Abstract
In vivo molecular dynamics in Halobacterium salinarum cells under stress conditions was measured by neutron scattering experiments coupled with microbiological characterization. Molecular dynamics alterations were detected with respect to unstressed cells, reflecting a softening of protein structures consistent with denaturation. The experiments indicated that the neutron scattering method provides a promising tool to study molecular dynamics modifications in the proteome of living cells induced by factors altering protein folds.
Keywords: protein folding, stress response, cell biology
1. Introduction
Many studies have addressed the effects of physico-chemical stresses on proteins in vitro. They revealed that protein molecular dynamics is much more sensitive to environment than protein structure [1]. A protein can be inactive, for example, in an environment in which its structure is perfectly stable, because of inappropriate dynamics [2]. By the very nature of the forces stabilizing tertiary structure, protein dynamics cannot be considered independently of solvent conditions. Thus, the crowded conditions as well as the presence of extensive molecular interactions that prevail in the cytosol of a living cell are believed to interfere significantly with protein folding and unfolding processes [3]. It follows that the consequences of environmental stress on dynamic state of a protein in the cytosol could be very different from that measured in vitro on a homogeneous population of purified protein, under dilute conditions. However, while many studies have addressed the effects of physico-chemical stresses and especially dehydration on protein molecular dynamics in vitro [4], few direct biophysical observations have been performed on whole cells in vivo and it is still difficult to appreciate the consequences of environmental stress on the molecular dynamic state of the whole proteome.
Incoherent neutron scattering is a method of choice to probe protein dynamics [5]. Neutrons are scattered by atomic nuclei providing information on the momentum and energy of their motions. The probed length and time scales are defined by the scattering vector (Q) range and energy resolution of the experiment, respectively. By suitably choosing these values, it has been possible to observe internal dynamics of a protein population, in vivo, without the requirement for specific protein labelling [6]. Experiments are based on ‘elastic temperature scans’, in which neutrons scattered in a very narrow energy window around the energy of the incident beam are observed as a function of sample temperature. The results of the experiment are atomic mean square displacements (MSD, in Å2 units) and an averaged effective force constant ( in N m–1 units), on the length and time scales selected by the experimental conditions (typically approximately ångström amplitude in pico to nanosecond time scale). These parameters inform on the mean flexibility and resilience of the protein structures, respectively [5]. The resilience has to be considered with respect to the free energy landscape, i.e. it includes both structural rigidity (the enthalpic term owing to internal forces) and conformational sampling (the entropic term). The neutron length–time window is well suited to examine the internal motions in unfolded polypeptides compared with those in folded proteins. Incoherent neutron scattering was used to probe the mean molecular dynamics properties of living cells [6–8]. Because of their relative abundance the macromolecular signal is dominated by the contribution of the proteins. Until now, the approach has been used in the case of adaptation of micro-organisms to temperature. It was found that evolution modifies in depth the molecular dynamics in psychrophile and thermophile microbes, with resilience values being a factor of two smaller or larger, respectively, than values found in mesophiles [7]. In these experiments, we noted a small difference that appeared when Escherichia coli cells were exposed to a heat stress. In this work, Halobacterium salinarum (Hs) was used as a model micro-organism to study, in vivo and in greater detail, the effects of temperature stress on molecular dynamics. Hs is not a thermophile, but it can tolerate heat stress better than most mesophilic cells [9]. Because of this, it was possible to explore a wider range of stress conditions than in E. coli (from 37°C to 65°C). This allowed the identification of a new Q range better suited to assess how a physical stress impacts the mean molecular dynamics properties of a microbial proteome within the cellular context, in vivo.
in N m–1 units), on the length and time scales selected by the experimental conditions (typically approximately ångström amplitude in pico to nanosecond time scale). These parameters inform on the mean flexibility and resilience of the protein structures, respectively [5]. The resilience has to be considered with respect to the free energy landscape, i.e. it includes both structural rigidity (the enthalpic term owing to internal forces) and conformational sampling (the entropic term). The neutron length–time window is well suited to examine the internal motions in unfolded polypeptides compared with those in folded proteins. Incoherent neutron scattering was used to probe the mean molecular dynamics properties of living cells [6–8]. Because of their relative abundance the macromolecular signal is dominated by the contribution of the proteins. Until now, the approach has been used in the case of adaptation of micro-organisms to temperature. It was found that evolution modifies in depth the molecular dynamics in psychrophile and thermophile microbes, with resilience values being a factor of two smaller or larger, respectively, than values found in mesophiles [7]. In these experiments, we noted a small difference that appeared when Escherichia coli cells were exposed to a heat stress. In this work, Halobacterium salinarum (Hs) was used as a model micro-organism to study, in vivo and in greater detail, the effects of temperature stress on molecular dynamics. Hs is not a thermophile, but it can tolerate heat stress better than most mesophilic cells [9]. Because of this, it was possible to explore a wider range of stress conditions than in E. coli (from 37°C to 65°C). This allowed the identification of a new Q range better suited to assess how a physical stress impacts the mean molecular dynamics properties of a microbial proteome within the cellular context, in vivo.
