Summary
How inflammation causes cancer is unclear. IL-15 is a pro-inflammatory cytokine elevated in human large granular lymphocyte (LGL) leukemia. Mice overexpressing IL-15 develop LGL leukemia. Here we show that prolonged in vitro exposure of wild type (WT) LGL to IL-15 results in Myc-mediated up regulation of aurora kinases, centrosome aberrancies, and aneuploidy. Simultaneously, IL-15 represses miR-29b via induction of Myc/NF-κBp65/Hdac-1, resulting in Dnmt3b overexpression and DNA hypermethylation. All this is validated in human LGL leukemia. Strikingly, adoptive transfer of WT LGL cultured with IL-15 led to malignant transformation in vivo. Drug targeting which reverses miR-29b repression, cures otherwise fatal LGL leukemia. We show how excessive IL-15 initiates cancer and demonstrate effective drug targeting for potential therapy of human LGL leukemia.
Introduction
There is now strong epidemiologic data to support the notion that chronic inflammation can increase the risk for malignant transformation of otherwise normal host cells (Ames et al., 1995). Deregulated cytokine production in the context of persistent infections can stimulate cells expressing cognate cytokine receptors to alter cell growth, cell differentiation and cell survival, putting a cell at increased risk for malignant transformation when exposed to DNA damaging agents (Dranoff, 2004). For example, overexpression of macrophage-migration inhibitory factor in the setting of chronic inflammation can lead to the functional inactivation of the tumor suppressor gene p53 (Hudson et al., 1999), and overexpression of IL-1 is associated with an increased risk of developing gastric cancer (El-Omar et al., 2000). While the alteration of cytokine gene expression has been shown to alter the risk of malignant transformation, cytokines have only rarely been directly implicated as a cause of cancer, and to date, mechanistic insights into this process have not been understood.
IL-15 is a pro-inflammatory cytokine, which is required for the genesis and homeostasis of natural killer (NK) cells or large granular lymphocytes (LGL) (Caligiuri, 2008). IL-15 utilizes the β (CD122) and γ (CD132) chains of the IL-2 receptor to transmit its growth and activation signals in LGL, yet presents to the βγ receptor complex in trans via its binding to a high affinity IL-15 receptor α chain (Dubois et al., 2002). IL-15 was first shown to be overexpressed in HTLV-1-associated human T cell leukemia, and interruption of its autocrine loop with an anti-CD122 mab prevented leukemic cell growth and induced leukemic cell death in vitro (Bamford et al., 1994; Grabstein et al., 1994) but not in vivo (Morris et al., 2006). Overexpression of murine IL-15 causes LGL leukemia with either a NK cell or TNK cell phenotype in IL-15 transgenic (Tg) mice (Fehniger et al., 2001), and mice overexpressing mutated HMGI-C express excessive IL-15 that causes NK lymphoma (Baldassarre et al., 2001). Interestingly, IL-15 has been reported to be overexpressed in human LGL leukemia and to date, most human cell lines isolated from patients with LGL leukemia are dependent on IL-2 or IL-15 for in vitro propagation (Zambello et al., 1997). Collectively, these experimental and clinical data suggest a central role for IL-15 in the genesis of LGL leukemia, a highly malignant and uniformly fatal disorder, yet the mechanism by which this cytokine induces malignant transformation of LGL is not known. In the current study, we attempt to unravel the mechanism of IL-15 induced LGL leukemia in mouse and man.
Results
Chronic in vitro exposure of normal LGLs to IL-15 results in leukemic transformation
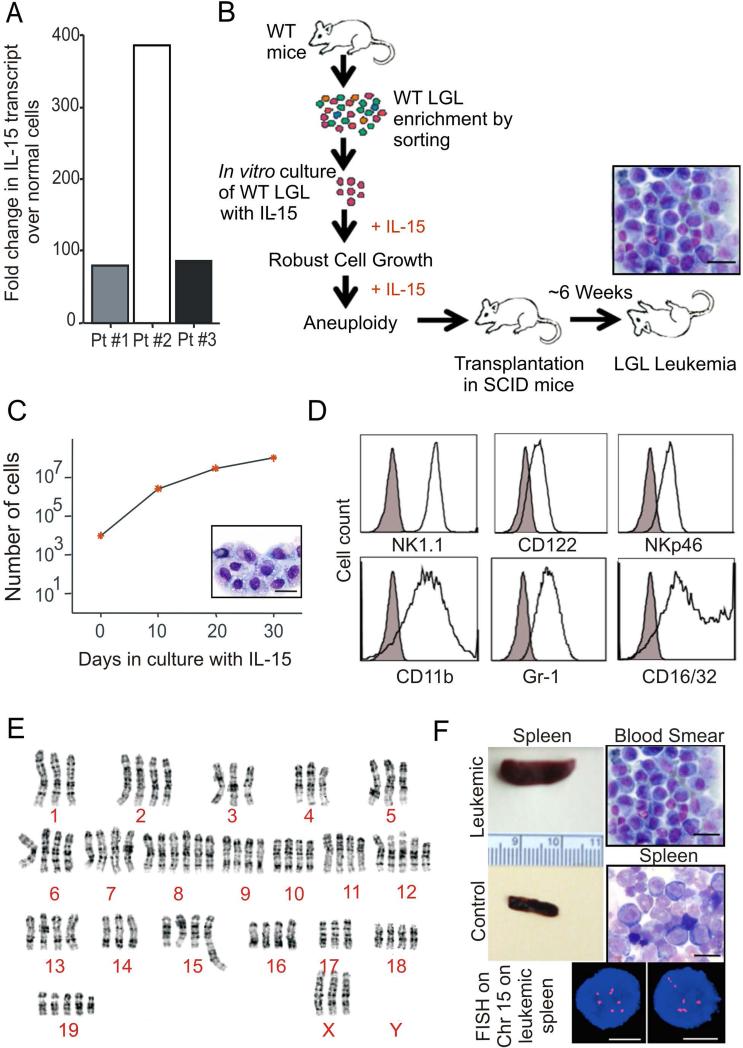
Since over expression of IL-15 as a single growth factor can initiate leukemic transformation of LGL in vivo, (Fehniger et al., 2001) and the neoplastic cells isolated from patients with LGL leukemia have higher expression of IL-15 than their normal LGL counterparts (Figure 1A), we first assessed the effects of IL-15 on wild type (WT) mouse LGL (> 95% CD3-NK1.1+) to determine if this transformation could occur in vitro (Figure 1B). Culture of 1 × 105 WT LGL with IL-15 at a concentration sufficient to saturate its cognate dimeric receptor induced robust growth of the LGL which has continued for over 18 months while maintaining the LGL phenotype (Figure 1C and 1D). At six months a karyotype was performed demonstrating striking aneuploidy (Figure 1E). While withdrawal of IL-15 led to eventual apoptosis of LGL in vitro (data not shown), adoptive transfer into SCID mice without exogenous IL-15 resulted in a dramatic increase in white blood cell (WBC) count (4.2 × 107/ml), splenomegaly and death from fatal leukemia in vivo (Figure 1B and 1F). Interphase fluorescence in situ hybridization (FISH) analysis of splenocytes from the leukemic mouse revealed multiple copies of chromosome 15 (Figure 1F) as was seen in the in vitro culture prior to adoptive transfer (Figure 1E), and in our original IL-15 Tg mice (Yokohama et al., 2010). Thus, chronic exposure of WT LGL to IL-15 alone contributes to robust growth and chromosomal instability (CIN) in vitro, and leukemic transformation in vivo.
Figure 1. Chronic exposure to IL-15 initiates robust expansion and chromosomal instability in WT LGL.
(A) Fold changes in expression of IL-15 mRNA in human LGL leukemia samples from three patients (Pt), normalized to 18S mRNA and then quantified relative to values of IL-15 measured in normal donors cells that were enriched for either CD56+ or CD8+ (n=4 each) and then arbitrarily set at 1. (B) Schema for the generation of in vitro robust expansion of WT LGL during culture in IL-15, and adoptive transfer followed by malignant transformation in vivo in the absence of exogenous IL-15. Scale bars, 10 μm (C) WT splenic NK1.1+ cells were sorted and incubated in triplicate with 100ng/ml rhIL-15. Cell growth was quantified as absolute number of cells (mean ± SEM) using enumeration in trypan blue exclusion dye. Wright-Giemsa stain of the in vitro cultured cells. Scale bars, 10 μm. (D) WT LGL were analyzed for their immuno-phenotype by FACS after six months of in vitro culture with IL-15. (E) Karyotype analysis was performed on metaphase spreads from WT LGL cultured in IL-15 for approximately six months, demonstrating marked aneuploidy. Composite karyotype of the in vitro transformed WT LGL cells was: 76-79<4n>,XXX,-X,-1,-3,del(4)(A1A2),-4,-5,+8,+8,+12,del(12)(A1.2B),-14,der(15)t(5;15)(B1;F2),-17,+19,+mar[cp7]/76 79,idem,+6[cp10]/76-79,idem,+19[cp3]. (F) ICR-SCID mice were intravenously injected with 1 × 107 WT LGL following approximately eight months of in vitro culture with IL-15. Splenomegaly (compared to WT control as shown) and the presence of neoplastic LGL, both at the feathered edge of peripheral blood smear and on the spleen cytospin preparation, are shown. Scale bars, 10 μm. A representative FISH image illustrating gain of several copies chromosome 15 (red, chromosome 15 probe; blue, DAPI counter-stain) as seen in WT LGL cultured in vitro for months in IL-15 (E) is also shown. Scale bars, 5 μm.
