Abstract
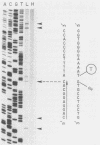
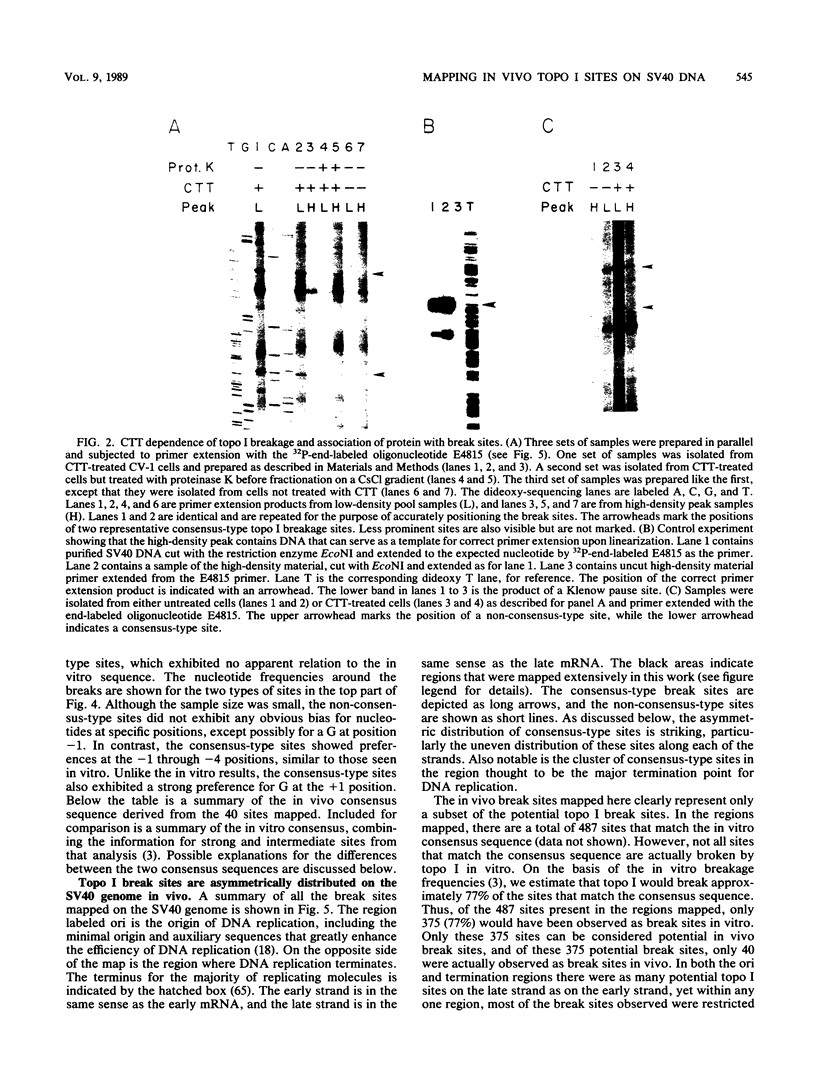
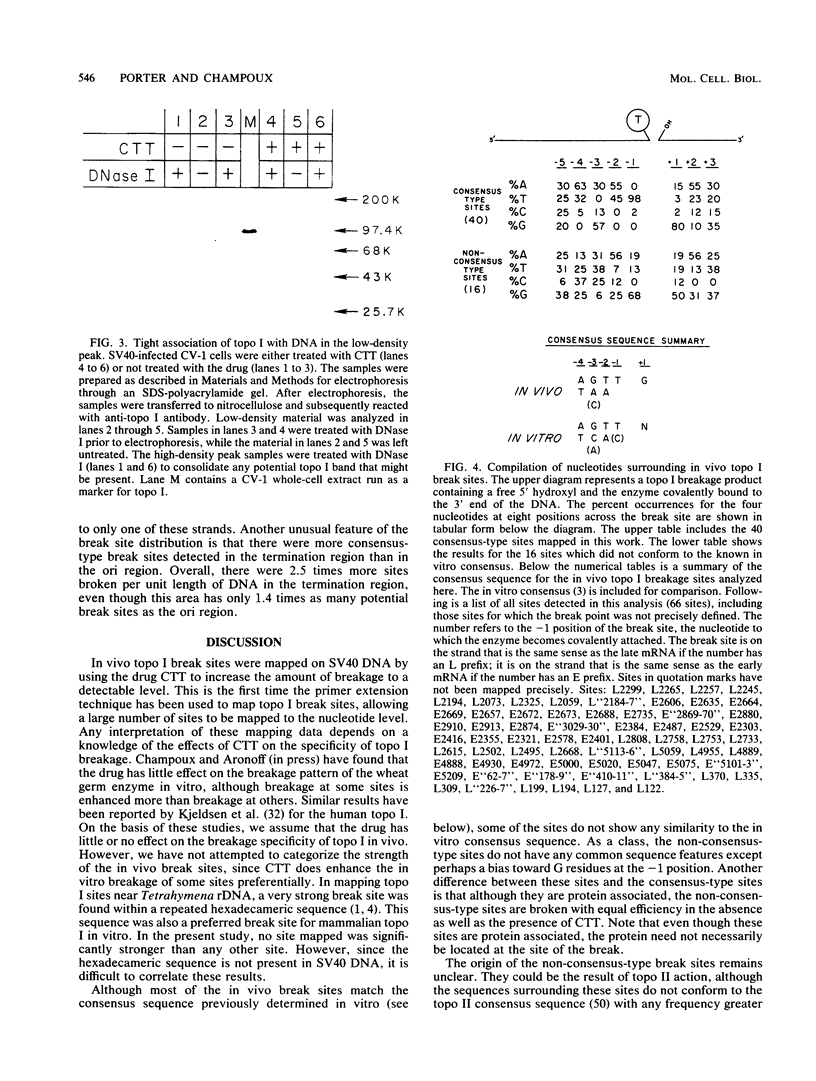
Complexes between simian virus 40 DNA and topoisomerase I (topo I) were isolated from infected cells treated with camptothecin. The topo I break sites were precisely mapped by primer extension from defined oligonucleotides. Of the 56 sites, 40 conform to the in vitro consensus sequence previously determined for topo I. The remaining 16 sites have an unknown origin and were detectable even in the absence of camptothecin. Only 11% of the potential break sites were actually broken in vivo. In the regions mapped, the pattern of break sites was asymmetric. Most notable are the clustering of sites near the terminus for DNA replication and the confinement of sites to the strand that is the template for discontinuous DNA synthesis. These asymmetries could reflect the role of topo I in simian virus 40 DNA replication and suggest that topo I action is coordinated spatially with that of the replication complex.
