Abstract
Effective chromosome organization is central to the functioning of any cell. In bacteria, this organization is achieved through the concerted activity of multiple nucleoid-associated proteins. These proteins are not, however, universally conserved, and different groups of bacteria have distinct subsets that contribute to chromosome architecture. Here, we describe the characterization of a novel actinobacterial-specific protein in Streptomyces coelicolor. We show that sIHF (SCO1480) associates with the nucleoid and makes important contributions to chromosome condensation and chromosome segregation during Streptomyces sporulation. It also affects antibiotic production, suggesting an additional role in gene regulation. In vitro, sIHF binds DNA in a length-dependent but sequence-independent manner, without any obvious structural preferences. It does, however, impact the activity of topoisomerase, significantly altering DNA topology. The sIHF–DNA co-crystal structure reveals sIHF to be composed of two domains: a long N-terminal helix and a C-terminal helix-two turns-helix domain with two separate DNA interaction sites, suggesting a potential role in bridging DNA molecules.
INTRODUCTION
The bacterial chromosome serves as the blueprint for all cellular activity. This chromosomal blueprint is, however, subject to paradoxical organization: it must be extensively constrained and compacted so as to be effectively contained within a cell, and yet must be flexible enough to allow for not only transcription of the appropriate genes at any given time but also replication and segregation during cell growth and cell division. This dynamic behaviour is mediated, in part, by a group of proteins collectively termed the ‘nucleoid-associated proteins’. These proteins bind DNA with varying degrees of specificity, and promote its compaction through DNA bending, bridging and wrapping (1). In addition to their structural function, many nucleoid-associated proteins also have a global impact on transcription, and their loss often results in pleiotropic phenotypic effects (1,2). Nucleoid-associated proteins have been most extensively studied in Escherichia coli, with IHF, HU and H-NS being among the best characterized. H-NS binds AT-rich DNA sequences, often within intergenic regions, and in such a way that the expression of flanking genes is repressed (3). It also oligomerizes with distantly bound H-NS molecules and effectively bridges disparate chromosomal DNA segments (4). Conversely, HU is a heterodimeric protein that binds DNA without any obvious sequence specificity and promotes DNA bending and wrapping (1,5). IHF is a structural homologue of the HU proteins. Like the HU proteins, IHF acts as a heterodimer, and its DNA binding leads to significant bending of the DNA, although unlike HU, it preferentially associates with specific DNA sequences (6).
While nucleoid-associated proteins are critical for chromosome organization in all bacteria, there is considerable diversity in the proteins that contribute to chromosome structure in different bacterial phyla. The actinobacteria, which include the filamentous Streptomyces and pathogenic Mycobacterium, for instance, encode HU-like proteins, but lack any obvious homologues to IHF or H-NS. Recent work in the mycobacteria has, however, revealed that these bacteria possess a functional equivalent to H-NS known as Lsr2 (7); there are two copies of Lsr2 encoded in most Streptomyces genomes sequenced to date. Lsr2 bears limited sequence and structural similarity to H-NS, but has analogous biochemical properties and is functionally interchangeable with H-NS in E. coli (7,8).
IHF had been investigated for its ability to stimulate phage DNA integration (hence its ‘integration host factor’ designation) prior to its characterization as a nucleoid-associated protein in E. coli. An equivalent activity has been observed for a protein in Mycobacterium tuberculosis and Mycobacterium smegmatis, whereby it facilitates bacteriophage DNA integration into the mycobacterial chromosome (9); this protein has subsequently been termed mIHF, for mycobacterial IHF. Like its E. coli namesake, mIHF is a small, heat-stable protein that promotes the integrative recombination of phage DNA with the chromosome by forming a stable intasomal complex with the phage integrase and attP DNA (9). Notably, mIHF is essential for M. smegmatis viability (10) and appears to be required for M. tuberculosis growth (11), suggesting that its function in the cell extends beyond phage DNA integration. Both IHF and mIHF accumulate to maximal levels during late exponential growth and entry into stationary phase (10,12,13). These proteins do not, however, share any obvious similarity at either a sequence or secondary structural level, they do not appear to bind similar DNA sequences (mIHF has no obvious sequence specificity while IHF exhibits preferred binding to a specific sequence), and IHF cannot function in place of mIHF (14), suggesting that their functional similarity may extend only to participation in phage DNA integration.
