (p=0.82). Brains from the second group (from animals trained with paired or unpaired protocols) were removed five hours after training and the LA was dissected from rats using the laser microdissection technique (Figure 1B). RNA was extracted, amplified and subjected to gene expression analysis using Affymetrix microarray. RNA from each rat was analyzed by a separate microarray (n=3 each group). Genes that were differentially expressed in all possible comparisons between the paired and unpaired animals were subjected for additional analysis, in which only those genes with average signal level > 100 and with fold change > 1.50 were filtered. Figure 2 shows the genes that were expressed at higher level in the paired animals when compared to the unpaired rats 5 hrs after fear conditioning in LA. Fold changes between paired and unpaired groups of these genes were significantly different (p<0.001 for all genes that were compared) from fold changes of the control genes beta-2-microglobulin (B2M; fold change=0.011) or cyclophilin A (PPIA; fold change=−0.011). The majority of the differentially expressed genes detected in the study encode proteins involved in the regulation of axonal growth and synaptic transmission.
Figure 2. Fear conditioning induces gene expression 5 hours after training.
The average of fold changes in LA obtained for all possible comparisons between paired animals and unpaired animals 5 hours after training (n=3 each) detected by microarray studies. Only genes with an average fold change above 1.5 are presented. Hpcal4-Hippocalcin-like 4; Hsp90β-heat shock 90kDa protein 1, beta; Kif2- kinesin heavy chain member 2, Lsamp- limbic system-associated membrane protein; NSF- N-ethylmaleimide sensitive fusion protein. Values are mean ± SEM. Fold changes detected for these genes between paired and unpaired groups were significantly different (p<0.001) from those detected for control genes beta-2-microglobulin (B2M; fold change=0.011) or cyclophilin A (PPIA; fold change=−0.011).
Temporal expression of genes in LA depends on whether the CS and US are presented in a paired or unpaired manner
To verify the findings obtained by the microarray method, the expression of the identified genes was also assessed by quantitative real time PCR (qPCR). In addition to the sister aliquots of the RNA samples used for the microarray analysis from animals sacrificed 5 hr after conditioning, a different set of samples was obtained from animals sacrificed 30 minutes after training. We chose 30 minutes since it has been shown that neuronal stimulation and learning induce gene expression at that time point (e.g. Ressler et al., 2002; Keeley et al., 2006). We explored whether the genes found to be differentially expressed between paired and unpaired groups after 5 hrs were also expressed 30 minutes following the training.
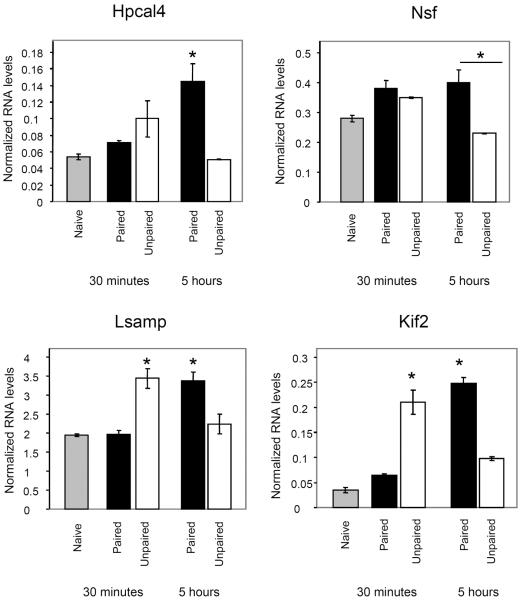
Table 1 shows that qPCR confirmed most of the changes in gene expression between the paired and unpaired groups 5 hrs after training in LA that were detected by Affymetrix microarray. We also found that genes were expressed in different temporal patterns (Figure 3). At the 30 minute time point Kif 2 (ANOVA F(4,9)= 45.72, p<0.001) and Lsamp (ANOVA F(4,9)= 10.957, p<0.003)) were induced by the unpaired stimulation (Unpaired vs. Naïve: Kif2- p<0.001; Lsamp- p<0.02) but not by the paired stimulus Furthermore, the level of their RNAs in unpaired group was significantly higher than in the paired group (Unpaired vs. Paired: Kif2-p<0.001; Lsamp-p<0.008). At 5 hrs after training the expression of Kif2, Lsamp, and Hpcal4 (ANOVA F(4,9)= 6.45, p< 0.01) was induced by the paired stimulation (Paired vs. Naïve: Kif2- p<0.001; Hpcal4- p<0.03; Lsamp= p<0.02) but not by the unpaired protocol. The level of the RNAs in paired group was also significantly higher than that in the unpaired group (Paired vs. Unpaired: Kif2-p<0.001; Hpcal4- p<0.02; Lsamp- p<0.04). The Nsf (ANOVA F(4,9)=7.83, p<0.005) gene transcript was significantly higher in the paired group when compared to the unpaired group 5 hrs after training (p<0.007). Both groups were not significantly different from the naïve group. The above results show that gene expression can be responsive specifically to one of the training protocols (e.g. paired only for Hpcal4) or to both paired and unpaired training where each protocol lead to different temporal pattern of gene expression (Kif2, Lsamp).
