Abstract
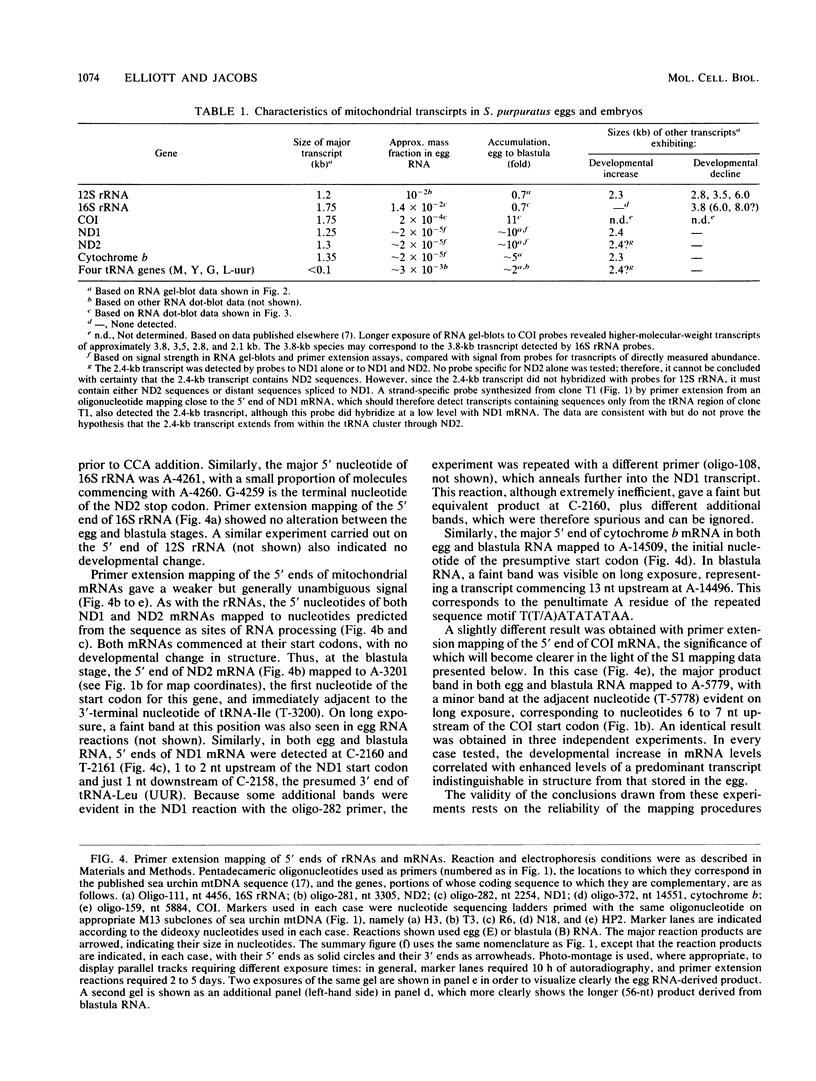
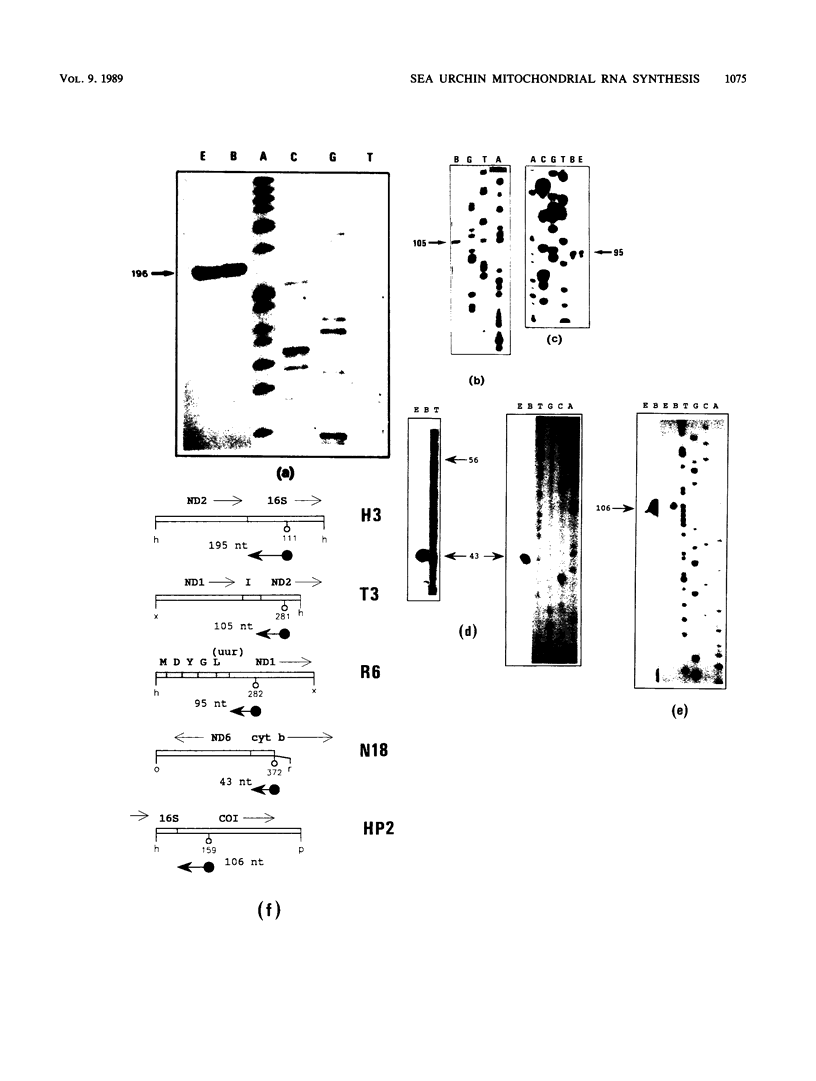
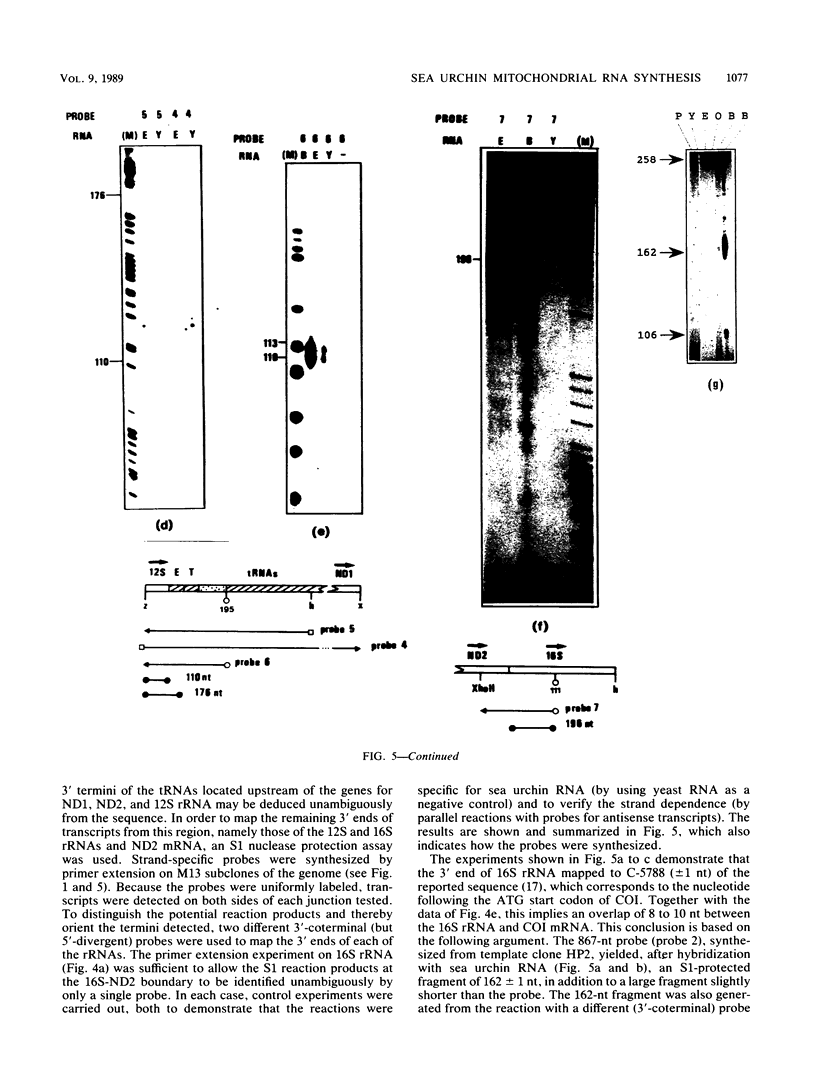
The structure and abundance of mitochondrial transcripts in sea urchin embryos were investigated by a combination of RNA blot-hybridization, S1 mapping, and primer extension assays. Between the egg and blastula stages, the relative abundance of mitochondrial rRNAs declined slightly, while that of mitochondrial mRNAs increased up to 10-fold. Fine mapping of the termini of the rRNAs and of the adjacent transcripts indicated that, although they appeared to be butt-joined at their 5' ends to the upstream transcripts, tRNA-Phe 5' to the small subunit (12S) rRNA and NADH dehydrogenase subunit 2 mRNA 5' to the large subunit (16S) rRNA, respectively, their 3' ends were found to overlap the 5' ends of the downstream transcripts. 12S rRNA was found to extend 7 to 13 nucleotides into the sequence of tRNA-Glu; 16S rRNA was shown to terminate 3 to 5 nucleotides inside the coding region of cytochrome oxidase subunit 1 (COI) and 8 to 10 nucleotides from the mapped 5' end of COI mRNA. The rRNAs and the downstream transcripts must therefore be synthesized by distinct pathways, either by alternative processing of the same primary transcript(s) or by processing of different precursors. In either case, the events which select the ribosomal 3' ends preclude the production of functional transcripts of the downstream genes from the same precursor molecule. No developmental alterations in transcript structure were detected. We propose that mitochondrial RNA levels are regulated in early development by the selection of alternate and mutually exclusive RNA-processing pathways.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angerer R. C., Davidson E. H. Molecular indices of cell lineage specification in sea urchin embryos. Science. 1984 Dec 7;226(4679):1153–1160. doi: 10.1126/science.6594757. [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Benson S., Sucov H., Stephens L., Davidson E., Wilt F. A lineage-specific gene encoding a major matrix protein of the sea urchin embryo spicule. I. Authentication of the cloned gene and its developmental expression. Dev Biol. 1987 Apr;120(2):499–506. doi: 10.1016/0012-1606(87)90253-3. [DOI] [PubMed] [Google Scholar]
- Biswas T. K., Ticho B., Getz G. S. In vitro characterization of the yeast mitochondrial promoter using single-base substitution mutants. J Biol Chem. 1987 Oct 5;262(28):13690–13696. [PubMed] [Google Scholar]
- Brandhorst B. P. Informational content of the echinoderm egg. Dev Biol (N Y 1985) 1985;1:525–576. doi: 10.1007/978-1-4615-6814-8_12. [DOI] [PubMed] [Google Scholar]
- Cabrera C. V., Jacobs H. T., Posakony J. W., Grula J. W., Roberts J. W., Britten R. J., Davidson E. H. Transcripts of three mitochondrial genes in the RNA of sea urchin eggs and embryos. Dev Biol. 1983 Jun;97(2):500–505. doi: 10.1016/0012-1606(83)90107-0. [DOI] [PubMed] [Google Scholar]
- Cantatore P., Roberti M., Morisco P., Rainaldi G., Gadaleta M. N., Saccone C. A novel gene order in the Paracentrotus lividus mitochondrial genome. Gene. 1987;53(1):41–54. doi: 10.1016/0378-1119(87)90091-6. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Clayton D. A. In vitro transcription of human mitochondrial DNA: accurate termination requires a region of DNA sequence that can function bidirectionally. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6277–6281. doi: 10.1073/pnas.83.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Rabinowitz M. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem. 1983 Nov 25;258(22):14025–14033. [PubMed] [Google Scholar]
- Feagin J. E., Jasmer D. P., Stuart K. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell. 1987 May 8;49(3):337–345. doi: 10.1016/0092-8674(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Gelfand R., Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981 Jun;1(6):497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
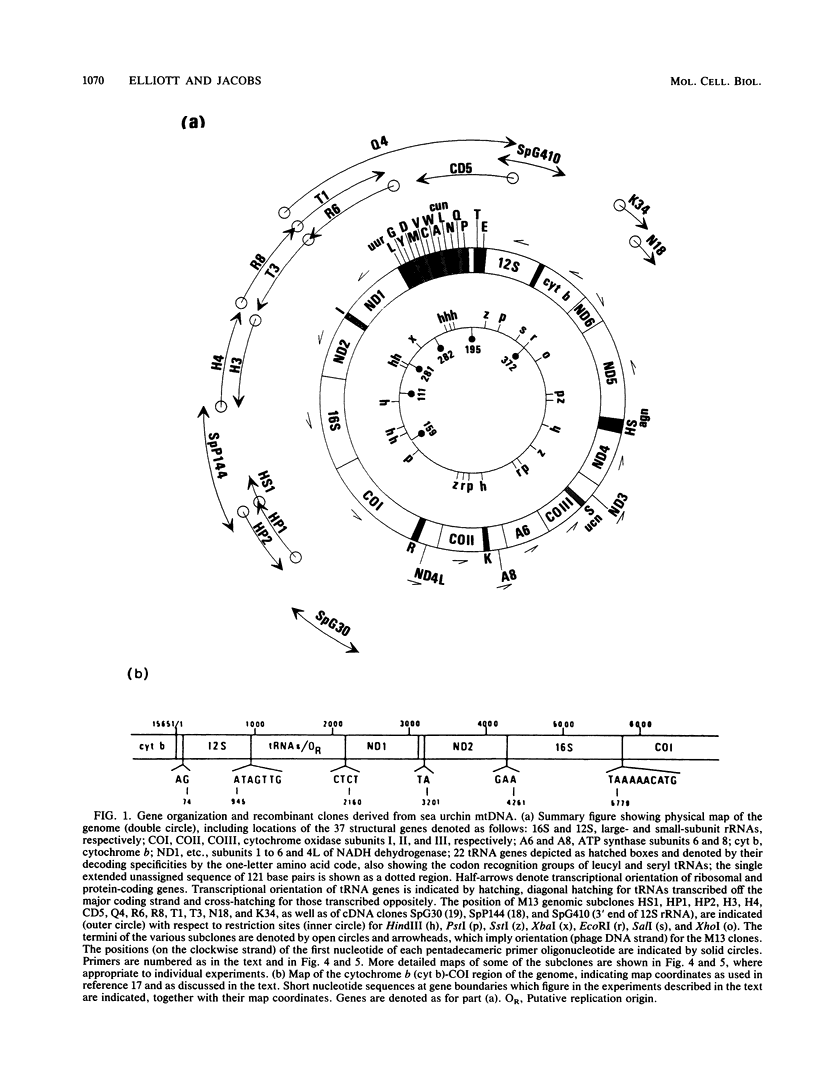
- Jacobs H. T., Elliott D. J., Math V. B., Farquharson A. Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol. 1988 Jul 20;202(2):185–217. doi: 10.1016/0022-2836(88)90452-4. [DOI] [PubMed] [Google Scholar]
- Jacobs H. T., Grimes B. Complete nucleotide sequences of the nuclear pseudogenes for cytochrome oxidase subunit I and the large mitochondrial ribosomal RNA in the sea urchin Strongylocentrotus purpuratus. J Mol Biol. 1986 Feb 20;187(4):509–527. doi: 10.1016/0022-2836(86)90330-x. [DOI] [PubMed] [Google Scholar]
- Jacobs H. T., Posakony J. W., Grula J. W., Roberts J. W., Xin J. H., Britten R. J., Davidson E. H. Mitochondrial DNA sequences in the nuclear genome of Strongylocentrotus purpuratus. J Mol Biol. 1983 Apr 25;165(4):609–632. doi: 10.1016/s0022-2836(83)80270-8. [DOI] [PubMed] [Google Scholar]
- Lee J. J., Calzone F. J., Britten R. J., Angerer R. C., Davidson E. H. Activation of sea urchin actin genes during embryogenesis. Measurement of transcript accumulation from five different genes in Strongylocentrotus purpuratus. J Mol Biol. 1986 Mar 20;188(2):173–183. doi: 10.1016/0022-2836(86)90302-5. [DOI] [PubMed] [Google Scholar]
- Levens D., Ticho B., Ackerman E., Rabinowitz M. Transcriptional initiation and 5' termini of yeast mitochondrial RNA. J Biol Chem. 1981 May 25;256(10):5226–5232. [PubMed] [Google Scholar]
- Montoya J., Christianson T., Levens D., Rabinowitz M., Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya J., Gaines G. L., Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983 Aug;34(1):151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman J. V., Schmidt M. R. RNA transcription and translation in sea urchin oocytes and eggs. Dev Biol. 1981 Jan 30;81(2):220–228. doi: 10.1016/0012-1606(81)90285-2. [DOI] [PubMed] [Google Scholar]
- Shott R. J., Lee J. J., Britten R. J., Davidson E. H. Differential expression of the actin gene family of Strongylocentrotus purpuratus. Dev Biol. 1984 Feb;101(2):295–306. doi: 10.1016/0012-1606(84)90143-x. [DOI] [PubMed] [Google Scholar]
- Wells D. E., Bruskin A. M., O'Brochta D. A., Raff R. A. Prevalent RNA sequences of mitochondrial origin in sea urchin embryos. Dev Biol. 1982 Aug;92(2):557–562. doi: 10.1016/0012-1606(82)90202-0. [DOI] [PubMed] [Google Scholar]