IL-15 induces centrosome aberrations in normal LGL
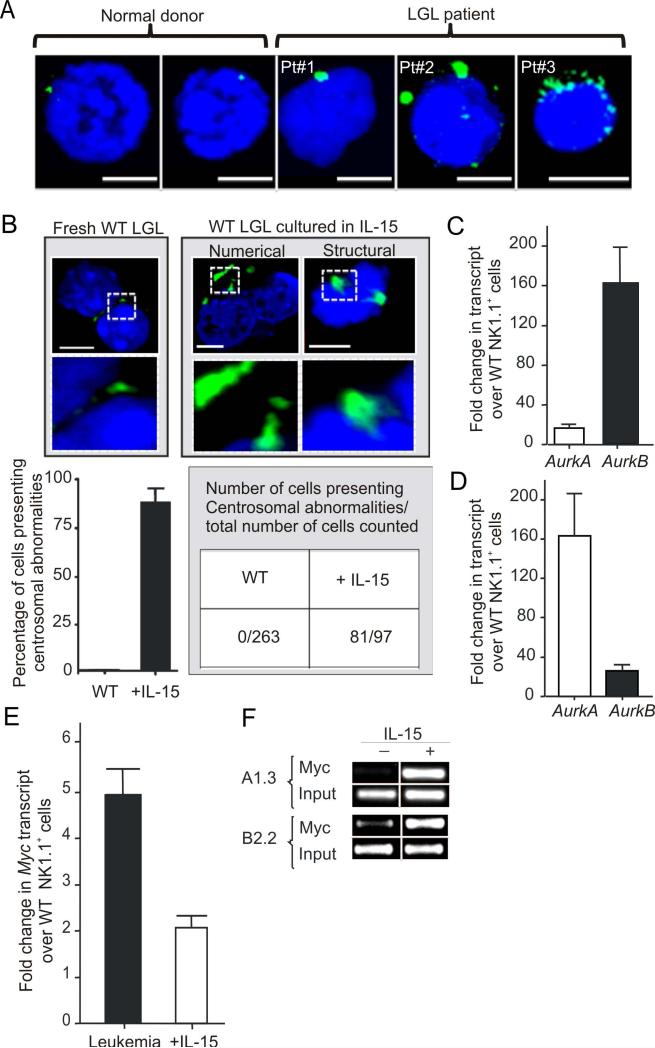
One proposed mechanism underlying CIN is centrosome mediated asymmetrical chromosome segregation resulting in aneuploidy (Ganem et al., 2009). Indeed we found numerical and structural centrosomal aberrations in primary cells obtained from patients with aggressive LGL leukemia (Figure 2A). We next assessed WT mouse LGL cultured for several months in IL-15 for centrosome aberrancies and compared them to fresh WT LGL. We found that the vast majority (81 of 97) of IL-15 cultured LGL counted had significant increases in centrosome numbers and/or size, while no such changes were noted in 263 fresh WT LGL (Figure 2B). Since altered regulation of centrosome replication can be caused by excessive aurora kinases encoded by AurkA and AurkB (Giet et al., 2005), we assessed the expression of these transcripts in LGL leukemic blasts from IL-15 Tg leukemic mice and found each to be significantly elevated in comparison to WT LGL from age matched WT mice, although AurkB was significantly higher than AurkA (Figure 2C). Similarly, WT LGL exposed to IL-15 for only 30 consecutive days also showed higher transcript levels of AurkA and AurkB compared to the fresh LGL, although AurkA was significantly higher than AurkB (Figure 2D). Interestingly, overexpression of AurkA can transform rodent fibroblasts while AurkB cannot (Bischoff et al., 1998; Kanda et al., 2005; Zhou et al., 1998). This may explain the relative abundance of AurkA earlier in this process. Indeed, we show that forced overexpression of AurkA in WT mouse LGL exposed to IL-15 in a short term culture resulted in quantitative and qualitative centrosome abnormalities as well as enhanced transformation in vitro (Figure S1A-B).
Figure 2. IL-15 induces centrosome aberration in normal LGLs.
(A) The size, structure and number of human centrosomes was determined by confocal microscopy and immunoflorescence staining with Pericentrin (green) in both freshly isolated normal human CD56+ or CD8+ LGL (top left) as well as in malignant cells from three patients with acute LGL leukemia. Cells were counterstained with DAPI (blue) for nuclear staining. Images shown are representative of slides with ~105 cells. Scale bars, 5 μm. (B) The size, structure and number of mouse centrosomes was determined by confocal microscopy and immunoflorescence staining with GTU88 (green) in both freshly isolated WT LGL (top left) as well as in WT LGL cultured in IL-15 for 6 months (top middle and right). Cells were counterstained with DAPI (blue) for nuclear staining. Images shown here are representative of 3 different slide preparation with ~105 cells. Graphical quantification of these centrosomal abnormalities is shown immediately below as percentage of cells (mean ± SEM) presenting centrosomal abnormalities from 3 independent slide preparations. Scale bars, 5 μm. (C) Relative fold changes (mean ± SEM) in mRNA expression of AurkA and AurkB in splenocytes from LGL leukemic mice relative to values measured in fresh WT LGL. Quantification was by Real-time RT-PCR (n = 3 each). Each measurement was normalized against the level of 18S mRNA, and then values for AurkA and AurkB in fresh WT LGL were arbitrarily set at 1. (D) Relative fold changes (mean ± SEM) in mRNA expression of AurkA and AurkB for WT LGL cultured in IL-15 for 30 days, relative again to values measured in fresh WT LGL which are arbitrarily set at 1 (n = 3 each). (E) Relative fold changes (mean ± SEM) in mRNA expression of Myc in splenotyces from LGL leukemic mice (n = 3), and in WT mouse LGL cultured in IL-15 for 30 days. Both measurements are relative to values of Myc measured in fresh WT LGL that are arbitrarily set at 1. (F) WT LGL were grown in vitro with IL-15 for approximately 6 months, and then harvested and starved for 24 hours and then divided up to be re-stimulated with either IL-15 or PBS for 4 hours. ChIP assay was performed using an anti-Myc antibody and PCR primers amplifying the AurkA and AurkB promoter 5’ regulatory regions.
See also Figure S1.
Since AurkA and AurkB are regulated by Myc (den Hollander et al., 2010), we measured and found increased transcript levels of Myc in both leukemic blasts as well as in the WT LGL cultured with IL-15 for 12 hours and for 30 days, and confirmed this at the protein level for both WT mouse and normal human LGL (Figures 2E, S1C and S1D). This elevation of AURKA, AURKB and MYC expression was confirmed in primary human LGL leukemia samples (Figure S1E). Chromatin immmuo-precipitation (ChIP) assays performed on WT mouse LGL which had been cultured in IL-15 for six months and then starved of IL-15 for 24 hours followed by re-stimulation with IL-15 or PBS for four hours demonstrated an increased binding of Myc within promoter regions of AurkA and AurkB (Figure 2F). Specific reduction of Myc by shRNA in IL-15-activated WT mouse LGL drastically reduced AurkA and AurkB expression (not shown). These data suggest that IL-15-mediated induction of Myc can lead to CIN in part via overexpression of AurkA and AurkB.
Using a luciferase construct, we next demonstrated that IL-15 induces Myc at least in part via activation of NF-kBp65 (Figure S1F). By EMSA, we showed binding at the putative NF-κB binding sites within the Myc promoter after 1 hour and 12 hours of exposure to IL-15 in both mouse (Figure S1G, left) and human LGL (Figure S1I, left). Supershift assay documented NF-κBp65 and p50 binding to the Myc promoter following LGL cell stimulation by IL-15 for 12 hours in both mouse (Figure S1G, right) and human LGL (Figure S1I, right). Finally, utilizing a ChIP assay, we demonstrated an enrichment of NF-κBp65 at the consensus binding site of the mouse Myc and human MYC promoter following LGL cell stimulation by IL-15 for 4 hours (Figure S1H and S1J). Thus, IL-15-mediated induction of Myc in normal LGL occurs, at least in part, via the activation of NF-κB.
