Abstract
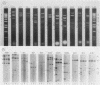
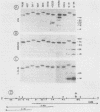
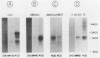
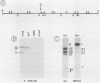
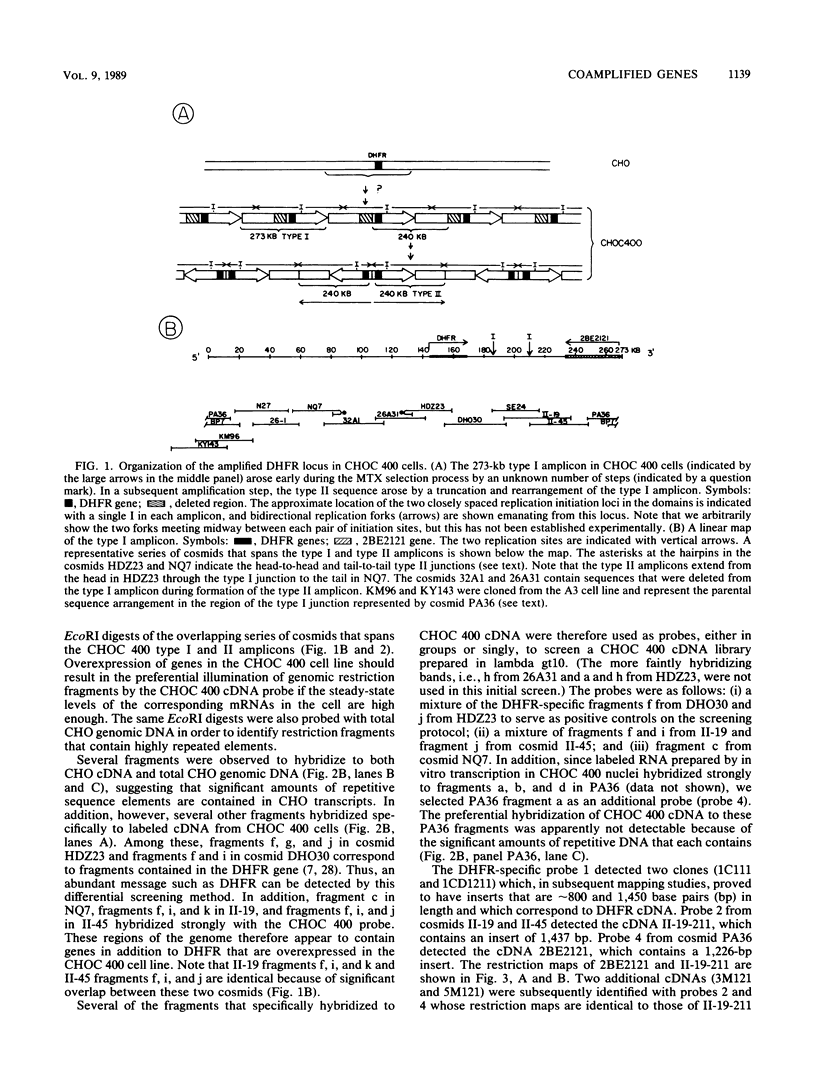
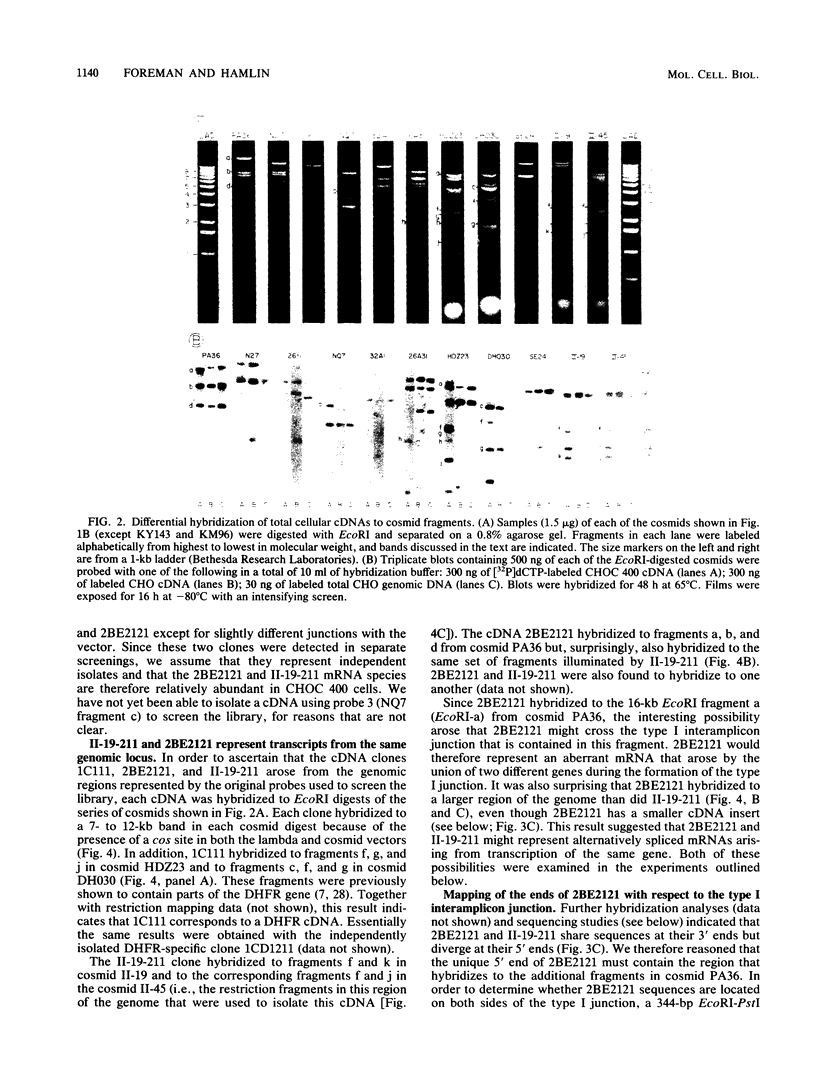
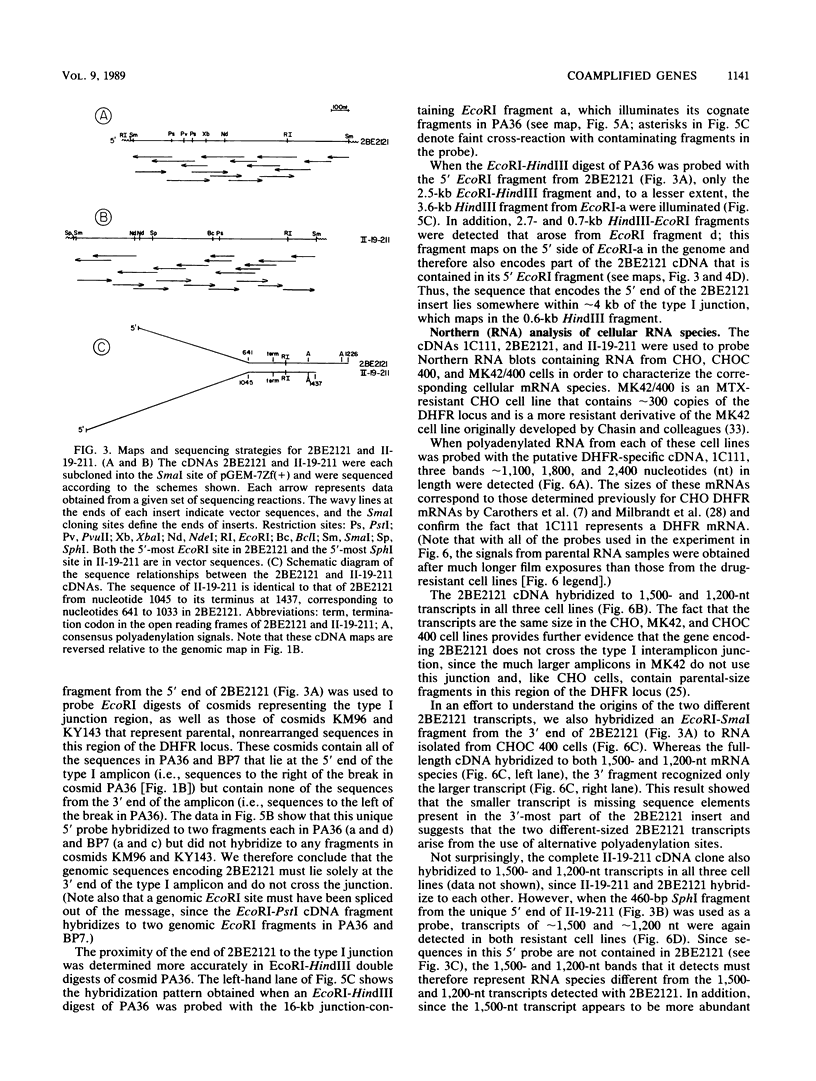
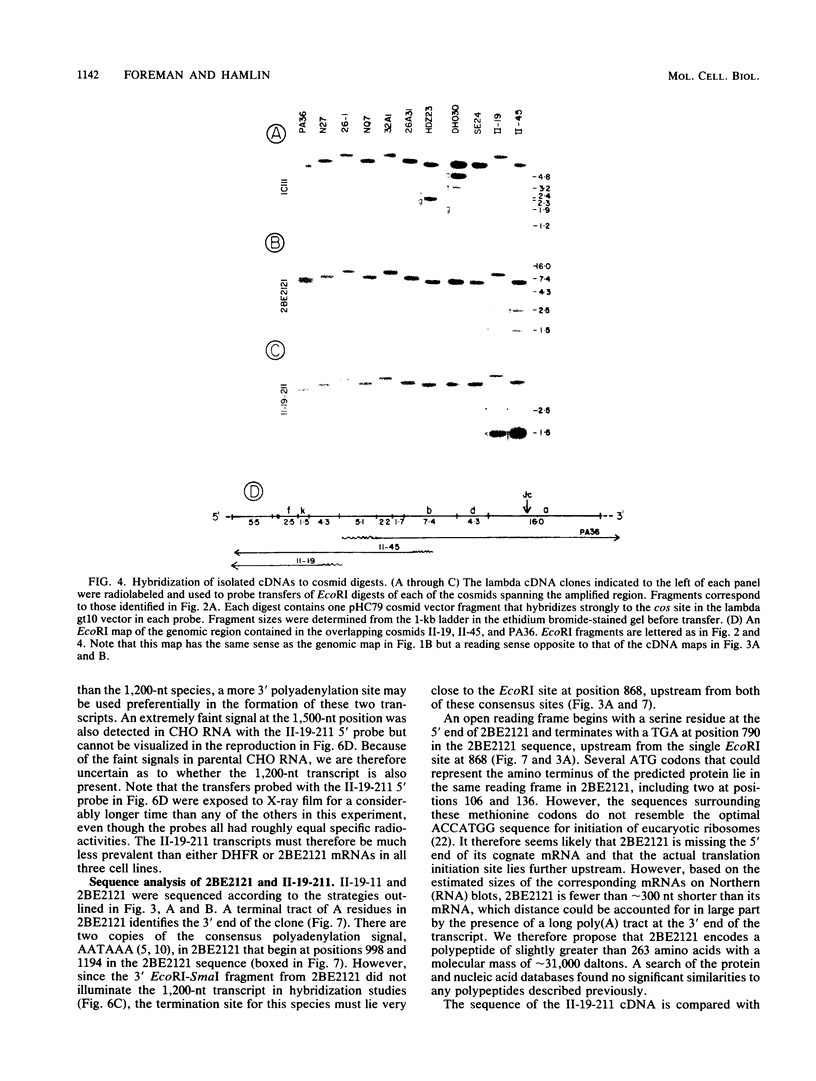
As part of an effort to characterize the spatial and functional relationships among genetic elements within the amplified dihydrofolate reductase (DHFR) domain in Chinese hamster cells, we have used a variation of the differential hybridization approach to identify cDNA clones whose genes are coamplified with DHFR in the methotrexate-resistant cell line, CHOC 400. Our initial screen was successful in isolating both DHFR and non-DHFR cDNAs. One of the non-DHFR cDNA clones, 2BE2121, hybridizes on Northern (RNA) blots to abundant 1,200- and 1,500-nucleotide (nt) transcripts which differ in the lengths of their 3' untranslated regions. The clone 2BE2121 contains a 789-nt open reading frame but does not appear to be related to any members of the protein or nucleic acid sequence databases. A second larger non-DHFR cDNA, II-19-211, was isolated that is transcribed from the same gene as 2BE2121 but contains only a small carboxyl-terminal portion of the open reading frame. II-19-211 may, therefore, represent either a splicing intermediate or an mRNA transcribed from a cryptic intragenic promoter. Hybridization to cosmids from the DHFR domain shows that 2BE2121 is encoded by a gene approximately 34 kilobases (kb) long. The 5'-most genomic fragment is less than 4 kb from an interamplicon