Probing the function of an essential protein like mIHF has its challenges, and thus we proposed to investigate the orthologous protein (SCO1480) in Streptomyces coelicolor. Streptomyces bacteria have an unusual life cycle, and because of this, some genes that are essential in the mycobacteria are not necessary to sustain growth of the streptomycetes. The streptomycetes and mycobacteria share a conserved genetic core, which encompasses most housekeeping and essential genes (15–17), and includes SCO1480/mIHF. They do, however, have very distinct life cycles. Mycobacteria are typically rod-shaped, grow by polar tip extension and divide by binary fission like many other bacteria (18). The streptomycetes also exhibit polarized tip growth, but instead of undergoing regular cell division, they grow filamentously, first establishing a branching vegetative mycelium and later raising aerial hyphae (19). These aerial hyphae undergo a defined maturation process that starts with coordinated septation which subdivides the hyphal filaments into pre-spore compartments and culminates with the differentiation of these compartments into chains of exospores (20). The streptomycetes are also distinguished by their prodigious secondary metabolic capabilities. Many of these metabolites have been co-opted for medical application and include a vast array of antibiotics (21). Secondary metabolism is intricately linked with the Streptomyces developmental program: aerial hyphae formation and antibiotic production are coordinately regulated, although the two processes are spatially segregated, with secondary metabolism occurring primarily in the vegetative hyphae (22). This unusual Streptomyces life cycle means that cell division and associated processes (e.g. chromosome segregation) are dispensable for viability, as cell division is only required during sporulation, while they would be essential in Mycobacterium.
Here, we probe the role of the mIHF orthologue, SCO1480—or ‘sIHF’ (23), in S. coelicolor, and provide evidence that these proteins constitute a new class of nucleoid-associated protein in the actinobacteria. We find that sIHF, unlike mIHF, is not essential for S. coelicolor viability, and instead is required for normal chromosome compaction, sporulation and secondary metabolism. We demonstrate that sIHF associates with the nucleoid in vivo, and apart from a preference for double-stranded over single-stranded DNA, it appears to bind DNA in a non-specific, but length-dependent manner. The crystal structure revealed that a monomer of sIHF associates with the minor groove of the DNA. sIHF interacts simultaneously with two DNA duplexes, and thus may have the capacity to bridge different DNA molecules. We have also found sIHF to impact the activity of topoisomerase in vitro, significantly affecting DNA topology. Taken together, these results suggest that sIHF and its orthologues are likely to function as novel actinobacterial-specific nucleoid-associated proteins.
MATERIALS AND METHODS
Bacterial strains and media
Streptomyces strains were grown on mannitol–soy flour (MS) agar, rich (glucose-containing) R2YE agar, or Difco nutrient agar (DNA), or in a 1:1 mixture of yeast extract–malt extract and tryptic soy broth (YEME-TSB) complex liquid medium (24). Strains were grown at 30°C for up to 10 days. Escherichia coli strains were grown at 37°C in liquid LB or SOB medium, or on LB agar plates, except for strains BW25113/pIJ790, which was grown at 30°C, and BT340, which was grown at either 30°C or 42°C. Strains and plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids
| Streptomyces strains | Genotype | Reference |
|---|---|---|
| M600 | SCP1− SCP2− | Chakraburtty and Bibb (25) |
| E310 | M600 ΔSCO1480(sIHF)::[aac(3)IV)-oriT] | This work |
| E310a | M600 ΔSCO1480(sIHF)::FRT | This work |
| Escherichia coli strains | Use | Reference |
| DH5α | Plasmid construction and general subcloning | Invitrogen |
| ET12567/pUZ8002 | Generation of methylation-free plasmid DNA | Paget et al. (26) |
| BW25113 | Construction of cosmid-based knockouts | Gust et al. (27) |
| BL21 DE3 pLysS Rosetta | Protein overexpression | Novagen |
| BT340 | DH5α carrying a ‘FLP recombination’ plasmid | Datsenko and Wanner (28) |