Table 1.
Folds changes in RNAs levels in LA between paired and unpaired groups 5 hrs after training.
| Gene | Affymetrix number | Fold change (Affymetrix) | Fold change (qPCR) |
|---|---|---|---|
| hippocalcin-like 4 (Hpcal4) | 1387639_at | 2.14 | 2.84 |
| hippocalcin-like 4 (Hpcal4) | 1375765_at | 2.5 | |
| N-ethylmaleimide sensitive factor (Nsf) | 1369689_at | 1.76 | 1.73 |
| N-ethylmaleimide sensitive factor (Nsf) | 1369690_at | 1.59 | |
| kinesin heavy chain (Kif2) | 1375508_at | 2.31 | 2.53 |
| limbic system-associated membrane protein (Lsamp) | 1370550_at | 1.56 | 1.5 |
| heat shock 90kDa protein 1, beta (Hsp90β) | 1375335_at | 1.74 | 1.22 |
| fetal Alzheimer antigen | 1374283_at | 1.8 |
Figure 3. Different temporal patterns of gene expression in LA in paired and unpaired groups.
The results of the qPCR analysis for the genes that were differentially expressed between the paired, unpaired and naive groups (n=3 paired or unpaired, n=2 Naive) reveal different temporal responses to paired or unpaired stimulation. The expression of a gene in LA depends on whether the CS and US stimuli are coupled or separated as well as on the time after training. Shown are mean ± SEM of values detected by qPCR analysis. *=p<0.05 between stimulus and naïve and between paired and unpaired groups. In NSF *=p<0.05 only between paired and unpaired groups.
To demonstrate that kif2, Lsamp, Hpcal4 and Nsf expression is induced in specific brain area we monitored the level of these genes, using qPCR, in paired and unpaired groups in olfactory bulb which serves as a control area. No differences were observed in gene expression between paired and unpaired groups 30 minutes (HpCal4 p=0.4; kif2 p=0.2: Lsamp p=0.1; Nsf p= 0.1) or 5 hrs (HpCal4 p=0.4; kif2 p=1.0: Lsamp p=1.0; Nsf p= 0.7) after training.
Discussion
In the present study we examined whether fear conditioning leads to expression of specific genes in LA. Toward that end we used the Affymetrix microarray to compare the level of gene expression in LA 5 hrs after training in animals that underwent associative fear conditioning (paired CS-US) as compared to non-associative training (same sensory simulation but in a non-overlapping pattern). We detected differential expression of genes encoding proteins that are involved in neuronal morphogenesis and synaptic transmission. We extended these findings by monitoring the temporal pattern of the expression of these genes at 30 minutes and 5 hrs after training using qPCR. We discovered that the patterns of expression depended on the training protocol and time after training, inasmuch as a given gene could be expressed late following fear conditioning but early following unpaired training.
