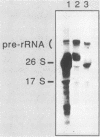
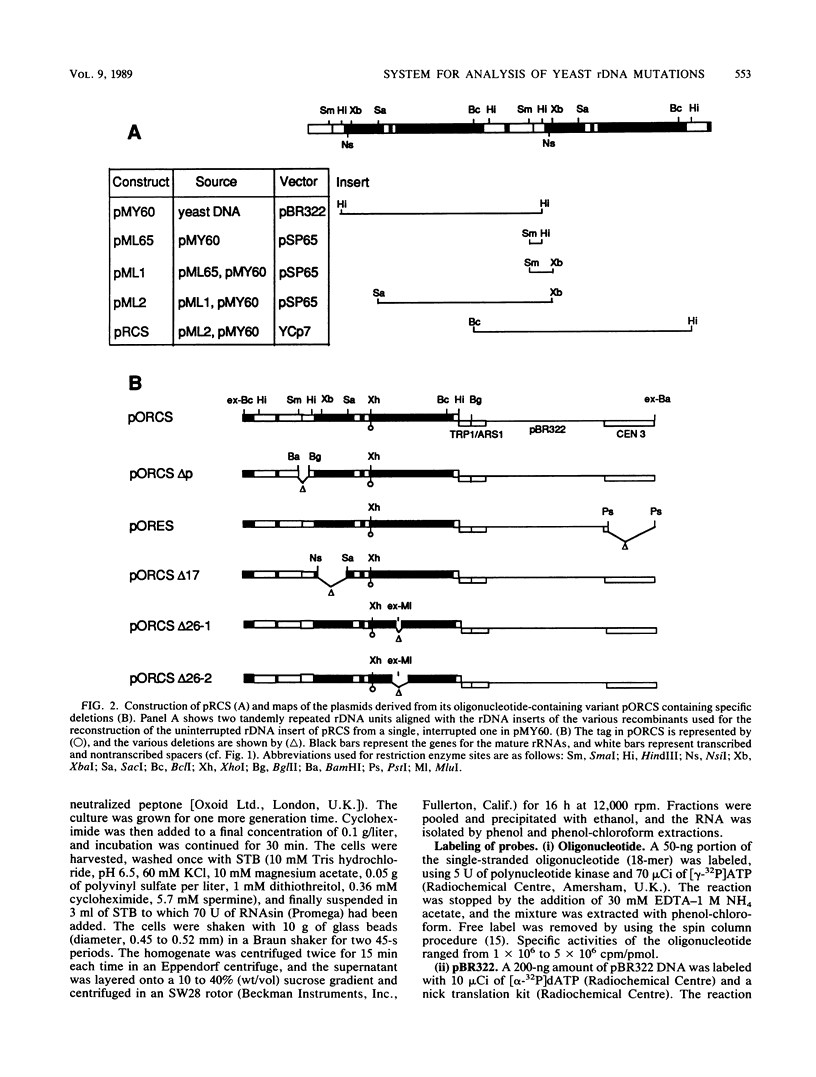
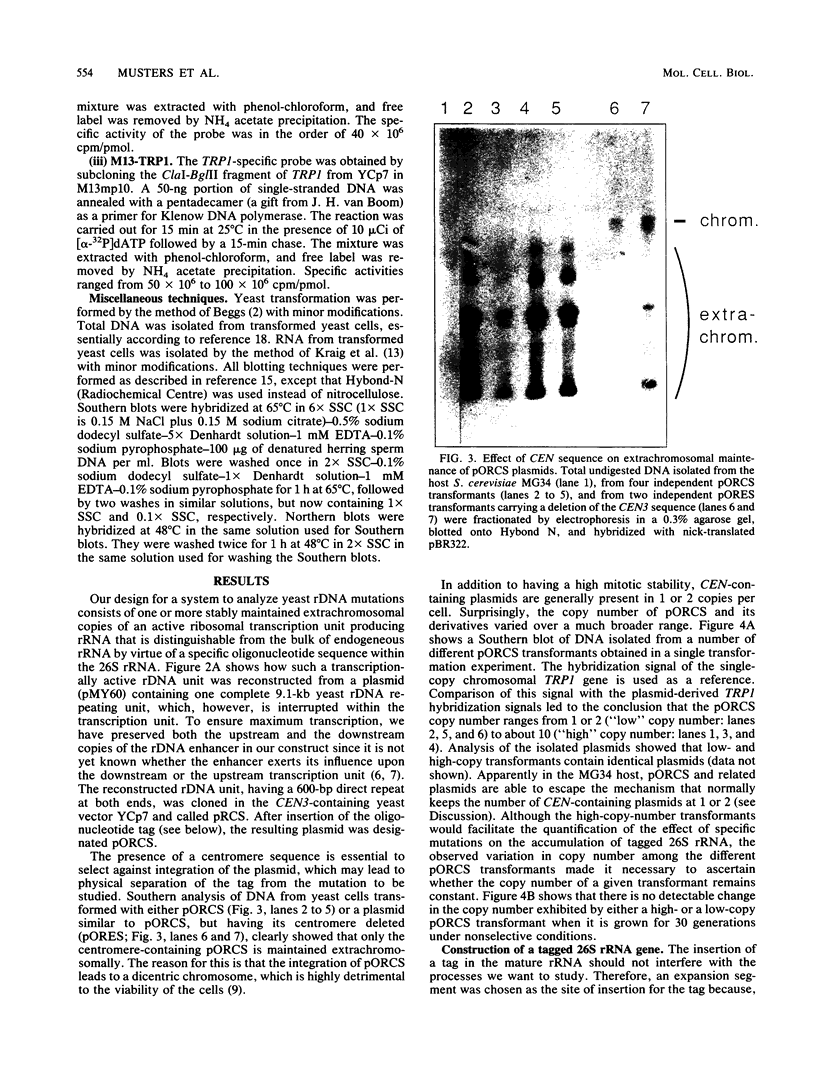
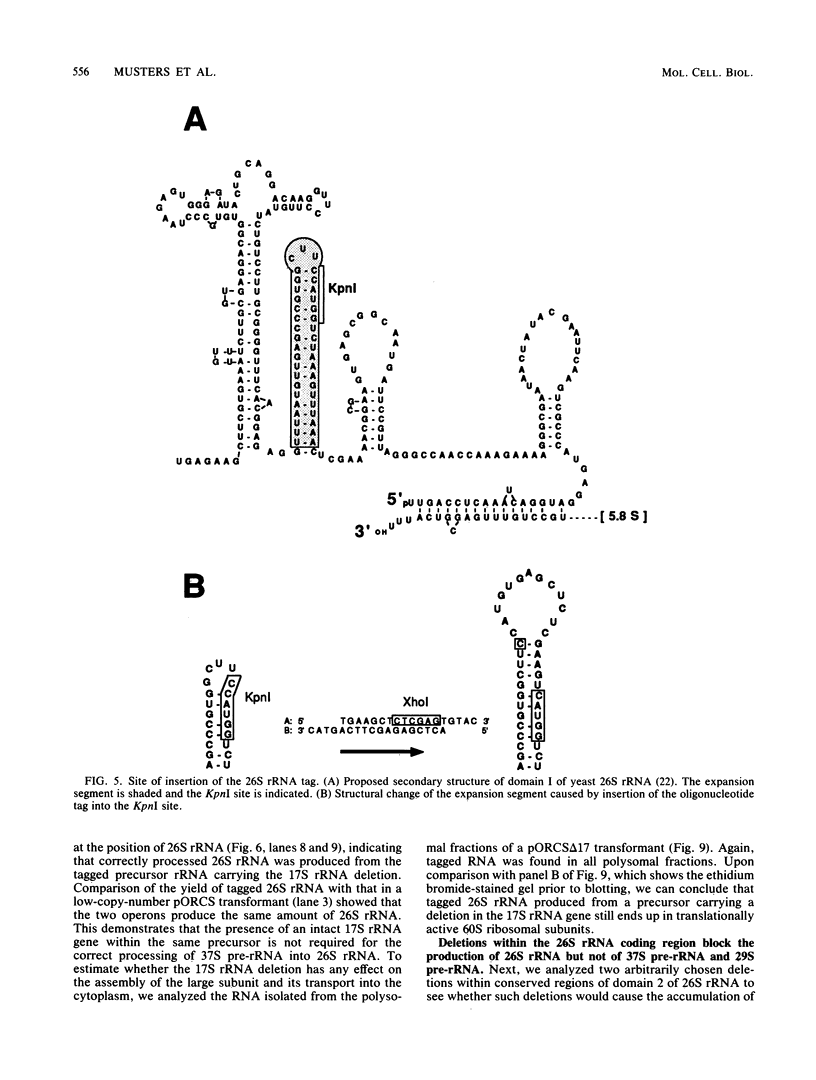
Abstract
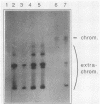
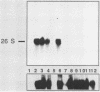
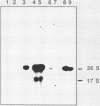
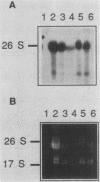
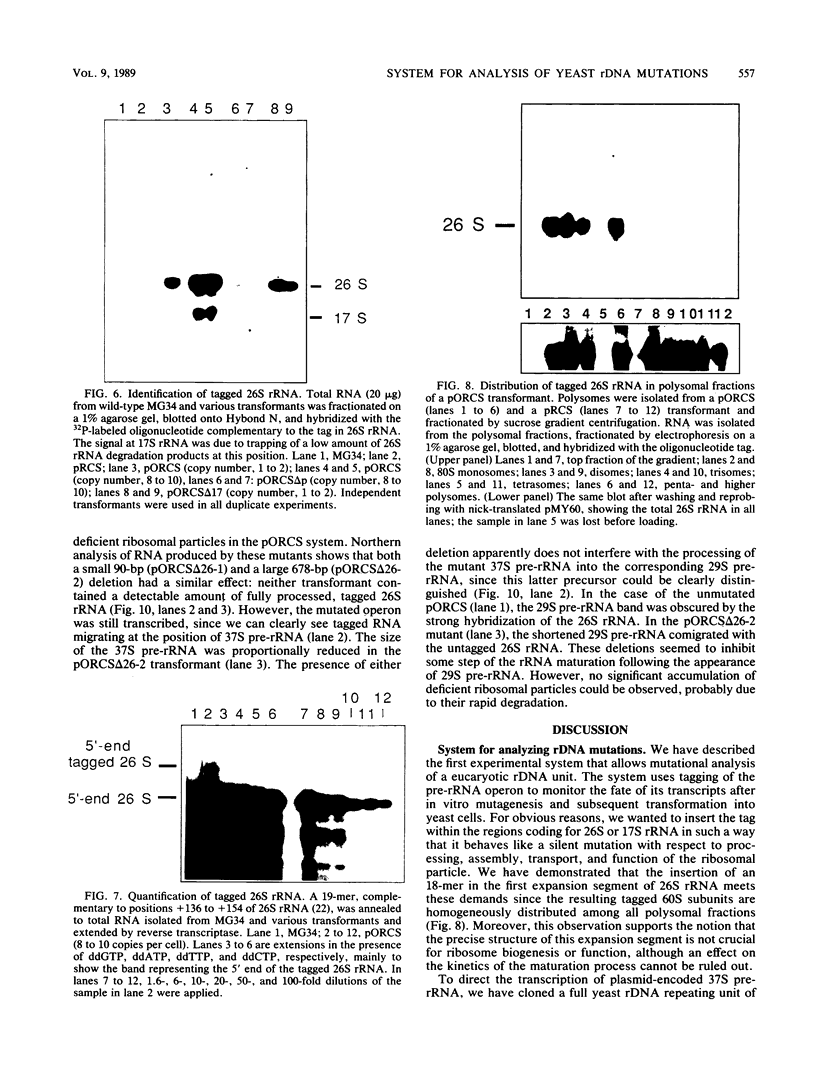
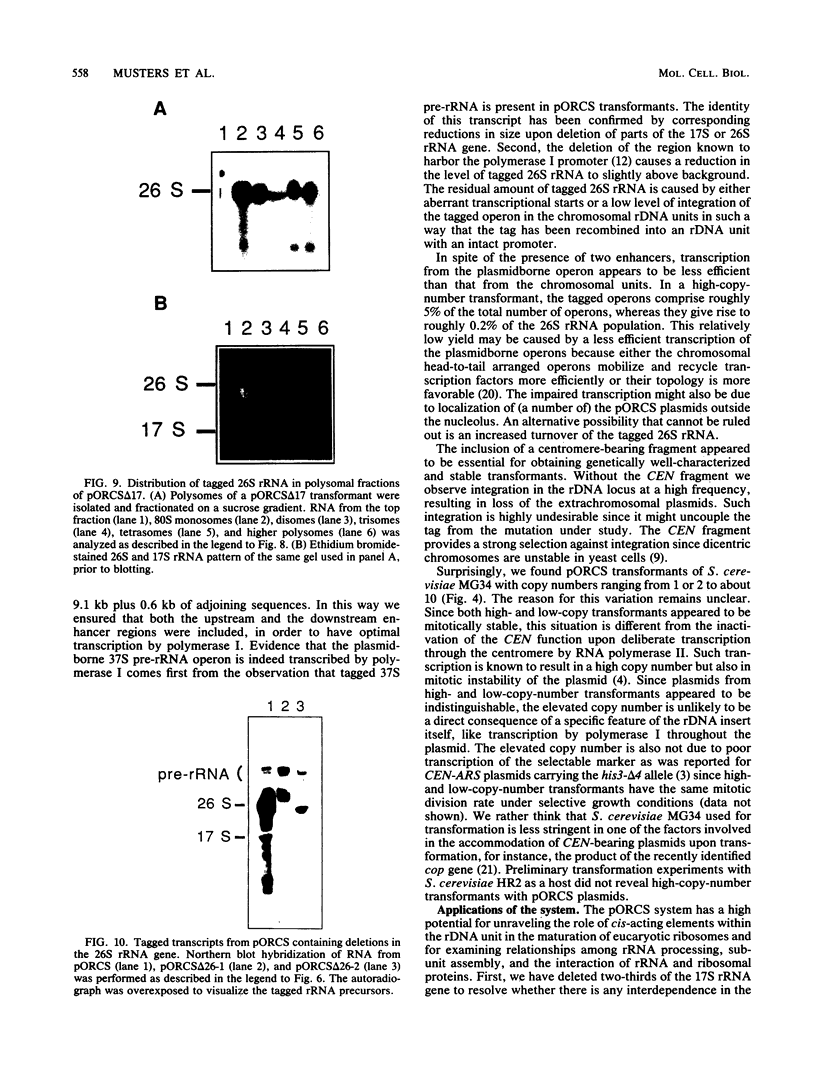
To develop a system for the analysis of eucaryotic ribosomal DNA (rDNA) mutations, we cloned a complete, transcriptionally active rDNA unit from the yeast Saccharomyces cerevisiae on a centromere-containing yeast plasmid. To distinguish the plasmid-derived ribosomal transcripts from those encoded by the rDNA locus, we inserted a tag of 18 base pairs within the first expansion segment of domain I of the 26S rRNA gene. We demonstrate that this insertion behaves as a neutral mutation since tagged 26S rRNA is normally processed and assembled into functional ribosomal subunits. This system