2. Results and discussion
2.1. Thermal stress response of Halobacterium salinarum in neutron experimental conditions
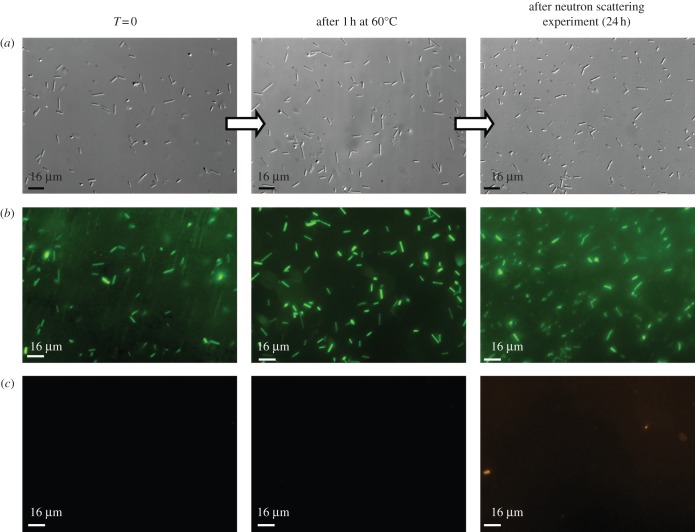
Microbiology experiments were performed in order to determine whether neutron experiment could be performed on stressed Hs R1 cells (Hs). The cells were cultivated at 37°C as described [10] and pelleted at mid-log phase. Halobacterium cells can maintain growth up to 50–55°C and stop dividing only at 60°C. The cells were exposed to 55°C and 60°C for 1 h, washed with a hypersaline physiological buffer and sealed in appropriate sample holders for 24 h under anaerobic conditions. The mortality rates were determined before and after heat shock, and after the neutron experiments by using light microscopy counts and live–dead staining protocols. Morphological analyses were also performed by confocal microscopy. In stressed and unstressed samples, the residual cell viability values were found to be over 95 per cent after 24 h of neutron measurements and no morphological changes could be detected (figure 1). Hs cells concentrate multi-molar amounts of K+ inside its cytosol to counterbalance the difference in osmotic pressure due to the external hypersaline conditions [11]. The process is driven by the membrane potential and is energy-dependent [12]. The biochemical activities and cellular functions are therefore adapted to function optimally in hypersaline conditions [13]. Thus, the measurement of intracellular K+ and Na+ concentration represent a good indicator of Hs cellular integrity. Inductively coupled plasma atomic emission spectroscopy (ICP) measurements performed on Hs cells showed that the high cytosol ion content values (3.7 M K+ and 1.1 M Na+) remained unchanged after 24 h of incubation in the sample holder, even after 1 h of a 60°C heat shock. We also tested the accumulation of the thermosome, a thermal stress protein, within the Hs cytosol in the different stress conditions, before and after neutron experiments. The thermosome is a type II chaperonin complex that represents the main protein quality control system in Archaea [14]. The thermosome acts on partially unfolded protein to promote their correct refolding in an energy-dependent manner [15]. Its abundance within Hs cells is associated with the accumulation of misfolded proteins substrates in the cytosol. The level of thermosome in the cells was immunodetected in Hs cell extracts (figure 2). These experiments showed that, for unstressed cells, the thermosome level remains constant after 24 h of neutron experiment. On the contrary, a significant increase in thermosome protein was observed in heat-shocked cells, before and after neutron experiments. These experiments showed that, in Hs, significant neutron measurements could be performed on intact cells, under stress conditions that generate the accumulation of misfolded proteins in the cytosol.
Figure 1.