We detected that 7 genes were differentially expressed between paired and unpaired groups out of all genes represented in microarray. The relatively low number of differentially expressed genes between the paired and unpaired groups detected 5 hrs after training may be a result of several factors: First, the tone or shock per se are known to induce gene expression in amygdala (e.g. Campeau et al., 1991). These genes will not be detected in a type of comparison performed in our study. Second, we have used a stringent analysis procedure, where only genes that were detected to be differentially expressed after all possible comparisons of the paired and unpaired animals were considered. This approach reduces false positives. Third, although we have dissected the LA specifically, this brain area contains many cells that do not participate in fear conditioning. These cells may express basal level of the same genes that are induced by fear conditioning and thereby could mask their detection. Dissection of specific cells that participate in fear memory in LA may increase signal to noise ratio and unveil new genes participating in fear conditioning.
Two genes involved in axonal morphology were found to be differentially expressed: the Kinesin heavy chain member 2 (Kif2) and limbic system-associated membrane protein (Lsamp). Kif2 is a member of the kinesin family of motor proteins involved in neuronal morphogenesis (Hirokawa & Takemura, 2004). It has been found to regulate microtubule depolymerization in neurons and, is suggested to control the length of the axons (e.g., Kif2a−/− mice show aberrantly elongated axonal branches (Homma et al., 2003)). Kif2 has also been shown to translocate proteins, such as the growth-cone-specific IGF-1 receptor, to growth cone as well as to play an important role in expansion of the nerve growth cone (Morfini et al., 1997; Pfenninger et al., 2003). Kif2 gene was shown to be expressed following NGF or BDNF application in tissue culture (Morfini et al., 1997; Pfenninger et al., 2003). Lsamp also encodes a protein that regulates axonal growth. Lsamp has been found on the surface of neurons and their growing axons (Horton & Levitt, 1988) and is expressed mainly in the limbic areas including the perirhinal, hippocampus, cigulate, amygdala, and limbic thalamic neurons (Pimenta et al., 1995). Lsamp has homophilic binding property and is suggested to mediate interactions between neuronal populations of the limbic system. It mediates neuronal growth (e.g. expression of Lsamp in neurons facilitates neurite outgrowth) and is involved in proper targeting of neuronal pathways (e.g. inhibition of its activity in vivo results in abnormal neuronal connectivity) (Keller et al., 1989; Pimenta et al., 1995). Thus, Lsamp, which is involved in formation of homophilic binding, could be important for proper targeting of the growing axons in LA (as observed in neuronal circuits of the developing hippocampus), while Kif2, which mediates axonal morphology, may possibly support alteration in axonal patterning in LA.
The role of presynaptic axons in fear conditioning is further supported by our finding of an increase in the expression of Hippocalcin like-4 (Hpcal4). Hpcal4 (also known as Visinin-like protein 2 (VILIP-2) or Neural visinin-like protein 2, (NVP-2)) is a calcium binding protein that belongs to the neuronal Ca2+ sensor superfamily (Burgoyne, 2007). Hpcal4 (VILIP-2) has been shown to slow the rate of inactivation of Cav2.1 calcium channels (Lautermilch et al., 2005; Few et al., 2005). Those channels conduct P/Q type calcium currents that initiate exocytosis of neurotransmitters (Takahashi & Momiyama, 1993; Regehr & Mintz, 1994). By regulating presynaptic Cav2.1 channels, Hpcal4 may be directly involved in shaping presynaptic calcium transients, thus influencing the time course and the amount of neurotransmitter release. Our finding that another gene with 89% similarity to mouse Hpcal4 (1375765_at) is also differentially expressed between the paired and unpaired groups suggests that regulation of calcium transients in LA after fear conditioning may require several calcium sensor proteins. The increase of Hpcal4 could represent cellular processes that accompany changes in axonal morphogenesis namely the need for an increase in presynaptic proteins or processes regulating vesicle release in preexisting presynapses.
Another gene that was differentially expressed between the paired and unpaired groups and also encodes a protein that is intimately involved in synaptic transmission is the N-ethylmaleimide sensitive fusion protein (NSF; Zhao et al., 2007). In presynapses, NSF is involved in vesicle fusion cycle by forming complexes with SNAP to catalyze the disassembly of cis-SNARE complexes for reactivation (Brunger, 2001). In postsynpases, NSF is involved in insertion of AMPA receptors into the synapse: Inhibition of NSF-GluR2 interactions leads to the decrease in synaptic currents (Lee et al., 2002). Furthermore, NSF regulates GABAA receptor levels in synapse and GABAB receptors desensitization (Goto et al., 2005; Pontier et al., 2006). Thus, changes in NSF expression may modulate both pre- and post-synaptic transmission in LA by regulating vesicle cycle, excitatory (AMPA) and/or inhibitory (GABA) transmission. This, in turn, may lead to alteration of synaptic efficacy after learning.