Full text
PDF



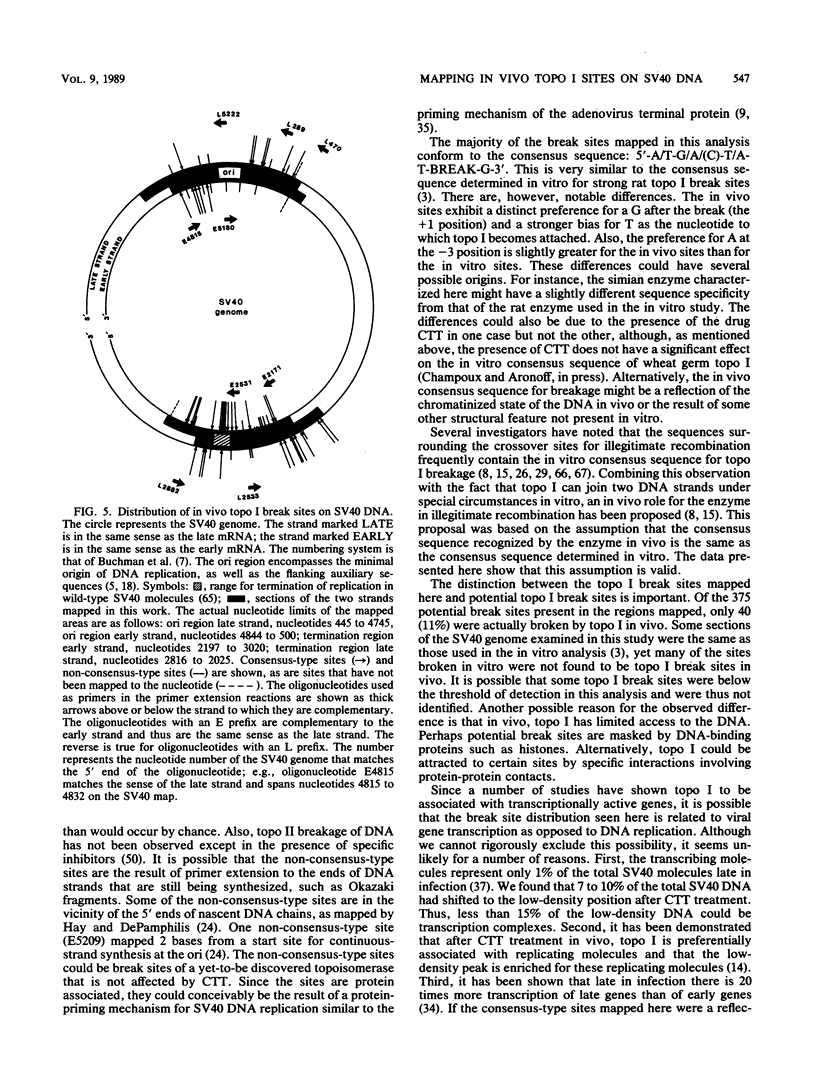



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen A. H., Gocke E., Bonven B. J., Nielsen O. F., Westergaard O. Topoisomerase I has a strong binding preference for a conserved hexadecameric sequence in the promoter region of the rRNA gene from Tetrahymena pyriformis. Nucleic Acids Res. 1985 Mar 11;13(5):1543–1557. doi: 10.1093/nar/13.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avemann K., Knippers R., Koller T., Sogo J. M. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol Cell Biol. 1988 Aug;8(8):3026–3034. doi: 10.1128/mcb.8.8.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Brill S. J., DiNardo S., Voelkel-Meiman K., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. 1987 Mar 26-Apr 1Nature. 326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Challberg M. D., Desiderio S. V., Kelly T. J., Jr Adenovirus DNA replication in vitro: characterization of a protein covalently linked to nascent DNA strands. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5105–5109. doi: 10.1073/pnas.77.9.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J Biol Chem. 1981 May 25;256(10):4805–4809. [PubMed] [Google Scholar]
- Champoux J. J. Evidence for an intermediate with a single-strand break in the reaction catalyzed by the DNA untwisting enzyme. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3488–3491. doi: 10.1073/pnas.73.10.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., McCoubrey W. K., Jr, Been M. D. DNA structural features that lead to strand breakage by eukaryotic type-I topoisomerase. Cold Spring Harb Symp Quant Biol. 1984;49:435–442. doi: 10.1101/sqb.1984.049.01.049. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Mechanism of the reaction catalyzed by the DNA untwisting enzyme: attachment of the enzyme to 3'-terminus of the nicked DNA. J Mol Biol. 1978 Jan 25;118(3):441–446. doi: 10.1016/0022-2836(78)90238-3. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Topoisomerase I is preferentially associated with isolated replicating simian virus 40 molecules after treatment of infected cells with camptothecin. J Virol. 1988 Oct;62(10):3675–3683. doi: 10.1128/jvi.62.10.3675-3683.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta V., Sollner-Webb B. Sites of topoisomerase I action on X. laevis ribosomal chromatin: transcriptionally active rDNA has an approximately 200 bp repeating structure. Cell. 1988 Feb 26;52(4):585–597. doi: 10.1016/0092-8674(88)90471-0. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Elgin S. C. Localization of specific topoisomerase I interactions within the transcribed region of active heat shock genes by using the inhibitor camptothecin. Mol Cell Biol. 1987 Jan;7(1):141–148. doi: 10.1128/mcb.7.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Pflugfelder G., Wang J. C., Lis J. T. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986 Feb 14;44(3):401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- Halligan B. D., Davis J. L., Edwards K. A., Liu L. F. Intra- and intermolecular strand transfer by HeLa DNA topoisomerase I. J Biol Chem. 1982 Apr 10;257(7):3995–4000. [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hogan A., Faust E. A. Nonhomologous recombination in the parvovirus chromosome: role for a CTATTTCT motif. Mol Cell Biol. 1986 Aug;6(8):3005–3009. doi: 10.1128/mcb.6.8.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985 Nov 25;260(27):14873–14878. [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell. 1988 Feb 26;52(4):535–544. doi: 10.1016/0092-8674(88)90466-7. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J. 1987 Aug;6(8):2401–2408. doi: 10.1002/j.1460-2075.1987.tb02518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. A nucleosome-free region in SV40 minichromosomes. Nature. 1980 May 22;285(5762):263–265. doi: 10.1038/285263a0. [DOI] [PubMed] [Google Scholar]