allows us to study the effect of subsequent mutations within the tagged rDNA unit on the biosynthesis and function of the rRNA. As a first application, we wanted to ascertain whether the assembly of a 60S subunit is dependent on the presence in cis of an intact 17S rRNA gene. We found that a deletion of two-thirds of the 17S rRNA gene has no effect on the accumulation of active 60S subunits derived from the same operon. On the other hand, deletions within the second domain of the 26S rRNA gene completely abolished the accumulation of mature 26S rRNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Gritz L., Tung L., Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3429–3435. doi: 10.1128/mcb.5.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Bitoun R., Zamir A. Spontaneous amplification of yeast CEN ARS plasmids. Mol Gen Genet. 1986 Jul;204(1):98–102. doi: 10.1007/BF00330194. [DOI] [PubMed] [Google Scholar]
- Chlebowicz-Sledziewska E., Sledziewski A. Z. Construction of multicopy yeast plasmids with regulated centromere function. Gene. 1985;39(1):25–31. doi: 10.1016/0378-1119(85)90103-9. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Musters W., Dekker A. F., Klootwijk J., Planta R. J. Deletion mapping of the yeast Pol I promoter. Curr Genet. 1985;10(4):253–260. doi: 10.1007/BF00365621. [DOI] [PubMed] [Google Scholar]
- Kraig E., Haber J. E., Rosbash M. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1199–1204. doi: 10.1128/mcb.2.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nam H. G., Fried H. M. Effects of progressive depletion of TCM1 or CYH2 mRNA on Saccharomyces cerevisiae ribosomal protein accumulation. Mol Cell Biol. 1986 May;6(5):1535–1544. doi: 10.1128/mcb.6.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta R. J., Raué H. A. Control of ribosome biogenesis in yeast. Trends Genet. 1988 Mar;4(3):64–68. doi: 10.1016/0168-9525(88)90042-x. [DOI] [PubMed] [Google Scholar]
- Pruitt S. C., Reeder R. H. Effect of topological constraint on transcription of ribosomal DNA in Xenopus oocytes. Comparison of plasmid and endogenous genes. J Mol Biol. 1984 Mar 25;174(1):121–139. doi: 10.1016/0022-2836(84)90368-1. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Saccharomyces cerevisiae mutants that tolerate centromere plasmids at high copy number. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7203–7207. doi: 10.1073/pnas.84.20.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeet M. P., van Heerikhuizen H., Klootwijk J., Fontijn R. D., Planta R. J. Evolution of yeast ribosomal DNA: molecular cloning of the rDNA units of Kluyveromyces lactis and Hansenula wingei and their comparison with the rDNA units of other Saccharomycetoideae. Mol Gen Genet. 1984;195(1-2):116–125. doi: 10.1007/BF00332733. [DOI] [PubMed] [Google Scholar]
- Ware V. C., Tague B. W., Clark C. G., Gourse R. L., Brand R. C., Gerbi S. A. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 1983 Nov 25;11(22):7795–7817. doi: 10.1093/nar/11.22.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]