Effect of thermal stress on the viability of Hs. The cell cultures were heat-shocked at different temperatures (from 40°C to 60°C) for 1 h under agitation. Here, we present the data for the 60°C experiment. After centrifugation, the stressed cell paste was enclosed in the sample holder and the mean molecular dynamic properties of the cell were measured during 24 h by neutron scattering. To assess for the cell mortality rates after the experiment, the pellet was resuspended in the initial volume of cultivation media. Haloarchaeal cells were stained with the LIVE/DEAD BacLight kit. (a) Differential interference contrast image of Hs. (b) Green fluorescence corresponds to viable Archaea with intact membranes. (c) Red fluorescence indicates non-viable Archaea. The viability rates were found to be over 95% after 1 h of stress and after the 24 h neutron experiment.
Figure 2.
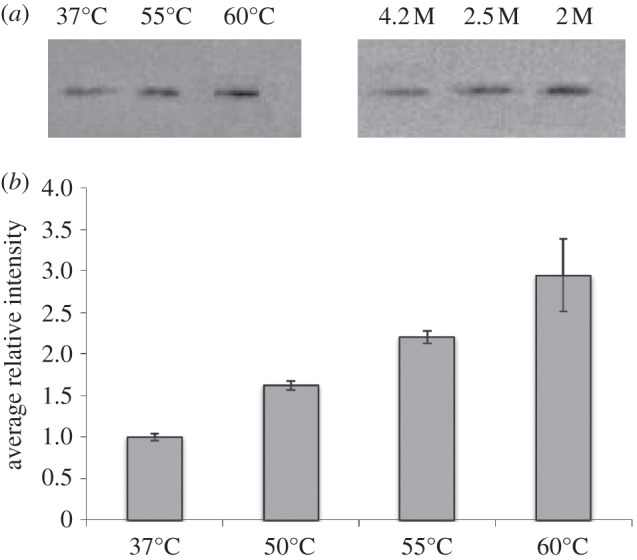
Modification of the thermosome accumulation levels in Halobacterium cells exposed to high temperature. Total protein extracts were analysed by western blots. (a) Immunodetection of the thermosome alpha subunit after 1 h of temperature stress. (b) Histograms showing the thermosome subunit accumulation after 24 h of neutron experiments performed with different heat-stressed samples. The temperatures values indicated below the histograms correspond to those applied to the Halobacterium cultures, during 1 h, prior to the 24 h neutron experiments. The band intensities were normalized as a function of the total protein amount determined by Bradford assays.
2.2. Neutrons detect protein denaturation in vivo
The energy and momentum changes measured in a neutron spectroscopy experiment are related to the time-scale and amplitude of atomic motions, respectively. In order to assess whether the effects of thermal stress on the in vivo molecular dynamics state could be detected, we performed neutron scattering experiments on unstressed and stressed Hs cells by using the IN13 spectrometer at the Institut Laue Langevin, as described in §3. In a complex system, different motion populations with different MSDs are observed, depending on the length and time scales. Because of this polydispersity, the most reliable analysis is via comparison of observations on the same system under different conditions on the same time and length scales. The time scale sampled on IN13 is approximately 0.1 ns, defined by the energy resolution of the instrument (8 µeV). The length scale is defined by the Q (scattering vector or momentum change modulus) range, which is quite broad on IN13: from below 0.3 Å−1 to above 6 Å−1, giving effective access to a length scale extending from a few ångströms at low Q to a fraction of an ångström at the high Q end. The most efficient and arguably most useful neutron spectroscopy method to measure dynamics is by analysis of the temperature-dependent elastic incoherent neutron scattering (EINS), in which the momentum change of elastically scattered neutrons is measured for different temperatures [1]. A Gaussian approximation analysis (see §3) provides the MSD of atomic motions as a function of temperature, in a given time and length scale. An effective force constant for the motions (resilience,  ) (N m–1) is calculated from the temperature dependence [5]. The condition for the Gaussian approximation to be valid is that the amplitude of the MSD is well contained within the length–time window defined by the energy resolution and Q range of the spectrometer. In practice, this means that the analysis is limited to the low Q end of the range on IN13, where the length window is sufficiently wide to accommodate MSD of a few ångströms. Hydrogen atoms dominate EINS, because of a scattering cross section that is more than an order of magnitude higher than for other atoms. In the length and time scales considered, hydrogen atoms in amino acid residues in a protein, for example, move with the groups to which they are bound and reflect well macromolecular internal motions as well as the larger fluctuation amplitudes involved in unfolding processes. In a complex system, hydrogen atoms in different groups have different motion amplitudes, so that the MSD is exactly that: the MSD of all hydrogen atoms. In the case of bacterial cells, the MSD was estimated to be dominated by hydrogen displacements in the proteome [7].