junction. The 3' end of the 2BE2121 gene lies approximately 75 kb downstream from the DHFR gene and approximately 25 kb downstream from the proximal replication initiation site, and the transcriptional polarity is opposite to that of the leading strand of replication. Thus, both the DHFR and 2BE2121 genes are exceptions to the theory that transcription proceeds in the same direction as the leading strand of the replication fork.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biedler J. L., Spengler B. A. A novel chromosome abnormality in human neuroblastoma and antifolate-resistant Chinese hamster cell lives in culture. J Natl Cancer Inst. 1976 Sep;57(3):683–695. doi: 10.1093/jnci/57.3.683. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Braunstein J. D., Schulze D., DelGiudice T., Furst A., Schildkraut C. L. The temporal order of replication of murine immunoglobulin heavy chain constant region sequences corresponds to their linear order in the genome. Nucleic Acids Res. 1982 Nov 11;10(21):6887–6902. doi: 10.1093/nar/10.21.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Stacy T. P. Identification of sequences in the herpes simplex virus thymidine kinase gene required for efficient processing and polyadenylation. Mol Cell Biol. 1985 Aug;5(8):2104–2113. doi: 10.1128/mcb.5.8.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Debatisse M., de Saint Vincent B. R., Buttin G. Expression of several amplified genes in an adenylate-deaminase overproducing variant of Chinese hamster fibroblasts. EMBO J. 1984 Dec 20;3(13):3123–3127. doi: 10.1002/j.1460-2075.1984.tb02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. Inhibition of splicing but not cleavage at the 5' splice site by truncating human beta-globin pre-mRNA. Proc Natl Acad Sci U S A. 1986 Feb;83(4):927–931. doi: 10.1073/pnas.83.