IL-15 increases global DNA methylation levels in LGL
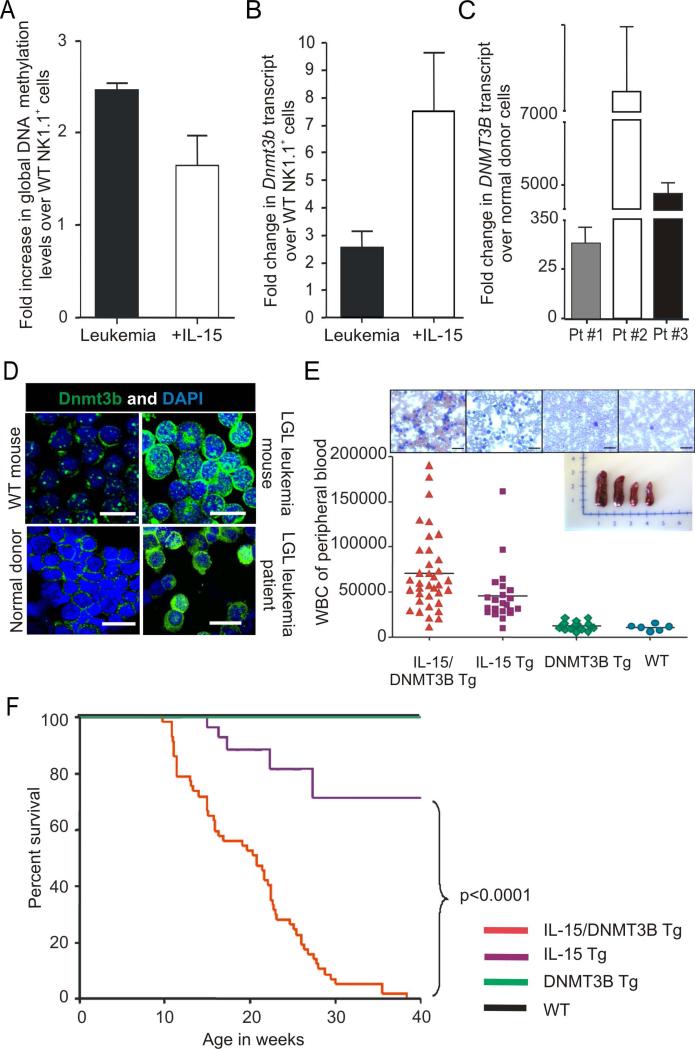
Hypermethylation of tumor suppressor and “stability” genes can also contribute to CIN (Esteller, 2006; Esteller, 2007), so we also pursued this in our mouse model. We previously showed that leukemic blasts from IL-15 Tg mice have an increase in methylation within the 5’ regulatory region of genes (Yu et al., 2005). We confirmed an increase in global DNA methylation (GDM) within LGL leukemic blasts from IL-15 Tg mice compared to age matched WT LGL controls. We also measured a GDM increase in WT mouse LGL cultured with IL-15 for 30 days, suggesting that indeed IL-15 is inducing methylation prior to leukemic transformation (Figure 3A). A similar increase in GDM was seen in our primary human LGL leukemia samples (Figure S2A). We investigated whether changes in the DNA methyltransferase level is a cause for IL-15-mediated hypermethylation by measuring the mRNA levels of Dnmt1, Dnmt3a and Dnmt3b in LGL leukemic blasts from IL-15 Tg mice and in WT LGL cultured with IL-15. Only Dnmt3b (mRNA and protein) was consistently elevated in both populations (Figure 3B and 3D). Further, only the expression of DNMT3B was consistently elevated in primary leukemic blasts from patients with LGL leukemia (Figure 3C-D), and was abundant in the nucleus of normal human LGL stimulated with IL-15 (Figure S2B). These data suggest that IL-15 may mediate hypermethylation of DNA in LGL at least in part via an induction of DNMT3B.
Figure 3. Aberrant DNA methylation and methytransferase activity contribute to IL-15 induced LGL leukemia.
(A) Relative fold increase in global DNA methylation (GDM) levels in splenocytes from LGL leukemic mice (n=4) and in WT LGL cultured in IL-15 (n=3) for 30 days relative to values of GDM measured in fresh WT LGL (n=4), which is arbitrarily set at 1. Quantification was done by mass spectrophotometery and shown as mean ± SEM. (B) Relative fold changes in mRNA expression of Dnmt3b, in splenocytes from LGL leukemic mice (n = 3) and in WT LGL cultured in IL-15 (n=3) for 30 days relative to values of Dnmt3b measured in fresh WT LGL (n=3). Quantification was by Real-time RT-PCR and shown as the mean ± SEM. Each measurement was normalized against the level of 18S mRNA, and then values of Dnmt3b for fresh WT LGL were arbitrarily set at 1. (C) Fold changes (mean ± SEM) in mRNA expression of DNMT3B in three human LGL leukemia patient (Pt) samples, relative to values of DNMT3B measured in normal donors cells that were enriched for either CD56+ or CD8+ (n=4 each) and arbitrarily set at 1. Each sample was normalized to 18S mRNA. (D) Confocal analysis of Dnmt3b protein expression in mouse and human LGL leukemia relative to normal LGL. Assay was done by immuno-labeling the cells with anti-Dnmt3b antibody. Data are representative of at least four independent mice, patients and normal donors. Cells were counterstained with DAPI (blue) for nuclear staining. Scale bars, 10 μm. (E) WBC counts of different genotypes of adult mice generated from mating IL-15 Tg and DNMT3B Tg parents. The horizontal bar in each lane indicates the mean WBC count for 6-36 mice per group. Also included are four representative images from Wright-Giemsa staining of blood smears for each of the four genotypes, and an image of four whole spleens obtained from mice of each genotype. (F) Comparative survival of DNMT3B Tg and WT mice (both 100%), IL-15 Tg and IL-15/DNMT3B Tg mice.
See also Figure S2.
De novo overexpression of DNMT3B augments the incidence of LGL leukemia in vivo
We created DNMT3B Tg mice driven by the Vav1 promoter (Figure S2C). In vitro, we demonstrated that LGL from these mice show enhanced transformation in the presence of IL-15, when compared to WT mouse LGL in the presence of IL-15 (Figure S2D). In contrast, mouse LGL with reduced expression of Dnmt3b showed a decrease in transformation in the presence of IL-15 when compared to WT mouse LGL in the same assay (Figure S2E). DNMT3B Tg mice were found to have a normal phenotype and life expectancy similar to their WT littermates (not shown). Mating IL-15 Tg and DNMT3B Tg parents generated four different genotypes: IL-15 Tg, DNMT3B Tg, IL-15/DNMT3B Tg and WT mice. IL-15/DNMT3B Tg mice showed an increase in WBC count significantly earlier than their IL-15 Tg counterparts, while DNMT3B Tg mice and WT mice had normal WBC counts (Figure 3E). IL-15/DNMT3B Tg mice showed a significantly shorter latency and 100% incidence of fatal LGL leukemia compared with IL-15 Tg mice (Figure 3F and S2F). The immunophenotype of IL-15/DNMT3B Tg leukemia was similar to that of IL-15 Tg leukemia (Yokohama et al., 2010) (not shown) but IL-15/DNMT3B Tg mice had lymphadenopathy (Figure S2G). Thus, excess expression of DNMT3B does not initiate leukemia but contributes to its progression upon chronic exposure with IL-15.
Both the IL-15 Tg and the IL-15/DNMT3B Tg mice had significantly greater CpG methylation, as measured by LINE-1 hypomethylation (Estecio et al., 2007; Ogino et al., 2008), when compared to the DNMT3B Tg and WT mice (Figure S2H). While the differences in these measurements as well as those seen in Figure S2A appear modest, they measure methylation across the entire genome, rather than methylation of specific tumor suppressor genes. We previously documented promoter methylation and silencing of the tumor suppressor Idb4 in leukemia from IL-15 Tg mice (Yu et al., 2005). Notably, Idb4 promoter methylation was more pronounced in LGL leukemia from IL-15/DNMT3B Tg mice compared to that of IL-15 Tg mice (Figure S2I). Culture of WT LGL with IL-15 for 30 days also resulted in methylation of the Idb4 promoter, albeit not as striking as seen in primary LGL leukemia samples from IL-15 Tg mice (Figure S2I). In contrast, LGL from DNMT3B Tg mice did not show any methylation at Idb4 promoter, similar to WT LGL (Figure S2I). Thus, overexpression of DNMT3B without IL-15 overexpression was insufficient to cause Idb4 promoter methylation. Collectively, these results indicate that the overexpression of DNMT3B is likely necessary but without overexpression of IL-15 is insufficient for the hypermethylation of DNA noted in LGL chronically exposed to this cytokine.