| Plasmids | Description and use | Reference |
| pIJ2925 | General cloning vector | Janssen and Bibb (29) |
| pIJ773 | Plasmid carrying the apramycin knockout cassette | Gust et al. (27) |
| pIJ790 | Temperature sensitive plasmid carrying λ-RED genes | Gust et al. (27) |
| pIJ8660 | Integrating plasmid vector carrying eGFP | Sun et al. (30) |
| pIJ82 | Hygromycin-resistant integrating plasmid vector | Gift from H. Kieser |
| pET15b | 6×His-tag protein fusion overexpression vector | Novagen |
| pMC153 | pIJ8660 + sIHF; sIHF-eGFP localization | This work |
| pMC154 | pIJ82 + sIHF; ΔsIHF complementation plasmid | This work |
| pMC155 | pET15b + sIHF; overexpression of sIHF | This work |
| pMC141 | pET15b + srtA; overexpression of SrtA | Duong et al. (31) |
| pET11a | Protein overexpression vector | Novagen |
| pAG8744 | pET11a + topA; overexpression of TopA | This work |
| St9C5 | Cosmid for sIHF knockout | Redenbach et al. (32) |
Creation of a SCO1480/sIHF knockout strain
An in-frame deletion of SCO1480/sIHF was generated using the ReDirect system described by Gust et al. (27). The sIHF-coding sequence (from start codon to stop codon) was initially replaced by an oriT-containing apramycin resistance cassette, before it was removed using Flp-mediated recombination, as described previously (27). Cassette removal was confirmed by testing the apramycin sensitivity of the mutant strain and further verified by PCR using primers SCO1480 ko 1/ko 2 (Supplementary Table S1). All subsequent phenotypic analyses were conducted using this in-frame deletion strain.
To complement the ΔsIHF-mutant phenotype, the sIHF-coding sequence was amplified, together with additional upstream (284 nt) and downstream (156 nt) sequences, using primers SCO1480 up and SCO1480 down (Supplementary Table S1). The PCR-amplified product was phosphorylated and cloned into the EcoRV site of the integrating vector pIJ82 (Table 1). The resulting construct was introduced into the ΔsIHF-mutant strain (E310a) via conjugation from E. coli strain ET12567/pUZ8002 (Table 1). pIJ82 alone was also introduced into both the mutant- and wild-type M600 strains as a control.
Construction of a sIHF–eGFP translational fusion
The sIHF-coding sequence, along with 284 nt of upstream sequence, was amplified using Fuse 1 and Fuse 2 primers (Supplementary Table S1). These primers included BamHI and NdeI sites, respectively, within their 5′-ends to allow the resulting amplified product to be digested with the corresponding enzymes and cloned into the equivalent sites upstream of encoding an enhanced green fluorescent protein (eGFP) in pIJ8660 (Table 1). The Fuse 2 primer also included an extra 30 nt encoding a flexible linker peptide (LPGPELPGPE) to facilitate proper folding and functioning of the sIHF–eGFP fusion protein. This translational fusion was introduced into the ΔsIHF-mutant strain by conjugation (24) and was determined to function in place of the wild-type sIHF by testing for complementation of the mutant phenotype (antibiotic production, spore pigmentation and spore size).
Light, fluorescence and scanning electron microscopy
Samples for light and fluorescence microscopy were obtained by growing wild type, mutant and complementation strains along the underside of a sterile coverslip inserted into MS agar at a 45° angle. After 5 days, the coverslip was removed from the agar and adherent cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) diluted in a SlowFade solution (1:250; Invitrogen) and mounted onto a microscope slide. All images were obtained with a Leica DMI 6000 B wide-field deconvolution microscope using the Leica HCS Plan Apo oil immersion objective (magnification: ×100; numerical aperture: 1.4; Leica Microsystems, Wetzlar, Germany). Spore lengths, nucleoid areas and nucleoid fluorescence were all determined using ImageJ software (33). When visualizing eGFP, control strains (harbouring empty pIJ8660) were first examined to adjust fluorescence levels such that there was no detectable GFP signal, before imaging the experimental strains. Scanning electron microscopy was conducted on strains grown for 5 days on MS agar medium. Samples were prepared and visualized as described previously (34).