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin J. L., Milbrandt J. D., Heintz N. H., Azizkhan J. C. DNA sequence amplification in mammalian cells. Int Rev Cytol. 1984;90:31–82. doi: 10.1016/s0074-7696(08)61487-4. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C. D., Leffak M. Polarity of DNA replication through the avian alpha-globin locus. Mol Cell Biol. 1986 Apr;6(4):976–984. doi: 10.1128/mcb.6.4.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney J. E., Hamlin J. L. Isolation of the amplified dihydrofolate reductase domain from methotrexate-resistant Chinese hamster ovary cells. Mol Cell Biol. 1987 Feb;7(2):569–577. doi: 10.1128/mcb.7.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney J. E., Ma C., Leu T. H., Flintoff W. F., Troutman W. B., Hamlin J. L. The dihydrofolate reductase amplicons in different methotrexate-resistant Chinese hamster cell lines share at least a 273-kilobase core sequence, but the amplicons in some cell lines are much larger and are remarkably uniform in structure. Mol Cell Biol. 1988 Dec;8(12):5268–5279. doi: 10.1128/mcb.8.12.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Looney J. E., Leu T. H., Hamlin J. L. Organization and genesis of dihydrofolate reductase amplicons in the genome of a methotrexate-resistant Chinese hamster ovary cell line. Mol Cell Biol. 1988 Jun;8(6):2316–2327. doi: 10.1128/mcb.8.6.