Our qPCR experiments confirmed the results obtained by the Affymetrix microarray analysis and revealed different temporal patterns of gene expression in LA following the training. Interestingly, the level of Kif2 and Lsamp RNAs in paired and unpaired groups depends on the time after training. Kif2 and Lsamp were found at higher level in LA of paired animals when compared to unpaired groups 5 hrs after training. In contrast, the level of Kif2 and Lsamp were higher at the unpaired group 30 minutes after training. This may represent a cellular mechanism where genes are induced shortly after stressful events (the US in unpaired), but if the stressful event (US) is coupled to a meaningful sensory stimulus (CS), these genes are expressed in a different pattern engaging more robustly with the later consolidation processes of fear memory. Consistently, exposure of rats to stressful situation (cat odor) for 30 minutes has previously been shown to induce rapid expression of Lsamp in the rat amygdala (including basolateral, central and medial nuclei) detected at the end of the trial (Kõks et al., 2004). In addition, rats with low exploratory activity in the elevated plus-maze (“anxious rats”) have shown an increased level of Lsamp gene in amygdala compared to high exploratory rats (Nelovkov et al., 2006). Furthermore, the basic neuroanatomical organization and sensory and motor development is normal in the Lsamp KO mice (Catania et al., 2008); however, these mice exhibit heightened response to novelty in several behavioral tests, which may reflect an altered response to environmental stressors. Lsamp may, therefore, mediate changes in the fine neuronal connectivity that subserve these behaviors. Collectively, these results suggest that expression of the Lsamp gene in amygdala mediates emotional stress associated with the foot shock (US) in the unpaired stimulation 30 minutes after training but not 5 hrs afterwards, as at this time the level of Lsamp in unpaired group is not different from controls. At that time point (5hr after training), Lsamp is induced only after paired (learning) stimulation.
Alternatively, changes in Kif2 and Lsamp may represent cellular mechanisms responding differently to stimuli that are close in time (paired CS-US) compared to stimuli that are dispersed in time (unpaired). These stimuli patterns lead to different behavioral responses. The paired CS-US stimulation leads to fear learning whereas the unpaired may lead to learning of safety (Rogan et al., 2005). In the later case the tone represents a safety situation when it is not coupled to a footshock. Thus, the differential expression of the genes in response to paired or unpaired stimuli may underlie cellular mechanisms mediating fear or safety learning.
Another possibility is that genes expressed in the unpaired group could mediate contextual fear memory formation in LA. It has been shown that tone-shock paired training induces fear memories to the tone whereas unpaired training do not (e.g. Lamprecht et al., 2002 and figure 1A). On the other hand unpaired training induces significantly increased contextual fear memory when compared to the paired group (Trifilieff et al., 2007). This study also shows that unpaired training induces an activation of ERK1/2 in LA. Thus, although we have habituated the rats to the context for 4 days before training, gene expression in LA in the unpaired group may result from context-shock association.
It would be interesting to examine the upstream molecular mechanisms whereby neurons use to translate the temporal relationships between the CS and US into differential gene expression. The difference in the patterns of Kif2 and Lsamp induction in response to the paired and unpaired stimulation might indicate that these training protocols engage different intracellular signaling pathways in amygdala neurons. Unpaired stimulation may activate transcription factors rapidly by posttranslation modifications (e.g. phosphorylation) that can, in turn, facilitate their binding to the Kif2 and Lsamp promoters leading to fast transcription of these genes. Paired stimulation, on the other hand, may induce more elaborated pathways leading first to the expression and translation of transcription factors that then bind to the genes' promoters leading to the late induction of Kif2 and Lsamp. These cellular pathways have been shown to be used for the induction of transcription factors followed by expression of late response genes (Ginty et al., 1992). Differential activation of signaling molecules at the synapse may engage different nuclear signaling pathways and, in turn, induce distinct patterns of gene expression (West et al., 2002). For example, stimuli leading to increased calcium influx at the synapse may result in different activation of transcription factors than stimuli leading to elevated cAMP levels at the synapse (e.g Kornhauser et al. 2002). Future studies will attempt to understand how the function of these genes at different time points in LA leads to neuronal and behavioral responses underlying fear conditioning.