IL-15 regulates expression of DNMT3B via repression of microRNA (miR)-29b
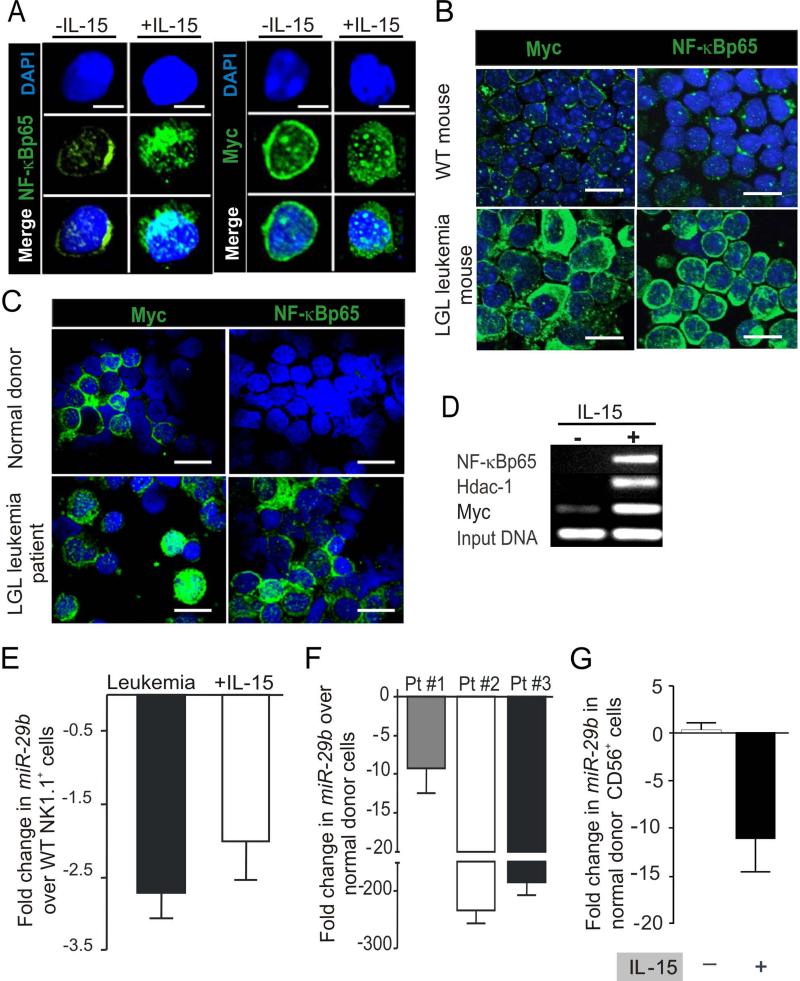
We have previously reported that Dnmt3b is a direct, negatively regulated target of, miR-29b and repression of miR-29b is induced by binding of Myc, Hdac-1 and NF-κBp65 to its promoter (Chang et al., 2008; Garzon et al., 2009b; Liu et al., 2010). We showed that short-term culture of WT mouse LGL with IL-15 induces Myc protein via NF-κB (Figure S1F-I). Similar data were obtained for Myc and NF-κB for both long-term cultures of WT mouse LGL in IL-15 (Figure 4A) as well as in mouse and human LGL leukemia (Figure 4B-C). Finally, we used a ChIP assay to show increased binding of Myc, Hdac-1 and NF-κBp65 repressors to the miR-29b 5’ regulatory region in WT mouse LGL first starved and then re-exposed to IL-15 (Figure 4D). Similar results were obtained in splenocytes from LGL leukemia in IL-15 Tg mice when compared to WT mouse splenocytes (Figure S3). As a result of these experiments, we postulated that IL-15 stimulation of WT LGL represses miR-29b via Myc, Hdac-1 and NF-κBp65.
Figure 4. Alteration of gene expression by IL-15 and in LGL leukemia.
(A) Confocal analysis of fresh WT mouse LGL and WT mouse LGL cultured in IL-15 for 30 days was performed by immuno-labeling the cells with anti-Myc and anti-NF-κBp65 as above. Scale bars, 5 μm. (B) Confocal analysis of spleen samples from LGL leukemic mice and fresh WT LGL for Myc and NF-κB was performed by immuno-labeling of the cells with the respective antibody. Cells were counterstained with DAPI (blue) for nuclear staining. Data are representative of at least five independent mice evaluated for protein expression. Scale bars, 10 μm. (C) Confocal analysis of enriched LGL from normal human donor blood samples compared with LGL leukemia samples from patients. Immuno-staining was done by incubating the cells with anti-MYC and an anti-NF-κBp65 antibody. Data are representative of at least four independent normal donors and LGL leukemia patients evaluated for protein expression. Scale bars, 10 μm. (D) In vitro cultured WT LGL were grown with IL-15 for approximately eight months, then harvested and starved for 24 hours and then divided up to be re-stimulated with either IL-15 or PBS for four hours. A ChIP assay was performed using the indicated antibody. The gel image shows the PCR product of the indicated gene performed on ChIP DNA with primers designed for amplification along the miR-29b promoter/enhancer region. The data are representative of three independent experiments. (E) Fold decrease (mean ± SEM) in miR-29b transcript levels in splenocytes from LGL leukemic mice (n=4) and in WT LGL cultured in IL-15 (n=4) for 12 hours relative to value of miR-29b measured in fresh WT LGL (n=4) which is arbitrarily set at 1. Each sample was normalized to U6. (F) Fold changes (mean ± SEM) in expression of miR-29b transcript levels in human LGL leukemia samples, normalized to U6 and then quantified relative to values of miR-29b measured in normal donor cells that were enriched for either CD56+ or CD8+ (n=4 each) and then arbitrarily set at 1. (G) Fold changes (mean ± SEM, n=3) in expression of miR-29b in CD3-CD56+ normal human donor LGL from IL-15 stimulated and PBS treated control for 12 hours relative to values of miR-29b measured in unstimulated normal donor LGL and arbitrarily set at 1. Each measurement was normalized against the level of U6.
See also Figure S3.
Indeed, when compared to fresh WT LGL, miR-29b expression was significantly decreased in LGL leukemia from IL-15 Tg mice (P < 0.02) as well as in WT mouse LGL stimulated with IL-15 for 12 hours (Figure 4E). We found similar striking results in human LGL leukemia (P < 0.0009) and in normal human LGL incubated with IL-15 (P < 0.003) (Figure 4F-G). Further, forced overexpression of miR-29b by 40-60-fold in IL-15-activated WT mouse LGL was associated with a proportional decrease in Dnmt3b expression (Figure 5A-B), while the reduction of Myc expression by shRNA in IL-15-activated WT mouse LGL significantly increased miR-29b expression and decreased Dnmt3b expression (not shown). Thus, the IL-15 mediated induction of Myc, NF-κB and Hdac-1 repressors in WT mouse LGL results in a decrease in miR-29b expression that in turn increases Dnmt3b expression.
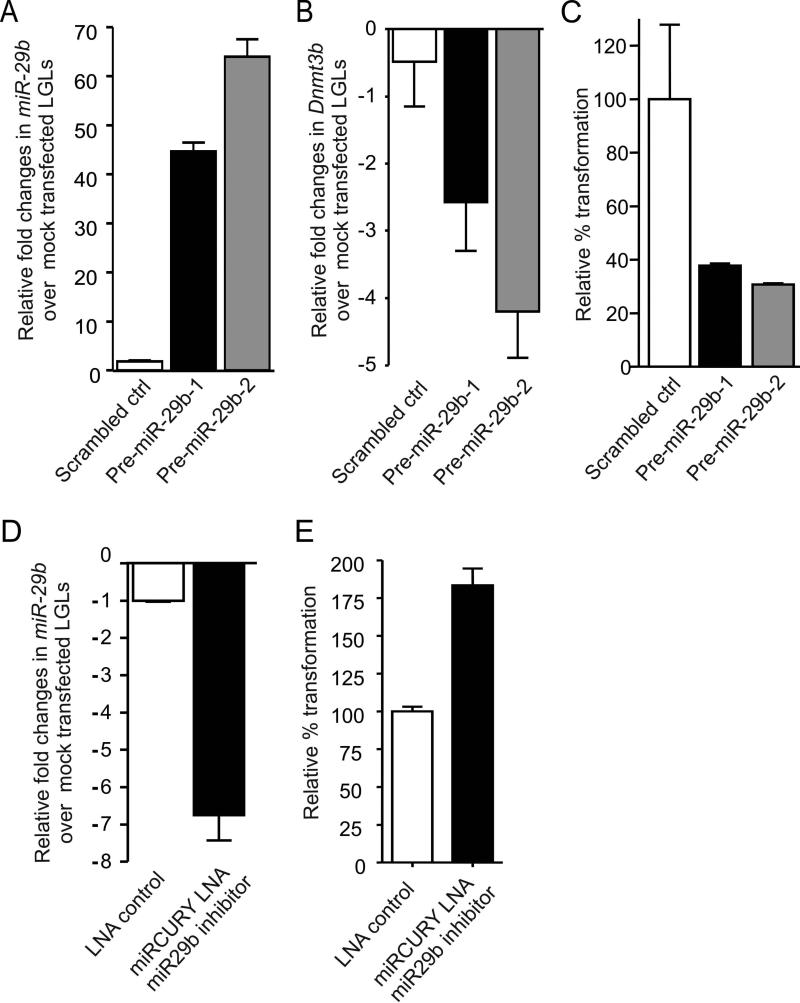
Figure 5. Mir-29b overexpression or repression alters LGL transformation.
(A) Relative fold overexpression (mean ± SEM) of miR-29b in WT mouse LGL transfected with scrambled control or pre-miR-29b, relative to the value measured in mock transfected LGL (not shown), which is arbitrarily set at 1. Each sample was normalized to U6. (B) Relative fold changes (mean ± SEM, n=3) in mRNA expression of Dnmt3b in LGL shown in (A). Each measurement was normalized against the level of 18S mRNA, and then values of Dnmt3b for mock-transfected LGL cells were arbitrarily set at 1. (C) LGL similarly transfected with scrambled control or pre-miR-29b as shown in (A) were then plated at 1×105/well in triplicate and cultured with IL-15 in a semisolid agar medium for 8-10 days after which CFU are detected and quantified in a cell transformation assay using a microtiter plate reader as described in Experimental Procedures. Percent transformation of pre-miR-29b transfected LGL relative to control (scrambled transfected LGL, arbitrarily set at 100%) is shown as mean ± SEM, n=3 each. (D) In vitro cultured mouse LGL were transfected with 50 pmole solution of LNA control or LNA miR-29b inhibitor oligonuleotide per manufacturer's instruction by electroporation. Cells were measured for miR-29b expression levels 24 hours post transfection. (E) WT mouse LGL were plated at 1×105/well in triplicate and cultured with IL-15 in a semisolid agar medium for 8-10 days after which CFU are detected and quantified in a cell transformation assay using a microtiter plate reader as described in Experimental Procedures. Percent transformation of cells relative to LNA control (arbitrarily set at 100%) is shown as mean ± SEM, n=3 each.
To further confirm the role of miR-29b in IL-15-mediated LGL leukemia, we overexpressed miR-29b in fresh WT LGL cells, which were then cultured in IL-15 for 8-10 days prior to assessment for early evidence of transformation in vitro. We noted that overexpression of miR-29b was inversely associated with IL-15-mediated LGL transformation compared to control transfected cells, i.e., WT LGL with overexpression of miR-29b showed significantly less IL-15-mediated transformation (Figure 5C). Further, a significant decrease of miR-29b expression in WT LGL cells transfected with miR-29b antagomir and cultured in IL-15 for 8-10 days resulted in a significant increase in their transformation compared with the same LGL transfected with a scrambled control (Figure 5D and E). Thus, collectively, we provide evidence in WT LGL for a mechanistic link between IL-15 mediated down-regulation of miR-29b, increased Dnmt3b, and enhanced transformation in short term cultures with IL-15, as well as increased methylation of the genome, including the tumor suppressor Idb4, in WT LGL cells chronically exposed to IL-15.
A formulation of a proteasomal inhibitor provides long-term disease free survival in leukemic mice
Since our previous data reveal that it is possible to reverse miR-29b repression in leukemia cells by targeting the transcriptional repression of Myc, Hdac-1 and NF-κBp65 using pharmacological proteasome inhibitors (Garzon et al., 2009b; Liu et al., 2010), we exposed LGL leukemia blasts from IL-15 Tg mice to the proteasome inhibitor bortezomib for two hours, and observed a 13,000-fold up-regulation of miR-29b when compared to blasts treated with PBS (n=3, p=.02; Figure 6A). This resurgence in miR-29b expression within two hours of treatment is followed by its return to levels seen in WT cells, suggestive of a block in the binding of Myc, Hdac-1 and NF-κBp65 to the miR-29b promoter. We also noted a significant (~50-fold) decrease in Dnmt3b transcript in bortezomib-treated LGL leukemic blasts at 48 and 72 hours, compared to PBS treated blasts (Figure 6B), which was validated at the protein level (Figure 6C). Targeting the transcriptional repressors of miR-29b also led to a significant re-expression of the tumor suppressor gene Idb4 compared to PBS treated blasts (Figure 6D). COBRA analysis confirmed this was due to reduction in Idb4 promoter methylation (Figure 6E). Notably, this in vitro treatment with bortezomib not only resulted in down-regulation of Dnmt3b, but also reduced expression of AurkA and AurkB transcript in leukemic blasts compared to PBS treated blasts (Figure S4A and S4B). Marked apoptosis of WT LGL in the presence of IL-15 as well of LGL leukemia was noted with the in vitro treatment with bortezomib, which may also be secondary to the drug's effect on the induction of pro-apoptotic Bid, as previously reported (Hodge et al., 2009).
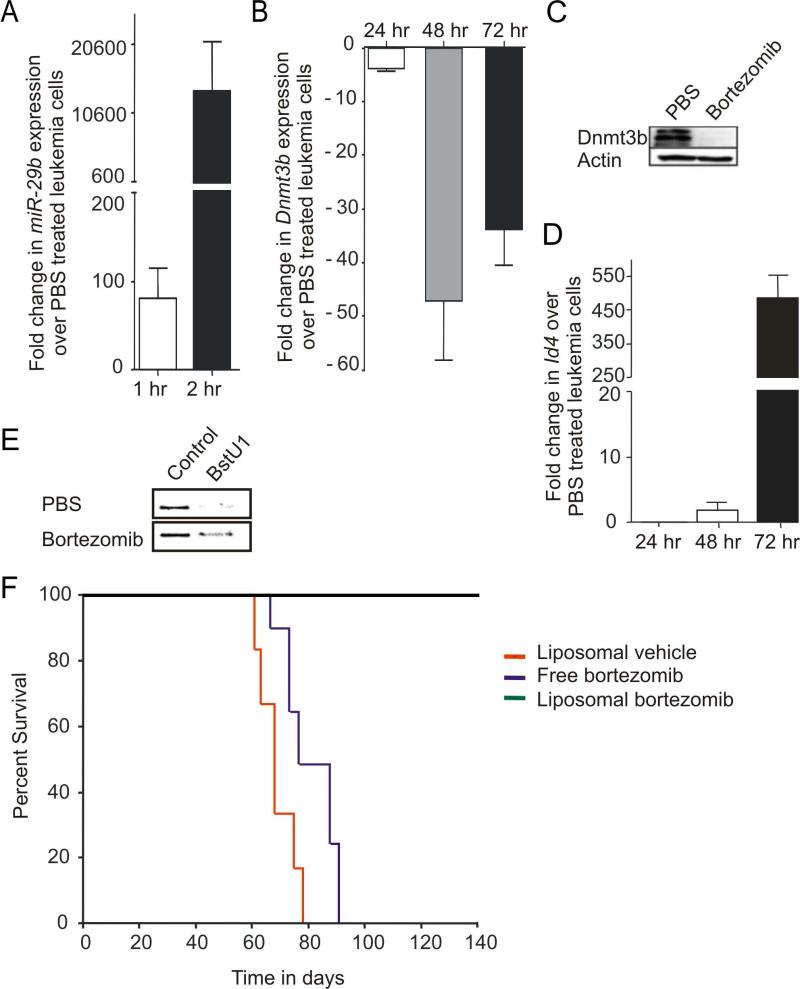
Figure 6. In vitro and in vivo targeting of miR-29b transcriptional repression.
(A) Fold increase in miR-29b transcript at 1 (white) and 2 (black) hours post in vitro bortezomib treatment (20μM). Both measurements normalized to U6 and then set relative to values of miR-29b measured in PBS treated LGL leukemia cells, which are arbitrarily set at 1. (B) Relative fold changes in mRNA expression of Dnmt3b in bortezomib (20nM) treated LGL leukemia samples at 24, 48 and 72 hour, normalized to 18S mRNA and then quantified relative to values of Dnmt3b in PBS treated LGL leukemia samples that are arbitrarily set at 1. Data for (A) and (B) are mean ± SEM (n=3). (C) Splenocytes from LGL leukemic mice were treated in vitro with either control (PBS) or bortezomib (20nM) for 24 hours and then immunoblotted for Dnmt3b and actin that was used as an internal control. (D) Relative fold changes (mean ± SEM) in mRNA expression of Idb4 in mouse LGL leukemia cells treated in vitro with bortezomib (20μM) at 24, 48 and 72 hour, normalized to 18S mRNA and then quantified relative to values of Idb4 in PBS treated LGL leukemia samples that are arbitrarily set at 1. (E) COBRA analysis for Idb4 promoter methylation in splenocytes from LGL leukemic mice that were treated in vitro for 72 hours with either control (PBS) or bortezomib (20nM). Amplification of the Idb4 promoter region and sequential COBRA analysis with a methylation sensitive restriction enzyme (BstU1) reveals only a partial loss of methylation at the Idb4 promoter in LGL leukemia cells treated with bortezomib (20μM), but not PBS. (F) Kaplan–Meier survival plot for ICR-SCID mice (n = 6–8/group) after intravenous injection of splenocytes from LGL leukemia mice. Disease-free survival in mice treated with empty liposomes, free-bortezomib and liposomal-bortezomib.
See also Figure S4.
We previously reported that in vivo treatment of ICR-SCID mice engrafted with primary LGL leukemia from IL-15 Tg mice with a DNA hypomethylating agent that targets Dnmt1 (i.e., decitabine) or in combination with a HDAC inhibitor (i.e., depsipeptide) was either ineffective or highly toxic, respectively (Yu et al., 2009). To test the efficacy of bortezomib we needed to first improve the in vivo pharmacodynamics of the drug, as the naked compound was toxic and ineffective. We therefore developed a liposomal preparation (Figure S4C). Four weeks after ICR-SCID mice were engrafted with a lethal dose of LGL leukemic blasts from IL-15 Tg mice, a twice weekly treatment with the liposomal bortezomib was initiated at the dose of 1.0 mg/kg/mouse for the first week, which was continued at the dose of 2.0 mg/kg/mouse for next two weeks while monitoring for any clinical signs of toxicity or leukemia related distress. In a randomized trial, control mice treated with a preparation of empty liposomes first succumbed to fatal LGL leukemia within 60-80 days following infusion of the LGL leukemic blasts, and mice treated with free bortezomib at a similar dose and schedule as liposomal bortezomib all died from leukemia generally within the same period of time. However, the mice engrafted with LGL leukemia and treated with liposomal-bortezomib showed 100% survival 130 days following infusion of the LGL leukemic blasts, without any evidence of toxicity (Figure 6F). Spleens from mice treated with liposomal-bortezomib were significantly reduced in weight (Figure S4D) and histologic examination showed a clearance of leukemic blasts from liver and spleen (not shown).