Antibiotic production assay
Equal numbers of wild type and mutant spores were used to inoculate 10 ml YEME-TSB liquid starter cultures that were grown for 2 days at 30°C. From these, an equivalent amount of biomass (∼0.25 g) was used to inoculate 100 ml of YEME-TSB liquid medium. Levels of actinorhodin and undecylprodigiosin were measured every 24 h after inoculation for 6 days and were quantified using techniques described previously (35,36). Calcium-dependent antibiotic (CDA) bioassays were performed as outlined previously (24,37).
sIHF overexpression, purification, antibody generation and immunoblot analysis
sIHF was PCR amplified from chromosomal DNA using primers SCO1480 Nde and SCO1480 Bam (Supplementary Table S1). The resulting PCR product was introduced into SmaI-digested and dephosphorylated pIJ2925 (Table 1). sIHF was then excised by digestion with NdeI and BamHI and cloned into a similarly digested pET15b vector (Table 1). It was then sequenced and introduced into E. coli Rosetta cells (Table 1). Overexpression of 6×His-sIHF was achieved by growing cultures at 37°C to mid-exponential phase, before adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and growing them overnight at 26°C. Cells were resuspended in binding buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole, pH 8.0) containing 1 mg/ml lysozyme and one complete mini EDTA-free protease inhibitor pellet (Roche), and incubated on ice for 30 min before being lysed by sonication. 6×His-sIHF was purified by passing the soluble protein extract over a Ni-NTA column (BioRad), washing with binding buffer supplemented with increasing concentrations of imidazole and eluting with 150 mM imidazole. The purified protein was then dialyzed into storage buffer (50 mM Tris, pH 7.5, 150 mM NaCl and 1 mM dithiothreitol [DTT]) and used for polyclonal antibody generation (Cedarlane Labs).
Protein samples were separated by sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (PAGE) and were either visualized by staining with Coomassie Brilliant Blue to ensure equal sample loading, or were transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked for 3 h at room temperature with Tris-buffered saline tween (TBST) 20, containing 10 mM Tris, pH 8.0, 100 mM NaCl and 0.05% Tween 20) containing 6% fat-free skim milk and subsequently incubated overnight at 4°C in TBST/6% skim milk containing crude polyclonal sIHF antibody (1:7000 dilution). The membrane was then washed three times with TBST, incubated for 30 min in TBST/6% skim milk containing goat anti-rabbit secondary antibody (1:3500 dilution) before again being washed three times with TBST.
Purification of untagged sIHF
The 6×His-sIHF was purified using a HiTrap Ni-chelating affinity column (GE Healthcare) equilibrated with 20 mM Tris, pH 8.0, 300 mM NaCl, 1.4 mM β-mercaptoethanol and 5% glycerol. Impurities were eliminated with a step gradient and 6×His-sIHF was eluted with 150 mM imidazole. Fractions containing 6×His-sIHF were pooled, diluted to adjust the salt concentration and loaded onto a 10/100 MonoS column (GE Healthcare) equilibrated with 20 mM Tris, pH 8.0, 100 mM NaCl, 1.4 mM β-mercaptoethanol and 5% glycerol. 6×His-sIHF was eluted using a linear salt gradient to 1 M NaCl. Fractions containing pure 6×His-sIHF were pooled, concentrated to 0.5 mg/ml and stored in storage buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1.4 mM β-mercaptoethanol and 5% glycerol). To cleave the histidine tag, 6×His-sIHF at 0.5 mg/ml was incubated with 0.03 units/µl of thrombin at room temperature for 60 min. The reaction was stopped with 1 mM benzamidine and sIHF was subsequently separated from the 6×His tag and thrombin over a 5/50 MonoS column (GE Healthcare).
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed using 8–15% native polyacrylamide gels and [γ-32P]dATP 5′-end-labelled probes. Increasing concentrations of sIHF (0–100 µM) were combined with 0.1 µM probe, 1 mg/ml bovine serum albumin (BSA) and binding buffer (10 mM Tris, pH 7.8, 5 mM MgCl2, 60 mM KCl and 10% glycerol) and incubated at room temperature for 10 min and then on ice for 30 min prior to adding a glycerol-based loading dye. To test binding specificity, increasing concentrations of poly(dI-dC) (0–5 µg; Roche) were added together with 10 µM sIHF to the EMSA reactions described above. Gels were exposed to Kodak Biomax XAR film at room temperature for ∼20 min before being developed. For probing sIHF binding to supercoiled (pUC19) versus linear (pUC19 digested with XbaI) DNA, 8 nM DNA was incubated with 0.08–17.3 µM sIHF in binding buffer as above, before reactions were separated on 1% TBE agarose gels and visualized by staining with ethidium bromide.