References
- Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci U S A. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DJ, Kim JJ, Sun W, Thompson RF, Helmstetter FJ. Acquisition of fear conditioning in rats requires the synthesis of mRNA in the amygdala. Behav Neurosci. 1999;113:276–282. doi: 10.1037//0735-7044.113.2.276. [DOI] [PubMed] [Google Scholar]
- Blair HT, Schafe GE, Bauer EP, Rodrigues SM, LeDoux JE. Synaptic plasticity in the lateral amygdala: a cellular hypothesis of fear conditioning. Learn. Mem. 2001;8:229–242. doi: 10.1101/lm.30901. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Structure of proteins involved in synaptic vesicle fusion in neurons. Annu Rev Biophys Biomol Struct. 2001;30:157–171. doi: 10.1146/annurev.biophys.30.1.157. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau S, Hayward MD, Hope BT, Rosen JB, Nestler EJ, Davis M. Induction of the c-fos proto-oncogene in rat amygdala during unconditioned and conditioned fear. Brain Res. 1991;565:349–352. doi: 10.1016/0006-8993(91)91669-r. [DOI] [PubMed] [Google Scholar]
- Catania EH, Pimenta A, Levitt P. Genetic deletion of Lsamp causes exaggerated behavioral activation in novel environments. Behav Brain Res. 2008;188:380–390. doi: 10.1016/j.bbr.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- Davis M, Whalen PJ. The amygdala: vigilance and emotion. Mol. Psychiatry. 2001;6:13–34. doi: 10.1038/sj.mp.4000812. [DOI] [PubMed] [Google Scholar]
- Dityatev AE, Bolshakov VY. Amygdala, long-term potentiation, and fear conditioning. Neuroscientist. 2005;11:75–88. doi: 10.1177/1073858404270857. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Byne W, Chin B, Haroutunian V. Ionotropic glutamate receptor mRNA expression in the human thalamus: Absence of change in schizophrenia. Brain Res. 2008;1214:23–34. doi: 10.1016/j.brainres.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y. The Neurobiology of Memory. Oxford Univ. Press; New York: 1989. [Google Scholar]
- Duvarci S, Nader K, LeDoux JE. De novo mRNA synthesis is required for both consolidation and reconsolidation of fear memories in the amygdala. Learn Mem. 2008;15:747–755. doi: 10.1101/lm.1027208. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS, LeDoux JE. Why we think plasticity underlying Pavlovian fear conditioning occurs in the basolateral amygdala. Neuron. 1999;23:229–232. doi: 10.1016/s0896-6273(00)80775-8. [DOI] [PubMed] [Google Scholar]
- Fanselow MS, Poulos AM. The neuroscience of mammalian associative learning. Annu Rev Psychol. 2005;56:207–234. doi: 10.1146/annurev.psych.56.091103.070213. [DOI] [PubMed] [Google Scholar]
- Few AP, Lautermilch NJ, Westenbroek RE, Scheuer T, Catterall WA. Differential regulation of CaV2.1 channels by calcium-binding protein 1 and visinin-like protein-2 requires N-terminal myristoylation. J Neurosci. 2005;25:7071–7080. doi: 10.1523/JNEUROSCI.0452-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginty DD, Bading H, Greenberg ME. Trans-synaptic regulation of gene expression. Curr Opin Neurobiol. 1992;2:312–316. doi: 10.1016/0959-4388(92)90121-z. [DOI] [PubMed] [Google Scholar]
- Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory--a molecular framework. Nature. 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- Goto H, Terunuma M, Kanematsu T, Misumi Y, Moss SJ, Hirata M. Direct interaction of N-ethylmaleimide-sensitive factor with GABA(A) receptor beta subunits. Mol Cell Neurosci. 2005;30:197–206. doi: 10.1016/j.mcn.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Hebb DO. The Organization of Behavior: a Neuropsychological Theory. Wiley; New York: 1949. [Google Scholar]
- Hirokawa N, Takemura R. Molecular motors in neuronal development, intracellular transport and diseases. Curr Opin Neurobiol. 2004;14:564–573. doi: 10.1016/j.conb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114:229–239. doi: 10.1016/s0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- Hong SJ, Li H, Becker KG, Dawson VL, Dawson TM. Identification and analysis of plasticity-induced late-response genes. Proc Natl Acad Sci U S A. 2004;101:2145–2150. doi: 10.1073/pnas.0305170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton HL, Levitt P. A unique membrane protein is expressed on early developing limbic system axons and cortical targets. J Neurosci. 1988;8:4653–4661. doi: 10.1523/JNEUROSCI.08-12-04653.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Keeley MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S, Abel T. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn. Mem. 2006;13:135–142. doi: 10.1101/lm.86906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller F, Rimvall K, Barbe MF, Levitt P. A membrane glycoprotein associated with the limbic system mediates the formation of the septo-hippocampal pathway in vitro. Neuron. 1989;3:551–561. doi: 10.1016/0896-6273(89)90265-1. [DOI] [PubMed] [Google Scholar]
- Kõks S, Luuk H, Nelovkov A, Areda T, Vasar E. A screen for genes induced in the amygdaloid area during cat odor exposure. Genes Brain Behav. 2004;3:80–89. doi: 10.1046/j.1601-183x.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- Konorski J. Conditioned Reflexes and Neuron Organization. Cambridge Univ. Press; Cambridge, UK: 1948. [Google Scholar]
- Kornhauser JM, Cowan CW, Shaywitz AJ, Dolmetsch RE, Griffith EC, Hu LS, Haddad C, Xia Z, Greenberg ME. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Lamprecht R, Farb CR, LeDoux JE. Fear memory formation involves p190 RhoGAP and ROCK proteins through a GRB2-mediated complex. Neuron. 2002;36:727–738. doi: 10.1016/s0896-6273(02)01047-4. [DOI] [PubMed] [Google Scholar]
- Lamprecht R, LeDoux J. Structural plasticity and memory. Nat Rev Neurosci. 2004;5:45–54. doi: 10.1038/nrn1301. [DOI] [PubMed] [Google Scholar]
- Lautermilch NJ, Few AP, Scheuer T, Catterall WA. Modulation of CaV2.1 channels by the neuronal calcium-binding protein visinin-like protein-2. J. Neurosci. 2005;25:7062–7070. doi: 10.1523/JNEUROSCI.0447-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux JE. Emotion Circuits in the Brain. Annu. Rev. Neurosci. 2000;23:155–184. doi: 10.1146/annurev.neuro.23.1.155. [DOI] [PubMed] [Google Scholar]
- Maren S, Ferrario CR, Corcoran KA, Desmond TJ, Frey KA. Protein synthesis in the amygdala, but not the auditory thalamus, is required for consolidation of Pavlovian fear conditioning in rats. Eur J Neurosci. 2003;18:3080–3088. doi: 10.1111/j.1460-9568.2003.03063.x. [DOI] [PubMed] [Google Scholar]
- Maren S. Synaptic mechanisms of associative memory in the amygdala. Neuron. 2005;47:783–786. doi: 10.1016/j.neuron.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu.. Rev. Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Mei B, Li C, Dong S, Jiang CH, Wang H, Hu Y. Distinct gene expression profiles in hippocampus and amygdala after fear conditioning. Brain Res Bull. 2005;67:1–12. doi: 10.1016/j.brainresbull.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Morfini G, Quiroga S, Rosa A, Kosik K, Cáceres A. Suppression of KIF2 in PC12 cells alters the distribution of a growth cone nonsynaptic membrane receptor and inhibits neurite extension. J Cell Biol. 1997;138:657–669. doi: 10.1083/jcb.138.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelovkov A, Areda T, Innos J, Kõks S, Vasar E. Rats displaying distinct exploratory activity also have different expression patterns of gamma-aminobutyric acid-and cholecystokinin-related genes in brain regions. Brain Res. 2006;1100:21–31. doi: 10.1016/j.brainres.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Pfenninger KH, Laurino L, Peretti D, Wang X, Rosso S, Morfini G, Cáceres A, Quiroga S. Regulation of membrane expansion at the nerve growth cone. J. Cell. 2003;116:1209–1217. doi: 10.1242/jcs.00285. [DOI] [PubMed] [Google Scholar]
- Pimenta AF, Zhukareva V, Barbe MF, Reinoso BS, Grimley C, Henzel W, Fischer I, Levitt P. The limbic system-associated membrane protein is an Ig superfamily member that mediates selective neuronal growth and axon targeting. Neuron. 1995;15:287–297. doi: 10.1016/0896-6273(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Pollak DD, Monje FJ, Zuckerman L, Denny CA, Drew MR, Kandel ER. An Animal Model of a Behavioral Intervention for Depression. Neuron. 2008;60:149–161. doi: 10.1016/j.neuron.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier SM, Lahaie N, Ginham R, St-Gelais F, Bonin H, Bell DJ, Flynn H, Trudeau LE, McIlhinney J, White JH, Bouvier M. Coordinated action of NSF and PKC regulates GABAB receptor signaling efficacy. EMBO J. 2006;25:2698–709. doi: 10.1038/sj.emboj.7601157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr WG, Mintz IM. Participation of multiple calcium channel types in transmission at single climbing fiber to Purkinje cell synapses. Neuron. 1994;12:605–613. doi: 10.1016/0896-6273(94)90216-x. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Paschall G, Zhou X-l, Davis M. Regulation of synaptic plasticity genes during consolidation of fear conditioning. J. Neurosci. 2002;22:7892–7902. doi: 10.1523/JNEUROSCI.22-18-07892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues SM, Schafe GE, LeDoux JE. Molecular mechanisms underlying emotional learning and memory in the lateral amygdala. Neuron. 2004;44:75–91. doi: 10.1016/j.neuron.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Rogan MT, Leon KS, Perez DL, Kandel ER. Distinct neural signatures for safety and danger in the amygdala and striatum of the mouse. Neuron. 2005;46:309–320. doi: 10.1016/j.neuron.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Sah P, Faber ES, Lopez De Armentia M, Power J. The amygdaloid complex: anatomy and physiology. Physiol. Rev. 2003;83:803–834. doi: 10.1152/physrev.00002.2003. [DOI] [PubMed] [Google Scholar]
- Schafe GE, LeDoux JE. Memory consolidation of auditory pavlovian fear conditioning requires protein synthesis and protein kinase A in the amygdala. J Neurosci. 2000;20:RC96. doi: 10.1523/JNEUROSCI.20-18-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafe GE, Nader K, Blair HT, LeDoux JE. Memory consolidation of Pavlovian fear conditioning: a cellular and molecular perspective. Trends. Neurosci. 2001;24:540–546. doi: 10.1016/s0166-2236(00)01969-x. [DOI] [PubMed] [Google Scholar]
- Stork O, Stork S, Pape H-S, Obata K. Identification of Genes Expressed in the Amygdala During the Formation of Fear Memory. Learn. Mem. 2001;8:209–219. doi: 10.1101/lm.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Momiyama A. Different types of calcium channels mediate central synaptic transmission. Nature. 1993;366:156–158. doi: 10.1038/366156a0. [DOI] [PubMed] [Google Scholar]
- Trifilieff P, Calandreau L, Herry C, Mons N, Micheau J. Biphasic ERK1/2 activation in both the hippocampus and amygdala may reveal a system consolidation of contextual fear memory. Neurobiol Learn Mem. 2007;88:424–434. doi: 10.1016/j.nlm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Tsien JZ. Linking Hebb's coincidence-detection to memory formation. Curr. Opin. Neurobiol. 2000;10:266–273. doi: 10.1016/s0959-4388(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat. Rev. Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Zhao C, Slevin JT, Whiteheart SW. Cellular functions of NSF: not just SNAPs and SNAREs. FEBS Lett. 2007;581:2140–9. doi: 10.1016/j.febslet.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]