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J. D., Azizkhan J. C., Hamlin J. L. Amplification of a cloned Chinese hamster dihydrofolate reductase gene after transfer into a dihydrofolate reductase-deficient cell line. Mol Cell Biol. 1983 Jul;3(7):1274–1282. doi: 10.1128/mcb.3.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J. D., Heintz N. H., White W. C., Rothman S. M., Hamlin J. L. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Schimke R. T., Urlaub G., Chasin L. A. Amplified dihydrofolate reductase genes are localized to a homogeneously staining region of a single chromosome in a methotrexate-resistant Chinese hamster ovary cell line. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5553–5556. doi: 10.1073/pnas.75.11.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured animal cells. Cell. 1984 Jul;37(3):705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983 Dec;35(2 Pt 1):421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- Smithies O. The control of globin and other eukaryotic genes. J Cell Physiol Suppl. 1982;1:137–143. doi: 10.1002/jcp.1041130421. [DOI] [PubMed] [Google Scholar]
- Soma G., Murata M., Kitahara N., Gatanaga T., Shibai H., Morioka H., Andoh T. Detection of a countertranscript in promyelocytic leukemia cells HL60 during early differentiation by TPA. Biochem Biophys Res Commun. 1985 Oct 15;132(1):100–109. doi: 10.1016/0006-291x(85)90994-5. [DOI] [PubMed] [Google Scholar]