Cloning, protein overexpression and purification of TopA
The S. coelicolor topA-coding sequence was amplified from genomic DNA using primers topA1 and topA2 (Supplementary Table S1), and subcloned into pET11a using the NdeI and BamHI restriction sites to generate pAG8744. pAG8744 was transformed into BL21(DE3), grown to an OD600 of 0.7 and protein production was induced by adding 1 mM IPTG and incubating the culture for 3 h at 37°C. Cell pellets were resuspended in lysis buffer (20 mM Tris, pH 7.5, 25 mM KCl, 1.4 mM β-mercaptoethanol and 5% glycerol), lysed by sonication and cleared by centrifugation at 40 000g for 40 min. The supernatant was loaded onto a HiTrap SP HP column (GE Healthcare) equilibrated with lysis buffer. TopA was eluted using a linear gradient to 500 mM KCl. Fractions containing TopA were pooled together, concentrated to 3.5 mg/ml and stored at −80°C in 20 mM Tris, pH 7.5, 1.4 mM β-mercaptoethanol, 100 mM KCl and 25% glycerol.
Topoisomerase activity assays
The effect of sIHF on plasmids in vivo was assayed using E. coli Rosetta cells containing the 6×His-sIHF overexpression plasmid. Cells were grown to an OD600 of ∼0.4, at which point expression of the sIHF fusion was induced using 1 mM IPTG. As a negative control, equivalent cultures were grown without IPTG induction. To test whether general protein overexpression led to changes in plasmid structure, the same experiment was conducted using pET15b containing srtA [pMC141; (31)], where srtA encodes an endopeptidase used to anchor proteins to the surface of Gram-positive cells. Plasmid DNA was extracted using the PureLink™ Quick Plasmid Miniprep Kit (Invitrogen) 8 h after induction, and the resulting DNA was separated by electrophoresis on a 0.8% agarose gel. Cell-free extracts for each culture (induced and uninduced) were also prepared and were separated on 12–18% polyacrylamide gels and stained with Coomassie Brilliant Blue to ensure both sIHF and SrtA were effectively expressed after 8 h of induction.
To assay the effect of sIHF or a negative control (lysozyme) on the activity of TopA, 5 µl of pUC19 (64 nM) was incubated with 1 µl of TopA (8 µg/ml), 0.6 µL of BSA (1 mg/ml), 8.4 µl of reaction buffer (50 mM Tris, pH 7.5, 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT and 0.06 mg/ml BSA) and 5 µl of either sIHF or lysozyme at 8.64, 17.28 or 34.56 µM (1:135, 1:270 and 1:540 [DNA:protein] molar excess, respectively). To ensure that neither sIHF nor lysozyme alone affected plasmid mobility, binding reactions were conducted as described above, only substituting storage buffer for TopA. Additional reactions at lower DNA:sIHF molar excesses were conducted using sIHF at lower concentrations (1:17, 1:34, 1:68, 1:135, 1:270 and 1:540). Reactions were incubated at room temperature for 30 min and quenched with 5 µl of stop buffer (6% SDS, 30% glycerol, 10 mM EDTA and 0.25% bromophenol blue). All samples were resolved on 1% TAE-agarose gels, stained with ethidium bromide and visualized with UV light.
Protein crystallization
Complementary oligonucleotides 20 bp OH1 and 20 bp OH2 (Supplementary Table S1) were purchased from IDT, resuspended in deionized water to a final concentration of 6 mM, and annealed to form a 19-bp duplex (3 mM) with a single overhang on each duplex end. sIHF:DNA complexes (1:1 ratio) were prepared by mixing equal volumes of sIHF (3 mM) and DNA (3 mM), incubated for 10 min at room temperature and subsequently stored at 4°C. Crystals were grown at 4°C in 19% PEG 3350, 210 mM KSCN, 5% ethylene glycol, 100 mM HEPES, pH 7.6 and reached their final dimensions in ