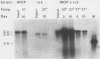
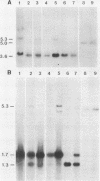
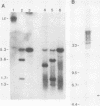
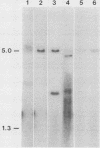
Abstract
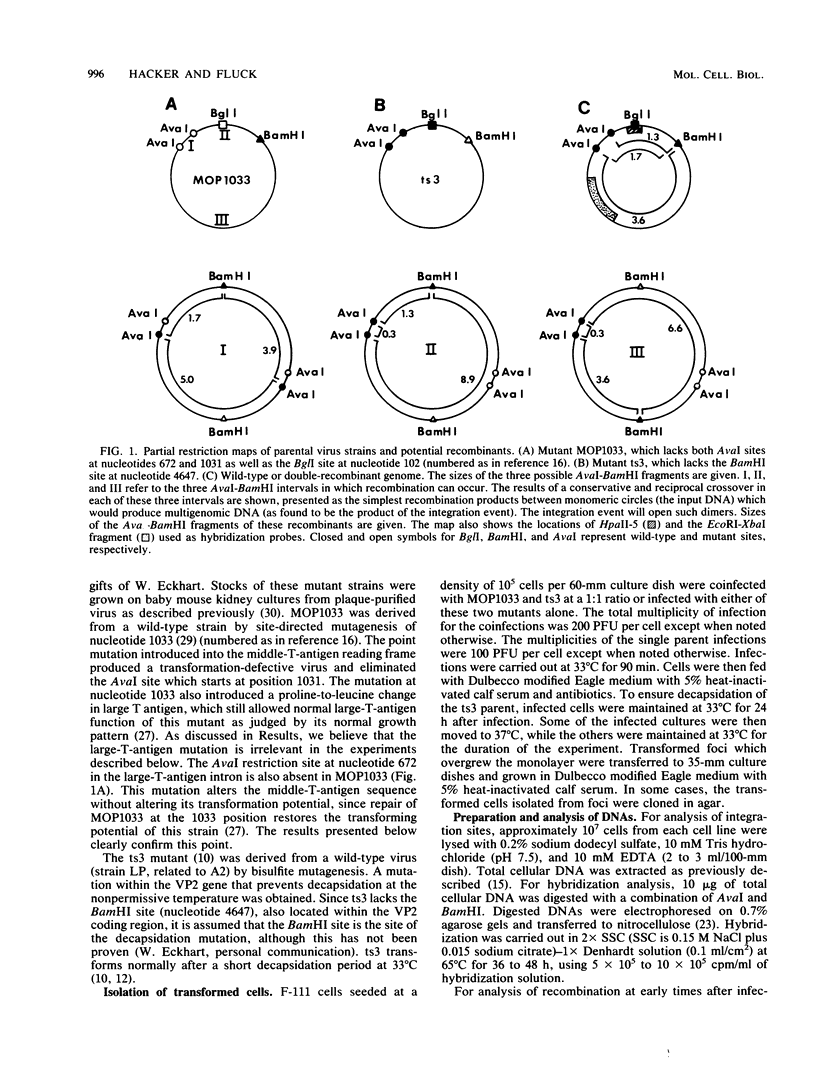
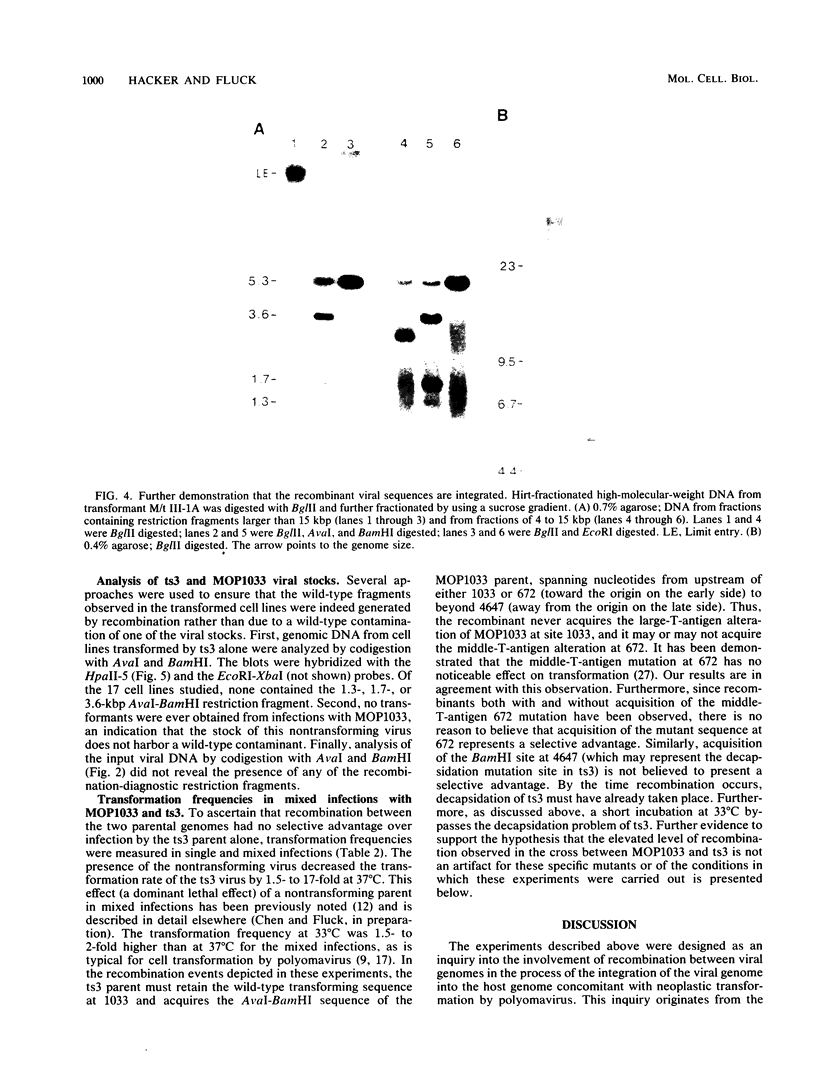
An unusually high incidence of interviral recombination was found in the process of integration of the polyomavirus genome concomitant with neoplastic transformation of nonpermissive cells. Transformants were isolated after mixed infections of Fischer rat cells with two mutants lacking restriction endonuclease sites and were analyzed for the presence of unselected integrated recombinant restriction fragments. A large fraction of the transformants isolated (38% of the 64 transformed cell lines studied) contained recombinant viral genomes that had undergone recombination in a 1.3-, 1.7-, or 3.6-kilobase-pair interval. More than 90% of these recombinant transformants showed evidence of crossovers in multiple intervals. To our knowledge, the recombination frequencies observed in these experiments represent the highest frequencies of homologous recombination reported for a mitotic mammalian system that does not involve transfection. In contrast to the elevated level of recombination in the integrated viral genomes, no evidence of recombination was obtained among the replicated unintegrated pool of viral genomes isolated from the same population of infected cells from which the recombinant transformants were derived. Either of two hypotheses can provide an explanation for the segregated recombination: either recombination occurs at elevated levels in a small, recombination-prone fraction of the population destined to become transformed, or recombination occurs only among those viral genomes which are engaged in the process of integration and thus interact with a recombinogenic host machinery (for example, the host scaffold). We favor the latter hypothesis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjursell G. Effects of 2'-deoxy-2'-azidocytidine on polyoma virus DNA replication: evidence for rolling circle-type mechanism. J Virol. 1978 Apr;26(1):136–142. doi: 10.1128/jvi.26.1.136-142.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Seidman M. M. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol Cell Biol. 1986 Jul;6(7):2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W., Rigby P. W. Fate of viral DNA in nonpermissive cells infected with simian virus 40. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6638–6642. doi: 10.1073/pnas.78.11.6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Pellegrini S., Basilico C. Deletion of the origin of replication impairs the ability of polyomavirus DNA to transform cells and to form tandem insertions. J Virol. 1984 Mar;49(3):984–987. doi: 10.1128/jvi.49.3.984-987.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Valle G., Fenton R. G., Basilico C. Polyoma large T antigen regulates the integration of viral DNA sequences into the genome of transformed cells. Cell. 1981 Feb;23(2):347–355. doi: 10.1016/0092-8674(81)90130-6. [DOI] [PubMed] [Google Scholar]
- Dulbecco R., Eckhart W. Temperature-dependent properties of cells transformed by a thermosensitive mutant of polyoma virus. Proc Natl Acad Sci U S A. 1970 Dec;67(4):1775–1781. doi: 10.1073/pnas.67.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Shaikh R., Benjamin T. L. An analysis of transformed clones obtained by coinfections with hr-t and ts-a mutants of polyoma virus. Virology. 1983 Oct 15;130(1):29–43. doi: 10.1016/0042-6822(83)90115-0. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):59–69. doi: 10.1128/mcb.5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A. E., Gilden R. V., Vernon M. L., Wolford R. G., Hugunin P. E., Huebner R. J. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friderici K., Oh S. Y., Ellis R., Guacci V., Fluck M. M. Recombination induces tandem repeats of integrated viral sequences in polyoma-transformed cells. Virology. 1984 Aug;137(1):67–73. doi: 10.1016/0042-6822(84)90009-6. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Hacker D. L., Fluck M. M. Viral DNA synthesis in nonpermissive rat F-111 cells and its role in neoplastic transformation by polyomavirus. Mol Cell Biol. 1989 Feb;9(2):648–658. doi: 10.1128/mcb.9.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayday A., Ruley H. E., Fried M. Structural and biological analysis of integrated polyoma virus DNA and its adjacent host sequences cloned from transformed rat cells. J Virol. 1982 Oct;44(1):67–77. doi: 10.1128/jvi.44.1.67-77.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Lania L., Griffiths M., Cooke B., Ito Y., Fried M. Untransformed rat cells containing free and integrated DNA of a polyoma nontransforming (Hr-t) mutant. Cell. 1979 Nov;18(3):793–802. doi: 10.1016/0092-8674(79)90132-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton D., Eckhart W. Mutation causing premature termination of the polyoma virus medium T antigen blocks cell transformation. J Virol. 1982 Mar;41(3):1014–1024. doi: 10.1128/jvi.41.3.1014-1024.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINOCOUR E. Purification of polyoma virus. Virology. 1963 Feb;19:158–168. doi: 10.1016/0042-6822(63)90005-9. [DOI] [PubMed] [Google Scholar]
- Williams T. J., Fried M. Inverted duplication-transposition event in mammalian cells at an illegitimate recombination join. Mol Cell Biol. 1986 Jun;6(6):2179–2184. doi: 10.1128/mcb.6.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberry L., Priehs C., Friderici K., Thompson M., Fluck M. Expression of proto-oncogenes in normal and papovavirus-transformed or -infected rat fibroblasts. Virology. 1985 Nov;147(1):154–168. doi: 10.1016/0042-6822(85)90235-1. [DOI] [PubMed] [Google Scholar]