- Srinivasan P. R., Tonin P. N., Wensing E. J., Lewis W. H. The gene for ornithine decarboxylase is co-amplified in hydroxyurea-resistant hamster cells. J Biol Chem. 1987 Sep 15;262(26):12871–12878. [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Trempe J. P., Lindstrom Y. I., Leffak M. Opposite replication polarities of transcribed and nontranscribed histone H5 genes. Mol Cell Biol. 1988 Apr;8(4):1657–1663. doi: 10.1128/mcb.8.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. J., Ting A. C., Nordstrom J. L., Zimmer W., O'Malley B. W. Processing of high molecular weight ovalbumin and ovomucoid precursor RNAs to messenger RNA. Cell. 1980 Nov;22(1 Pt 1):219–230. doi: 10.1016/0092-8674(80)90170-1. [DOI] [PubMed] [Google Scholar]
- Van der Bliek A. M., Van der Velde-Koerts T., Ling V., Borst P. Overexpression and amplification of five genes in a multidrug-resistant Chinese hamster ovary cell line. Mol Cell Biol. 1986 May;6(5):1671–1678. doi: 10.1128/mcb.6.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Urlaub G., Chasin L. A. Polyadenylation of Chinese hamster dihydrofolate reductase genomic genes and minigenes after gene transfer. Somat Cell Mol Genet. 1987 Sep;13(5):491–504. doi: 10.1007/BF01534491. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Vitto L., Rubnitz J. Co-amplification of rRNA genes with CAD genes in N-(phosphonacetyl)-L-aspartate-resistant Syrian hamster cells. Mol Cell Biol. 1983 Nov;3(11):2066–2075. doi: 10.1128/mcb.3.11.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- de Bruijn M. H., Van der Bliek A. M., Biedler J. L., Borst P. Differential amplification and disproportionate expression of five genes in three multidrug-resistant Chinese hamster lung cell lines. Mol Cell Biol. 1986 Dec;6(12):4717–4722. doi: 10.1128/mcb.6.12.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]