Discussion
There are previous reports documenting the increased risk of cancer in association with aberrant cytokine signaling, and a few murine models where their overexpression of cytokines (i.e., IL-9 or IL-15) induces malignant transformation (Fehniger et al., 2001; Renauld et al., 1994; Waldmann and Tagaya, 1999), yet mechanistic data regarding the latter remains incomplete (Hodge et al., 2009; Yu et al., 2005). The crucial role of IL-15 in the survival and proliferation of LGL leukemia has also been well documented in both humans and mice (Fehniger et al., 2001; Yokohama et al., 2010; Zambello et al., 1997). Abnormal expression of IL-15 has been described in patients with other lymphoid malignancies and autoimmune diseases including rheumatoid arthritis, multiple sclerosis, psoriasis, inflammatory bowel disease, and also in diseases associated with human T cell lymphotropic virus I (HTLV-I) (Asadullah et al., 2000; Azimi et al., 1998; Azimi et al., 1999; Carroll et al., 2008; D'Auria et al., 1999; Trentin et al., 1997). Zambello et al., previously showed that membrane-bound IL-15 is expressed on proliferating blast cells in LGL leukemia patients (Zambello et al., 1997), and most if not all human LGL leukemia cell lines require activation via the IL-15Rβγ for propagation in vitro, suggesting that IL-15 is required for initiation and maintenance of leukemogenesis in vivo.
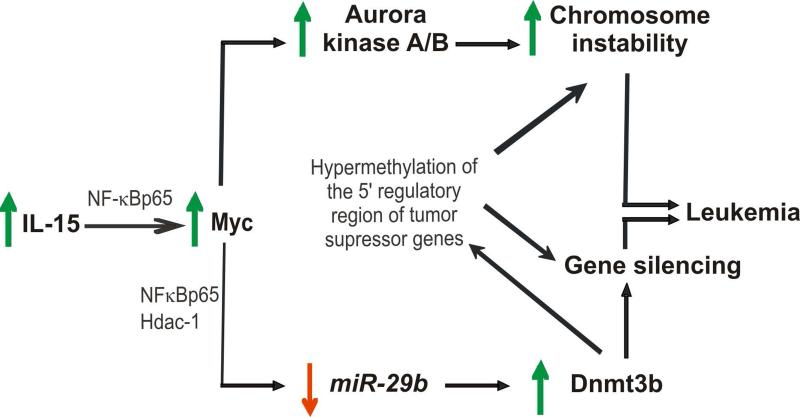
Here, we provide the evidence that IL-15 alone can immortalize WT LGL in vitro that, upon adoptive transfer in vivo, undergo fulminant leukemic transformation in the absence of exogenous IL-15. This oncogenic effect is the result of two, distinct pathways by which IL-15 contributes to malignant transformation: first via the induction of CIN and marked aneuploidy; and second via an induction of DNA hypermethylation which can contribute both to CIN (Esteller, 2006; Esteller, 2007; Ganem et al., 2009) and to silencing of tumor suppressor genes. We show that IL-15 induces Myc, Hdac-1 and NF-κBp65, which mediate down regulation of miR-29b and consequent overexpression of Dnmt3b, thereby hastening the onset of LGL leukemia (Figure 7). Concurrent upregulation of IL-15, MYC, AURKA, AURKB, NF-κBp65, and downregulation of miR-29 followed by upregulation of DNMT3B and increased GDM, as well as centrosome aberrancies were also demonstrated in primary patient LGL leukemic blasts compared with normal controls.
Figure 7.
Schematic of the proposed network of IL-15-mediated transformation of WT LGL to LGL leukemia.
There is currently no curative treatment for LGL leukemia and the disease is fatal in most circumstances. Treatment options include methotrexate, cyclosporine A, or cyclophosphamide and monotherapy with corticosteroids, (Sokol and Loughran, 2006; Utecht and Kolesar, 2008) and are often associated with significant hematologic and other toxicities (Sokol and Loughran, 2006). Relatively little data exist on effective alternative therapies for LGL leukemia. Targeting IL-15 or the β chain component of its receptor as a means of immunotherapy has been suggested, but has thus far not been successful in clinical trials for LGL leukemia patients (Morris et al., 2006). Furthermore, our own experimental therapy with the Dnmt inhibitor decitabine targeting the aberrant epigenetic changes observed in our preclinical model of LGL leukemia resulted in only transient responses and associated hematologic toxicity, and the combination of decitabine with a histone deacetylase inhibitor resulted in fatal toxicity (Yu et al., 2009). These data therefore emphasize the need for approaches aimed at the pathogenic mechanisms responsible for initiation and maintenance of LGL leukemia. To pursue this goal, we designed a therapeutic strategy that used liposomal bortezomib to concurrently and effectively target upregulation of Myc, activation of AurkA, AurkB, NF-κBp65, and downregulation of miR-29 that were shown to play a pivotal role in inducing CIN and aberrant Dnmt expression and in turn leukemia. Garzon et al., have previously shown that decreased miR-29b expression can also contribute to cancer progression by a concomitant increased expression of the anti-apoptotic Mcl-1 (Garzon et al., 2009a), and Hodge et al., have shown that IL-15 can lower the pro-apoptotic Bid protein in LGL leukemia via proteasome-mediated degradation (Hodge et al., 2009). STAT3 also lies downstream of IL-15 receptor activation and LGL leukemia from our IL-15 Tg mice display constitutive activation if STAT3 (our data, not shown). The latter has been shown to induce anti-apoptotic Mcl-1 expression in patients with LGL leukemia (Epling-Burnette et al., 2001), and a recent study has uncovered activating STAT3 mutations in a subset of patients with chronic LGL leukemia (Johnston et al., 1995; Koskela et al., 2012). Hence it is possible that some of the effects shown to result from excessive IL-15 signaling in this report, as well as other unknown effects, are mediated at least in part by an activated STAT3. Extending and integrating these findings, therapeutic strategies for IL-15-mediated cancers could be also provided through the use of aurora kinase inhibitors, NF-κB inhibitors, STAT3 inhibitors, and/or synthetic miRNAs that target DNA methyltransferases (Chan et al., 2010).
Our therapeutic effectively targets the proteasomal degradation pathway and transcriptional repressors/activators in vivo, thereby inducing a long-term disease free remission as a single agent in this otherwise fatal and refractory malignancy. The correlative data we provide from human LGL leukemia samples document their overexpression of IL-15 as well as the two aberrant pathways that emanate from chronic stimulation by IL-15. Collectively this offers a strong rationale for a therapeutic intervention with liposomal bortezomib in patients with LGL leukemia.
Experimental Procedures
Generation of transgenic mice
The Institutional Animal Care and Use Committee (IACUC) of The Ohio State University (OSU) approved all procedures involving animals. Expression of the human DNMT3B in Tg mouse is driven by the Vav promoter, which restricts expression to cells of hematopoietic lineage. The Vav-transgenic system is well established for mouse models of hematopoietic disease (Ogilvy et al., 1998). The transgenic construct, including a HA-tag, was injected into mouse pronuclei and multiple offspring were obtained with variable numbers of the transgene (Figure S2C) but with normal survival (Figure 3F). IL-15 Tg mice were generated and maintained as described previously (Fehniger et al., 2001).
In vitro culture of LGL cells
To expand LGL cells in vitro approximately 1×105 FACS sorted splenic NK1.1+ cells were cultured in 96-well plates at a density of 1×105/100 μl RPMI medium supplemented with β-mercaptoethanol, antibiotics and 100ng/ml of rhIL-15. Typically culture medium was refreshed every 48-72 hours, and cells then transferred to 6 well plates and split back to original density in culture when they reached a density of 1.6 × 106/100 μl.
Antibody staining and flow cytometry
Spleen, bone marrow, and peripheral blood samples were harvested from moribund mice following sacrifice and single cell suspensions were prepared as described previously (Fehniger et al., 2001). The experiments were performed according to the Institutional Animal Care and Use Committee (IACUC) guidelines. The following fluorochrome-conjugated monoclonal antibodies (mAb) were purchased from BD Pharmingen, San Jose, CA and used for flow cytometry: anti-CD3 (clone 145-2C11), anti-NK1.1 (clone PK136), anti-CD4 (clone RM4-5) and anti-CD8a (clone 53-6.7). Human LGL enrichment antibodies, anti-CD56 (clone N901) and anti-CD3 (clone UCHT-1) were purchased from Beckman Coulter (Beckman Coulter, Inc., Brea CA).
Enrichment of LGL
Normal human peripheral blood LGLs were enriched from leukopacks purchased from the American Red Cross (Columbus, OH) to > 70% cell purity utilizing methods previously described (Park et al., 2009). Briefly, mononuclear cells were obtained using Ficoll-Paque Plus (GE Healthcare, Little Chalfont, United Kingdom) and CD56+ cells were enriched using a Rosette separation cocktail (StemCell Technologies, Vancouver, BC) following the manufacturer's instructions. Enriched LGLs were then sorted with BD FACSAria™ cell sorter using anti-CD56 and anti-CD3. Sorted cells were >95% pure for CD56+CD3- population. Patient samples were obtained from the OSU Leukemia Tissue Bank following informed consent. The OSU Institutional Review Board approved all experiments performed with human materials.
Total RNA and DNA isolation
Single cell suspension was spun down and the cell pellet was resuspended in TRIzol® Lysis reagent (Invitrogen, Carlsbad, CA) processed further using miRNeasy Mini Kit and Reagents (Qiagen, Valencia, CA). On-column DNA digestion was performed to eliminate any residual DNA. Total RNA was eluted using RNase-free water, and was quantified by NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). DNA isolations were performed using QIAamp DNA Micro Kit (Qiagen), according to manufacturer's protocols with the exception of the elution step, which was done with water instead of Buffer AE.
First strand synthesis for RT-PCR and quantitative Taqman PCR
cDNA was generated from approximately 100 to 500 ng of total RNA, using the SuperScript First Strand Synthesis kit for RT-PCR (Invitrogen), according to the manufacturer's protocol. RT reaction was performed in a DNA Engine Dyad Peltier Thermal Cycler (Bio Rad, Hercules, CA) using the following program: 25°C for 10 minutes (min), 42°C for 50 min, and 70°C for 10 min. For first strand synthesis of miR-29b and U6 approximately 100ng of RNA was processed using the Applied Biosystems mir-RT kit (Applied Biosystems, Austin, TX). Total RNA was added to a mix of dNTP mix, RT enzyme, 10x RT buffer, RNase inhibitor, and each desired Taqman 5x RT primer. RT reaction was performed using the following temperature conditions: 16°C for 30 min, 42°C for 45 min, and 85°C for 5 min and stored at 4°C until needed for quantitative PCR. Quantitative RTPCR was performed using either 1 μl of cDNA or its dilutions. Briefly, 1 μl of sample was added to a 20 μl total reaction volume containing 20x RT Taqman Assay® for desired gene or miRs, 2X Taqman Universal Fast PCR Master Mix® (Applied Biosystems). Reaction was performed in 96 well Fast Optic Plates, and run using the 7900HT Fast Real-Time PCR System (Applied Biosystems). Taqman probe ID for the genes can be provided upon request.
Chromatin Immunoprecipitation (ChIP) assays and quantitative ChIP PCR for miR-29b and the AurkA and AurkB promoters
Approximately 2 × 107 cells were harvested from leukemic and WT mice and the ChIP assay were performed as described previously (Liu et al., 2010). For IL-15 stimulation experiments, WT LGL cultured with IL-15 were first starved of IL-15 for 24 hours, washed twice with cold RPMI medium and then re-exposed to either PBS or IL-15 for 4 hours. The following antibodies were purchased from AbCam (Cambridge, England) and used for ChIP: anti-HDAC-1 (Ab7028) and anti-NF-κB p65 (Ab7970). Anti-c-Myc antibody was purchased from Santa Cruz Biotechnology. ChIP PCR was performed using Power SYBR PCR mix (Applied Biosystems) or AmpliTaq Gold® 360 PCR Master Mix using ChIP DNA which was added to 25 μl total reaction volume containing 2X PCR mix, 10μM of miR-29b forward and reverse primers (Sigma-Aldrich, St. Louis, MO). Primer sequence can be provided upon request. Fold change in binding was compared using input DNA as control. PCR for AurkA and AurkB was performed as described previously (den Hollander et al., 2010).
Immuno fluorescence staining and confocal microscopy
Approximately 1-2 × 105 freshly harvested cells were spun onto a microscope slide using Shandon Cytospin 4 cytofuge (Thermo Fisher Scientific, Waltham, MA). The slides were fixed immediately with ice-cold acetone for 10 minutes and washed twice with PBS and 20% FBS. Blocking was performed by incubating the cells in protein-free blocking solution (Dako Denmark A/S, Glostrup, Denmark) for 30 minutes. Cells were then stained with a 1:100 dilution of goat polyclonal to rabbit IgG anti-c-Myc, anti-Dnmt3b (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-NF-κBp65 (AbCam) at room temperature for 60 minutes. Slides were washed twice with PBS containing 20% FBS before incubating the cells with 1:40 dilution of Texas Red®-X goat anti-rabbit IgG (Invitrogen, Carlsbad, CA) at room temperature for 60 minutes in dark. Cells were than washed with PBS containing 20% FBS and mounted with VECTASHIELD® Mounting Medium with DAPI. Pericentrin (Abcam) and GTU-88 (Vector Laboratories, Burlingame, CA) antibodies were used to perform centrosomal staining in acetone fixed cells. Briefly, cells were mounted on Poly-L-Lysine coated slides and fixed in ice-cold acetone for 5 minutes. Cells were incubated with either Pericentrin (for human samples) or GTU88 (for mouse samples) antibody for 1 hour at room temperature followed by incubation with secondary antibody. Images were acquired using Olympus FV1000-Filter Confocal microscope (Olympus, Center Valley, PA) and analyzed with Olympus Fluoview software (version 2.0).
In vitro drug studies
Cells were harvested from the spleens of leukemic mice. Cells were cultured in RPMI 1640 containing 10% FBS, at 37°C with 5% CO2 and 100 Units/ml of rhIL-2. Leukemic cells were incubated with 20nM bortezomib or PBS for an incubation time that varied with the experiment performed. Viable cells were measured by trypan blue exclusion. Cells were harvested and washed with PBS prior to DNA and RNA isolation.
Bisulphite conversion and COBRA Analysis of Idb4 promoter
Bisulphite conversion and COBRA Analysis of Idb4 promoter was performed as described (Yu et al., 2005).
Transfection of primary murine LGL cells
Transfections were performed using murine Dnmt3b and AurkA SureSilencing™ shRNA plasmids purchased from SABiosciences’ (Qiagen, Valencia, CA). Overexpression plasmids were purchased from Origene (Origene, Rockville, MD). Single cell suspension of LGL cells were transfected using the Amaxa Nucleofection system (Lonza Cologne GmbH, Cologne, Germany). Briefly, approximately 5×106 cells were transfected according to the manufacturer's protocol for Amaxa Mouse Macrophage Nucleofector® Kit. Cells were assessed for the expression of GFP 12-24 hours after transfection.
In vitro transformation assay
Normal activated T cells can form microscopically visible colonies in semisolid medium as early as 4-6 days following exposure to the transforming T cell leukemia virus, HTLV1. These lymphocytes proved to be transformed in that their proliferation subsequently became independent of growth factor. In the absence of infection by HTLV1, normal activated T cells do not transform and do not form colonies in semisolid medium (Aboud et al., 1987). We used a slightly modified colony forming unit (CFU) assay to assess for transformation of WT mouse LGL exposed to IL-15 and forced overexpression of AurkA or Dnmt3b or under-expression of miR-29b.
WT NK1.1+CD3- LGL were activated and expanded in vitro with IL-15 and transfected by electroporation with 5 μg of GFP-plasmid DNA control construct or a GFP-experimental vector construct as indicated in the figure legends. Twenty-four hours after transfection, cells were sorted for GFP expression (~95% purity) and seeded at the density of 0.5×105 - 1×105 cells/well in 96 well plates in a semisolid agar medium without or with a supplement of IL-15 (100ng/ml). The semisolid agar colony formation assay was performed per manufacturer's instruction using the Cytoselect™ 96-Well Cell Transformation Assay Kit (Cell Biolabs Inc, San Diego, CA). Briefly, following an 8-10 day incubation period, cell colonies are solubilized, lysed, detected and quantified by the provided MTT solution (Invitrogen, Carlsbad CA) in a microtiter plate reader.
CFUs was absent from wells seeded with WT mouse LGL transfected with the GFP-plasmid DNA control construct but without IL-15. CFUs quantified in wells seeded with IL-15 and WT mouse LGL transfected with the GFP-plasmid DNA control construct were labeled the control group and arbitrarily assigned a transformation score of 100%. CFU quantified in wells seeded with IL-15 and WT mouse LGL transfected with GFP plus the experimental vector construct were labeled the experimental group and were scored as a percent transformation relative to the control group.
In vivo drug studies
Age and sex matched ICR-SCID mice were used to study in vivo transplantability of the leukemia cells. Approximately 2 × 106 splenocytes obtained from IL-15 Tg leukemic mice were intravenously injected into recipient mice. Upon engraftment of leukemia (measured by increased peripheral WBC count), mice were given two doses of either bortezomib or liposomal bortezomib at the concentration of 1.0mg/week/kg of body weight, followed by two doses of 2.0mg/week/kg of body weight for next two weeks. Empty liposomes were injected as control. All the experimental mice were given drug or placebo via tail vein injection for three consecutive weeks.
Supplementary Material
SIGNIFICANCE.
Our best opportunity for curing cancer will come from a complete understanding of its causes. Inflammation has long been associated with the progression of cancer but few if any studies have shown how inflammation can initiate cancer. Here we show how an excess of a single proinflammatory cytokine, IL-15, causes chromosomal instability and DNA hypermethylation, leading to an aggressive acute leukemia of large granular lymphocytes, a rare yet uniformly fatal form of cancer. We show how novel drug targeting of the aberrant pathways induced by excessive IL-15 can cure this cancer. Thus, elucidating the molecular mechanisms involved in the genesis of cancer that results from chronic inflammation has merit for curing such cancers.
Acknowledgments
This work is supported by National Cancer Institute grants CA16058, CA95426, CA68458, CA09338 (M.A. Caligiuri); CA140158 (M.A. Caligiuri and G. Marcucci); CA102031 (G. Marcucci); CA149623 (S. Liu); NSF EEC-0914790 (R.J. Lee).
Footnotes
These authors contributed equally to this work.
References
- Aboud M, Golde DW, Bersch N, Rosenblatt JD, Chen IS. A colony assay for in vitro transformation by human T cell leukemia viruses type I and type II. Blood. 1987;70:432–436. [PubMed] [Google Scholar]
- Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc Natl Acad Sci U S A. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadullah K, Haeussler-Quade A, Gellrich S, Hanneken S, Hansen-Hagge TE, Docke WD, Volk HD, Sterry W. IL-15 and IL-16 overexpression in cutaneous T-cell lymphomas: stage-dependent increase in mycosis fungoides progression. Exp Dermatol. 2000;9:248–251. doi: 10.1034/j.1600-0625.2000.009004248.x. [DOI] [PubMed] [Google Scholar]
- Azimi N, Brown K, Bamford RN, Tagaya Y, Siebenlist U, Waldmann TA. Human T cell lymphotropic virus type I Tax protein trans-activates interleukin 15 gene transcription through an NF-kappaB site. Proc Natl Acad Sci U S A. 1998;95:2452–2457. doi: 10.1073/pnas.95.5.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi N, Jacobson S, Leist T, Waldmann TA. Involvement of IL-15 in the pathogenesis of human T lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis: implications for therapy with a monoclonal antibody directed to the IL-2/15R beta receptor. J Immunol. 1999;163:4064–4072. [PubMed] [Google Scholar]
- Baldassarre G, Fedele M, Battista S, Vecchione A, Klein-Szanto AJ, Santoro M, Waldmann TA, Azimi N, Croce CM, Fusco A. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc Natl Acad Sci U S A. 2001;98:7970–7975. doi: 10.1073/pnas.141224998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford RN, Grant AJ, Burton JD, Peters C, Kurys G, Goldman CK, Brennan J, Roessler E, Waldmann TA. The interleukin (IL) 2 receptor beta chain is shared by IL-2 and a cytokine, provisionally designated IL-T, that stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci U S A. 1994;91:4940–4944. doi: 10.1073/pnas.91.11.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. Embo J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll HP, Paunovic V, Gadina M. Signalling, inflammation and arthritis: Crossed signals: the role of interleukin-15 and -18 in autoimmunity. Rheumatology (Oxford) 2008;47:1269–1277. doi: 10.1093/rheumatology/ken257. [DOI] [PubMed] [Google Scholar]
- Chan KK, Liu Z, Xie Z, Chiu M, Wang H, Chen P, Dunkerson S, Liu S, Triantafillou G, Garzon R, et al. A novel ultrasensitive hybridization-based ELISA method for 2-methoxyphosphorothiolate microRNAs and its in vitro and in vivo application. Aaps J. 2010;12:556–568. doi: 10.1208/s12248-010-9214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria L, Bonifati C, Cordiali-Fei P, Leone G, Picardo M, Pietravalle M, Giacalone B, Ameglio F. Increased serum interleukin-15 levels in bullous skin diseases: correlation with disease intensity. Arch Dermatol Res. 1999;291:354–356. doi: 10.1007/s004030050421. [DOI] [PubMed] [Google Scholar]
- den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, Kremer M, Graf N, Scheerer M, Hall MA, et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116:1498–1505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, Li Y, Wang J-M, Yang-Yen H-F, Karras J, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Fehniger TA, Suzuki K, Ponnappan A, VanDeusen JB, Cooper MA, Florea SM, Freud AG, Robinson ML, Durbin J, Caligiuri MA. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J Exp Med. 2001;193:219–231. doi: 10.1084/jem.193.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009a;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009b;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, et al. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- Hodge DL, Yang J, Buschman MD, Schaughency PM, Dang H, Bere W, Yang Y, Savan R, Subleski JJ, Yin XM, et al. Interleukin-15 enhances proteasomal degradation of bid in normal lymphocytes: implications for large granular lymphocyte leukemias. Cancer Res. 2009;69:3986–3994. doi: 10.1158/0008-5472.CAN-08-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Bacon CM, Finbloom DS, Rees RC, Kaplan D, Shibuya K, Ortaldo JR, Gupta S, Chen YQ, Giri JD, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc Natl Acad Sci U S A. 1995;92:8705–8709. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A, Kawai H, Suto S, Kitajima S, Sato S, Takata T, Tatsuka M. Aurora-B/AIM-1 kinase activity is involved in Ras-mediated cell transformation. Oncogene. 2005;24:7266–7272. doi: 10.1038/sj.onc.1208884. [DOI] [PubMed] [Google Scholar]
- Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, Lagstrom S, Clemente MJ, Olson T, Jalkanen SE, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. The New England journal of medicine. 2012;366:1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, Hickey CJ, Yu J, Becker H, Maharry K, et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Janik JE, White JD, Fleisher TA, Brown M, Tsudo M, Goldman CK, Bryant B, Petrus M, Top L, et al. Preclinical and phase I clinical trial of blockade of IL-15 using Mikbeta1 monoclonal antibody in T cell large granular lymphocyte leukemia. Proc Natl Acad Sci U S A. 2006;103:401–406. doi: 10.1073/pnas.0509575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvy S, Elefanty AG, Visvader J, Bath ML, Harris AW, Adams JM. Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood. 1998;91:419–430. [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Giovenzana C, Hughes TL, Yu J, Trotta R, Caligiuri MA. The Axl/Gas6 pathway is required for optimal cytokine signaling during human natural killer cell development. Blood. 2009;113:2470–2477. doi: 10.1182/blood-2008-05-157073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld JC, van der Lugt N, Vink A, van Roon M, Godfraind C, Warnier G, Merz H, Feller A, Berns A, Van Snick J. Thymic lymphomas in interleukin 9 transgenic mice. Oncogene. 1994;9:1327–1332. [PubMed] [Google Scholar]
- Sokol L, Loughran TP., Jr. Large Granular Lymphocyte Leukemia. Oncologist. 2006;11:263–273. doi: 10.1634/theoncologist.11-3-263. [DOI] [PubMed] [Google Scholar]
- Trentin L, Zambello R, Facco M, Sancetta R, Agostini C, Semenzato G. Interleukin-15: a novel cytokine with regulatory properties on normal and neoplastic B lymphocytes. Leuk Lymphoma. 1997;27:35–42. doi: 10.3109/10428199709068269. [DOI] [PubMed] [Google Scholar]
- Utecht KN, Kolesar J. Bortezomib: a novel chemotherapeutic agent for hematologic malignancies. American journal of health-system pharmacy : AJHP : official journal of the American Society of Health-System Pharmacists. 2008;65:1221–1231. doi: 10.2146/ajhp070272. [DOI] [PubMed] [Google Scholar]
- Waldmann TA, Tagaya Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu Rev Immunol. 1999;17:19–49. doi: 10.1146/annurev.immunol.17.1.19. [DOI] [PubMed] [Google Scholar]
- Yokohama A, Mishra A, Mitsui T, Becknell B, Johns J, Curphey D, Blaser BW, Vandeusen JB, Mao H, Yu J, Caligiuri MA. A novel mouse model for the aggressive variant of NK cell and T cell large granular lymphocyte leukemia. Leuk Res. 2010;34:203–209. doi: 10.1016/j.leukres.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Ershler M, Yu L, Wei M, Hackanson B, Yokohama A, Mitsui T, Liu C, Mao H, Liu S, et al. TSC-22 contributes to hematopoietic precursor cell proliferation and repopulation and is epigenetically silenced in large granular lymphocyte leukemia. Blood. 2009;113:5558–5567. doi: 10.1182/blood-2009-02-205732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nature genetics. 2005;37:265–274. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- Zambello R, Facco M, Trentin L, Sancetta R, Tassinari C, Perin A, Milani A, Pizzolo G, Rodeghiero F, Agostini C, et al. Interleukin-15 triggers the proliferation and cytotoxicity of granular lymphocytes in patients with lymphoproliferative disease of granular lymphocytes. Blood. 1997;89:201–211. [PubMed] [Google Scholar]
- Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.