) (N m–1) is calculated from the temperature dependence [5]. The condition for the Gaussian approximation to be valid is that the amplitude of the MSD is well contained within the length–time window defined by the energy resolution and Q range of the spectrometer. In practice, this means that the analysis is limited to the low Q end of the range on IN13, where the length window is sufficiently wide to accommodate MSD of a few ångströms. Hydrogen atoms dominate EINS, because of a scattering cross section that is more than an order of magnitude higher than for other atoms. In the length and time scales considered, hydrogen atoms in amino acid residues in a protein, for example, move with the groups to which they are bound and reflect well macromolecular internal motions as well as the larger fluctuation amplitudes involved in unfolding processes. In a complex system, hydrogen atoms in different groups have different motion amplitudes, so that the MSD is exactly that: the MSD of all hydrogen atoms. In the case of bacterial cells, the MSD was estimated to be dominated by hydrogen displacements in the proteome [7].
2.3. Effects of temperature stress on the molecular dynamics state of the Hs proteome
The Gaussian approximation may be valid in different Q ranges to yield different MSD values, depending on the dominant amplitudes in the corresponding length scales. See, for example, the dynamics analysis of bacteriorhodopsin as a function of hydration in purple membranes of Hs [16]. In this study, two motion populations were identified, a hydration-dependent large amplitude population at low Q (attributed to residues in outer loops of bacteriorhodopsin) and a small amplitude population at higher Q (attributed to vibrational internal motions of the residues in the membrane inserted helices). A low Q range (0.5 Å−1 ≤ Q ≤ 1.5 Å−1) was chosen in the current analysis (full double arrow in figure 3). It is sensitive to higher amplitude MSD values that would result, for example, from macromolecular unfolding processes in the stressed cells. Contributions from internal vibrational motions appeared in a higher Q range. Interestingly, no differences in NSD were detected between stressed and unstressed samples, suggesting that vibrational internal motions were less affected by the stress.
Figure 3.
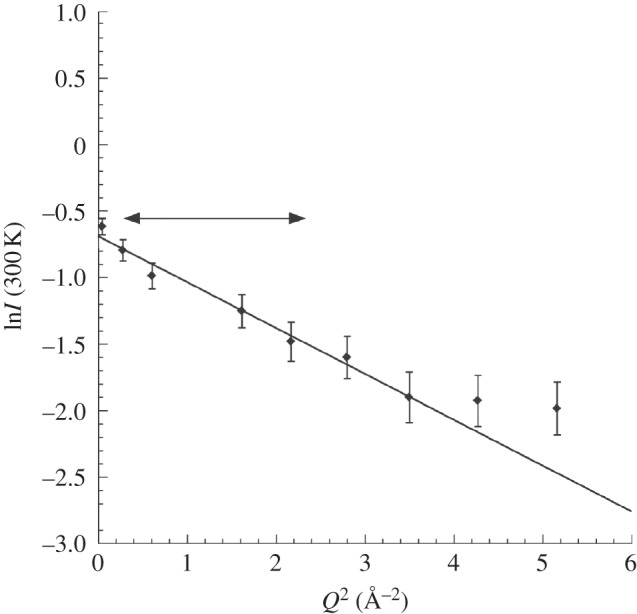
Natural logarithm of the intensity versus Q2 for unstressed Hs at 300 K, measured on IN13 with an energy resolution of 8 µeV (see text). The double arrow indicates the Q range used in the analysis, in which significant molecular dynamics perturbations were observed between stressed and unstressed cells.
A clear increase in macromolecular flexibility was detected for the two heat-stressed Hs cellular samples: the MSD values for the 55°C and 60°C stressed samples were shifted upwards progressively, compared with the unstressed control (figure 4a). The effective force constant  expressing macromolecular resilience was also calculated from the slope of the MSD temperature dependence as described in [6] (figure 4b). The
expressing macromolecular resilience was also calculated from the slope of the MSD temperature dependence as described in [6] (figure 4b). The 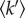 values indicated less rigid macromolecular states, which is likely to reflect perturbation in the folding state of a significant part of the Hs proteome. Unfolded proteins have been measured to display larger fluctuations and lower resilience than folded states [17]. EINS studies on purified enzymes have shown that increased flexibility corresponds to the thermal unfolded state of the proteins. Russo et al. [18] measured major losses in rigidity associated with the thermal unfolding of a small beta protein and Koutsopoulos et al. [19] showed a clear correlation between the heat denaturation of an endoglucanase from Pyrococcus furiosus and increased atomic fluctuation associated with lower resilience values [18,19]. Unfolded proteins in the cell may aggregate. However, on the time scale of internal motions, it is not unlikely that such aggregates display a lower rigidity compared with the folded state, which has a compact core [3]. Still, more experimental data on isolated proteins or cell extracts would be required to support this hypothesis. From these studies, it can be inferred that the increased MSD and lower resilience that we observed in vivo for the temperature-stressed cells correspond to proteome unfolding. The neutron experiments provided therefore a direct measurement of the thermal stress effect on the protein dynamics in the cytosolic context of an intact cell.
values indicated less rigid macromolecular states, which is likely to reflect perturbation in the folding state of a significant part of the Hs proteome. Unfolded proteins have been measured to display larger fluctuations and lower resilience than folded states [17]. EINS studies on purified enzymes have shown that increased flexibility corresponds to the thermal unfolded state of the proteins. Russo et al. [18] measured major losses in rigidity associated with the thermal unfolding of a small beta protein and Koutsopoulos et al. [19] showed a clear correlation between the heat denaturation of an endoglucanase from Pyrococcus furiosus and increased atomic fluctuation associated with lower resilience values [18,19]. Unfolded proteins in the cell may aggregate. However, on the time scale of internal motions, it is not unlikely that such aggregates display a lower rigidity compared with the folded state, which has a compact core [3]. Still, more experimental data on isolated proteins or cell extracts would be required to support this hypothesis. From these studies, it can be inferred that the increased MSD and lower resilience that we observed in vivo for the temperature-stressed cells correspond to proteome unfolding. The neutron experiments provided therefore a direct measurement of the thermal stress effect on the protein dynamics in the cytosolic context of an intact cell.
Figure 4.
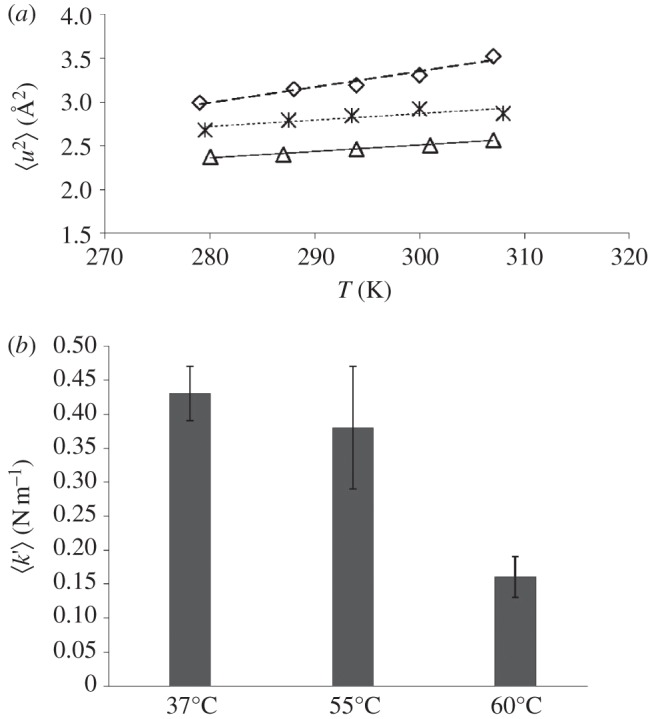
Dynamic response in live Hs to thermal stress. (a) Mean square amplitudes 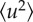 for large amplitude fluctuations plotted against temperature T (triangles) for unstressed cells or (asterisks and diamonds) cells that have been transferred at 55°C or 60°C during 1 h prior to neutron experiments. (b) Histograms of the effective mean force constants
for large amplitude fluctuations plotted against temperature T (triangles) for unstressed cells or (asterisks and diamonds) cells that have been transferred at 55°C or 60°C during 1 h prior to neutron experiments. (b) Histograms of the effective mean force constants 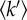 from different cell samples. The temperatures values indicated below the histograms correspond to those applied to the Halobacterium cultures, during 1 h, prior to the 24 h neutron experiments.
from different cell samples. The temperatures values indicated below the histograms correspond to those applied to the Halobacterium cultures, during 1 h, prior to the 24 h neutron experiments.
2.4. Conclusions
The observations reported in this paper represent a direct insight into the folded state of a proteome exposed to temperature stress conditions. They reveal that neutron spectroscopy can detect alterations in the mean molecular dynamics state of the proteome within a living cell in response to environmental changes. Despite the crowded intracellular environment and the induction of protein quality control systems, the dynamic state of a large fraction of the proteome is strongly perturbed under thermal stress. Interestingly, the cells could easily recover when replaced in optimal growth conditions. The experiments demonstrated the sensitivity of the neutron scattering method to probe in vivo the molecular dynamic alterations related to stress response.
3. Material and methods
3.1. Halobacterium salinarum cell culture and growth conditions
Hs R1 cultures were grown at 37°C, with shaking at 150 r.p.m., in standard medium containing 4.2 M NaCl [10]. Cells were grown to mid-log phase (optical density at 600 nm (OD600) = 0.8). For thermal stress, the cultures were transferred at the desired temperatures and cultivated under agitation for 1 h.
3.2. Sample preparation, neutron scattering experiments and analysis
For each sample, 2 l of Hs cultures were centrifuged at 4000g. The cell pellets were gently suspended with a brush in 500 ml of washing buffer containing 50 mM Tris–HCl pH 7.6; 4.2 M NaCl; 70 mM KCl and 80 mM MgSO4. The operation was repeated twice. A total of 540 mg of cell paste was taken from the pellet with a spatula to fill up completely a 0.3 mm path length gold-plated aluminium sample holders. The device was hermetically closed with an indium seal and mounted on the IN13 back spectrometer at the Institut Laue Langevin (http://www.ill.eu/html/instruments-support/instruments-groups/instruments/in13/).
Experiments on the cells were performed in H2O. The energy resolution and Q range of a neutron spectrometer define a window on a length and time scales. Motions confined within the window will contribute to observable elastic scattering (i.e. scattering without energy change) [1]. On IN13, the window corresponds to an MSD of approximately 2 Å2 in 0.100 ns. Internal and unfolding motions are within the window. Bulk water diffusion, however, covers approximately 25 Å2 in 0.100 ns, well outside the window [1]. IN13 data, therefore, are poorly sensitive to H2O diffusion. This was an important observation that made possible the comparison of protein dynamics in H2O and D2O, which revealed a significant solvent isotope effect on dynamics [20].
All measurements were repeated on samples from at least three different cultures. The IN13 spectrometer has an energy resolution of 8 μeV and is sensitive to motions that occur on a time scale up to about 0.1 ns. The EINS signal was measured and analysed over the scattering range 0.5 Å−1 < Q < 1.5 Å−1 (Q = 4π sinθ/λ for elastic scattering, where 2θ is the scattering angle and λ is the incident neutron wavelength). The elastic intensities were corrected for sample holder and buffer scattering, normalized to the scattering of vanadium (purely incoherent scattering) and corrected for sample absorption by using the ILL data reduction program LAMP (information on the program is available on the ILL website at http://www.ill.fr). For each sample, the elastic intensity I(Q) was obtained as a function of temperature, T, rising from 280 to 310 K, and lnI(Q) was plotted against Q2. The MSD of the scattering nuclei, 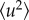 , was calculated from the slope of the straight-line fit to the experimental data according to the Gaussian approximation that is valid for
, was calculated from the slope of the straight-line fit to the experimental data according to the Gaussian approximation that is valid for 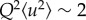 [5]:
[5]:
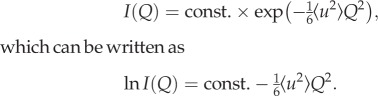 |
3.1 |
The value of the root MSD quantifies the global flexibility.
The mean resilience, 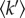 , was extracted from the slope of
, was extracted from the slope of 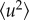 as a function of temperature T, using the following relation:
as a function of temperature T, using the following relation:
| 3.2 |
where 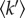 is in N m–1,
is in N m–1, 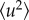 is in Å2 and T is in degrees K.
is in Å2 and T is in degrees K.
The value of 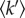 corresponds to a mean effective force constant and defines the average atomic resilience in a free energy potential [5,21].
corresponds to a mean effective force constant and defines the average atomic resilience in a free energy potential [5,21].
3.3. Viability assays
Serial dilutions were performed on cultures and resuspended cell paste in order to examine the influence of temperature on the Hs intracellular dynamics. Viability tests were performed before and after stress exposure, and before and after the 24 h neutron experiments. A Neubauer hemocytometer was used for cell count, and the LIVE/DEAD BacLight bacterial viability kit (Invitrogen detection technologies) was used to assess the mortality rate in the different physiological conditions as described [22]. Suspensions of Hs (4 × 108 cells ml−1) were pre-stained with LIVE/DEAD kit and pictures were taken with an epifluorescence microscope (IX 81 Olympus) coupled to a QImaging Retiga-SRV CCD digital camera. Fluorescence and contrast images were analysed using Volocity (Perkin Elmer). Survival was calculated as the number of viable cells following treatment divided by the number of viable untreated cells.
3.4. ICP measurement of intracellular K+ concentration
The K+ concentration in Hs cells was determined using induced coupled plasma spectrometry (ICP). After stress and neutron experiments, 1 ml of cell suspension was centrifuged. The pellets were washed with K+ free isotonic buffers and the cells were lysed by sonication in 10 ml of H2O. Analyses were performed with an ICP-AES Perkin Elmer, Optima 3300 DV plasma-mass spectrometer at the Equipe Géochimie de l'Environnement, LGIT/CNRS in Grenoble, France. A calibration curve was generated from KCl solutions. The total cell number and the average cell volume were determined in each sample by confocal microscopy. These values were used to calculate the intracellular K+ concentration. All measurements were performed in triplicate on at least three different experiments.
3.5. Protein immunodetection
One millilitre of OD 0.8 Halobacterium culture was pelleted and resuspended in 0.2 ml of distilled water. The cells were lysed and homogenized by flushing several times through a needle (diameter 0.6 mm) connected to a syringe. Cell debris was eliminated by centrifugation at 13000 r.p.m. for 10 min. The total protein amount in the supernatant was determined by using a Bradford quantification assay (BioRad). An equal volume of sample buffer was added to the extract. After boiling at 90°C for 5 min, 15 µl of this sample was subjected to 12 per cent SDS-polyacrylamide gels and the resolved proteins were transferred to nitrocellulose membranes (Hybond-P. GE healthcare). The blots were probed with anti-TF55 and anti MalDH antibodies obtained as described in [23]. All polysera were used at a 1 : 10 000 dilution. Immunoreactive bands were visualized by chemiluminescence according to the supplier's protocol (ECL detection kit; GE healthcare). The band intensities were measured by using the ImageJ software and were normalized by the total protein amount. The mean intensity values obtained for the 37°C unstressed condition were arbitrary fixed to ‘1’ (figure 2).
Acknowledgements
This work was funded by grants form the CNRS interdisciplinary program 'Environnement Planétaire et Origine du Vivant’ (EPOV) and ESF/Eurocore/EURODEEP. V.M. was supported by a PhD grant from the French ministry for Research and Technology. We thank Françoise Lacroix and Jean-Philippe Kleman (Institut de Biologie Structurale, Grenoble) for the support and access to the Cell Imaging Platform.
References
- 1.Zaccai G. 2011. Neutron scattering perspectives for protein dynamics. J. Non Cryst. Solids 357, 615–621 10.1016/j.jnoncrysol.2010.06.060 (doi:10.1016/j.jnoncrysol.2010.06.060) [DOI] [Google Scholar]
- 2.Zaccai G. In press The ecology of protein dynamics. Curr. Phys. Chem. 3 [Google Scholar]
- 3.Cino EA, Karttunen M, Choy W-Y. 2012. Effects of molecular crowding on the dynamics of intrinsically disordered proteins. PLoS ONE 7, e49876. 10.1371/journal.pone.0049876 (doi:10.1371/journal.pone.0049876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarek M, Tobias DJ. 2002. Role of protein-hydrogen bond dynamics in the protein dynamical transition. Phys. Rev. Lett. 88, 138101. 10.1103/PhysRevLett.88.138101 (doi:10.1103/PhysRevLett.88.138101) [DOI] [PubMed] [Google Scholar]
- 5.Zaccai G. 2000. How soft is a protein? A protein dynamics force constant measured by neutron scattering. Science 288, 1604–1607 10.1126/science.288.5471.1604 (doi:10.1126/science.288.5471.1604) [DOI] [PubMed] [Google Scholar]
- 6.Jasnin M, Moulin M, Haertlein M, Zaccai G, Tehei M. 2008. In vivo measurement of internal and global macromolecular motions in Escherichia coli. Biophys. J. 95, 857–864 10.1529/biophysj.107.124420 (doi:10.1529/biophysj.107.124420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tehei M, Franzetti B, Madern D, Ginzburg M, Ginzburg BZ, Giudici-Orticoni M, Bruschi M, Zaccai G. 2004. Adaptation to extreme environments: macromolecular dynamics in bacteria compared in vivo by neutron scattering. EMBO Rep. 5, 66–70 10.1038/sj.embor.7400049 (doi:10.1038/sj.embor.7400049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadler AM, van Eijck L, Demmel F, Artmann G. 2011. Macromolecular dynamics in red blood cells investigated using neutron spectroscopy. J. R. Soc. Interface 8, 590–600 10.1098/rsif.2010.0306 (doi:10.1098/rsif.2010.0306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamieh H, Marty V, Guetta D, Perollier A, Franzetti B. 2012. Stress regulation of the PAN-proteasome system in the extreme halophilic archaeon Halobacterium. Extremophiles 16, 215–225 10.1007/s00792-011-0421-0 (doi:10.1007/s00792-011-0421-0) [DOI] [PubMed] [Google Scholar]
- 10.Oesterhelt D, Stoeckenius W. 1974. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 31, 667–678 [DOI] [PubMed] [Google Scholar]
- 11.Ginzburg M, Sachs L, Ginzburg BZ. 1970. Ion metabolism in a Halobacterium. I. Influence of age of culture on intracellular concentrations. J. Gen. Physiol. 55, 187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11, 1641–1650 10.1101/gr.190201 (doi:10.1101/gr.190201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madern D, Ebel C, Zaccai G. 2000. Halophilic adaptation of enzymes. Extremophiles 4, 91–98 10.1007/s007920050142 (doi:10.1007/s007920050142) [DOI] [PubMed] [Google Scholar]
- 14.Klumpp M, Baumeister W. 1998. The thermosome: archetype of group II chaperonins. FEBS Lett. 430, 73–77 10.1016/S0014-5793(98)00541-9 (doi:10.1016/S0014-5793(98)00541-9) [DOI] [PubMed] [Google Scholar]
- 15.Lund P. 2011. Insights into chaperonin function from studies on archaeal thermosomes. Biochem. Soc. Trans. 39, 94–98 10.1042/BST0390094 (doi:10.1042/BST0390094) [DOI] [PubMed] [Google Scholar]
- 16.Lehnert U, Reat V, Weik M, Zaccai G, Pfister C. 1998. Thermal motions in bacteriorhodopsin at different hydration levels studied by neutron scattering: correlation with kinetics and light-induced conformational changes. Biophys. J. 75, 1945–1952 10.1016/S0006-3495(98)77635-0 (doi:10.1016/S0006-3495(98)77635-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabiani E, Stadler AM, Madern D, Koza MM, Tehei M, Hirai M, Zaccai G. 2009. Dynamics of apomyoglobin in the alpha-to-beta transition and of partially unfolded aggregated protein. Eur. Biophys. J. 38, 237–244 10.1007/s00249-008-0375-z (doi:10.1007/s00249-008-0375-z) [DOI] [PubMed] [Google Scholar]
- 18.Russo D, Perez J, Zanotti JM, Desmadril M, Durand D. 2002. Dynamic transition associated with the thermal denaturation of a small beta protein. Biophys. J. 83, 2792–2800 10.1016/S0006-3495(02)75288-0 (doi:10.1016/S0006-3495(02)75288-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koutsopoulos S, van der Oost J, Norde W. 2005. Temperature-dependent structural and functional features of a hyperthermostable enzyme using elastic neutron scattering. Proteins 61, 377–384 10.1002/prot.20606 (doi:10.1002/prot.20606) [DOI] [PubMed] [Google Scholar]
- 20.Tehei M, Madern D, Pfister C, Zaccai G. 2001. Fast dynamics of halophilic malate dehydrogenase and BSA measured by neutron scattering under various solvent conditions influencing protein stability. Proc. Natl Acad. Sci. USA 98, 14 356–14 361 10.1073/pnas.251537298 (doi:10.1073/pnas.251537298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bicout DJ, Zaccai G. 2001. Protein flexibility from the dynamical transition: a force constant analysis. Biophys. J. 80, 1115–1123 10.1016/S0006-3495(01)76089-4 (doi:10.1016/S0006-3495(01)76089-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leuko S, Legat A, Fendrihan S, Stan-Lotter H. 2004. Evaluation of the LIVE/DEAD BacLight kit for detection of extremophilic archaea and visualization of microorganisms in environmental hypersaline samples. Appl. Environ. Microbiol. 70, 6884–6886 10.1128/AEM.70.11.6884-6886.2004 (doi:10.1128/AEM.70.11.6884-6886.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamieh H, Guetta D, Franzetti B. 2008. The two PAN ATPases from Halobacterium display N-terminal heterogeneity and form labile complexes with the 20S proteasome. Biochem. J. 411, 387. 10.1042/BJ20071502 (doi:10.1042/BJ20071502) [DOI] [PubMed] [Google Scholar]