- Kjeldsen E., Mollerup S., Thomsen B., Bonven B. J., Bolund L., Westergaard O. Sequence-dependent effect of camptothecin on human topoisomerase I DNA cleavage. J Mol Biol. 1988 Jul 20;202(2):333–342. doi: 10.1016/0022-2836(88)90462-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laub O., Aloni Y. Transcription of simian virus 40. V. Regulattion of simian virus 40 gene expression. J Virol. 1975 Nov;16(5):1171–1183. doi: 10.1128/jvi.16.5.1171-1183.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichy J. H., Horwitz M. S., Hurwitz J. Formation of a covalent complex between the 80,000-dalton adenovirus terminal protein and 5'-dCMP in vitro. Proc Natl Acad Sci U S A. 1981 May;78(5):2678–2682. doi: 10.1073/pnas.78.5.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopis R., Perrin F., Bellard F., Gariglio P. Quantitation of transcribing native simian virus 40 minichromosomes extracted from CV1 cells late in infection. J Virol. 1981 Apr;38(1):82–90. doi: 10.1128/jvi.38.1.82-90.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- McCoubrey W. K., Jr, Champoux J. J. The role of single-strand breaks in the catenation reaction catalyzed by the rat type I topoisomerase. J Biol Chem. 1986 Apr 15;261(11):5130–5137. [PubMed] [Google Scholar]
- Mengeritsky G., Trifonov E. N. Nucleotide sequence-directed mapping of the nucleosomes of SV40 chromatin. Cell Biophys. 1984 Mar;6(1):1–8. doi: 10.1007/BF02788576. [DOI] [PubMed] [Google Scholar]
- Milavetz B. Analysis of the origin-specific nucleosome-free region in SV40 encapsidation intermediates. Virology. 1986 Sep;153(2):310–313. doi: 10.1016/0042-6822(86)90034-6. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Cereghini S., Yaniv M. Fine structure of the regulatory region of simian virus 40 minichromosomes revealed by DNAase I digestion. J Mol Biol. 1982 Sep 15;160(2):133–146. doi: 10.1016/0022-2836(82)90171-1. [DOI] [PubMed] [Google Scholar]
- Shero J. H., Bordwell B., Rothfield N. F., Earnshaw W. C. High titers of autoantibodies to topoisomerase I (Scl-70) in sera from scleroderma patients. Science. 1986 Feb 14;231(4739):737–740. doi: 10.1126/science.3003910. [DOI] [PubMed] [Google Scholar]
- Snapka R. M., Powelson M. A., Strayer J. M. Swiveling and decatenation of replicating simian virus 40 genomes in vivo. Mol Cell Biol. 1988 Feb;8(2):515–521. doi: 10.1128/mcb.8.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapka R. M. Topoisomerase inhibitors can selectively interfere with different stages of simian virus 40 DNA replication. Mol Cell Biol. 1986 Dec;6(12):4221–4227. doi: 10.1128/mcb.6.12.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Stahl H., Koller T., Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May 5;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Varshavsky A. A nuclease-hypersensitive region forms de novo after chromosome replication. Mol Cell Biol. 1987 Oct;7(10):3822–3825. doi: 10.1128/mcb.7.10.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzner J. R., Muller M. T. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988 Jun 24;16(12):5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A. F., Schütz G. Camptothecin-induced in vivo topoisomerase I cleavages in the transcriptionally active tyrosine aminotransferase gene. Cell. 1987 Sep 25;50(7):1109–1117. doi: 10.1016/0092-8674(87)90177-2. [DOI] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986 May 23;45(4):555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Tack L. C., Proctor G. N. Two major replicating simian virus 40 chromosome classes. Synchronous replication fork movement is associated with bound large T antigen during elongation. J Biol Chem. 1987 May 5;262(13):6339–6349. [PubMed] [Google Scholar]
- Tapper D. P., Anderson S., DePamphilis M. L. Distribution of replicating simian virus 40 DNA in intact cells and its maturation in isolated nuclei. J Virol. 1982 Mar;41(3):877–892. doi: 10.1128/jvi.41.3.877-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapper D. P., DePamphilis M. L. Discontinuous DNA replication: accumulation of Simian virus 40 DNA at specific stages in its replication. J Mol Biol. 1978 Apr 15;120(3):401–422. doi: 10.1016/0022-2836(78)90427-8. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer P., Dohan C., Jr, Reznikoff C. Inactivating and mutagenic effects of nitrosoguanidine on simian virus 40. Proc Natl Acad Sci U S A. 1970 Jul;66(3):745–752. doi: 10.1073/pnas.66.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash C., Voelkel K., DiNardo S., Sternglanz R. Identification of Saccharomyces cerevisiae mutants deficient in DNA topoisomerase I activity. J Biol Chem. 1984 Feb 10;259(3):1375–1377. [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Vishwanatha J. K., Yamaguchi M., DePamphilis M. L., Baril E. F. Selection of template initiation sites and the lengths of RNA primers synthesized by DNA primase are strongly affected by its organization in a multiprotein DNA polymerase alpha complex. Nucleic Acids Res. 1986 Sep 25;14(18):7305–7323. doi: 10.1093/nar/14.18.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P. DNA topoisomerases: enzymes that control DNA conformation. Curr Top Microbiol Immunol. 1985;114:19–102. doi: 10.1007/978-3-642-70227-3_2. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Recent studies of DNA topoisomerases. Biochim Biophys Acta. 1987 Jun 6;909(1):1–9. doi: 10.1016/0167-4781(87)90040-6. [DOI] [PubMed] [Google Scholar]
- Weaver D. T., Fields-Berry S. C., DePamphilis M. L. The termination region for SV40 DNA replication directs the mode of separation for the two sibling molecules. Cell. 1985 Jun;41(2):565–575. doi: 10.1016/s0092-8674(85)80029-5. [DOI] [PubMed] [Google Scholar]
- Wilkie T. M., Palmiter R. D. Analysis of the integrant in MyK-103 transgenic mice in which males fail to transmit the integrant. Mol Cell Biol. 1987 May;7(5):1646–1655. doi: 10.1128/mcb.7.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. J., Fried M. Inverted duplication-transposition event in mammalian cells at an illegitimate recombination join. Mol Cell Biol. 1986 Jun;6(6):2179–2184. doi: 10.1128/mcb.6.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yang L., Wold M. S., Li J. J., Kelly T. J., Liu L. F. Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A. 1987 Feb;84(4):950–954. doi: 10.1073/pnas.84.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang J. C., Liu L. F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]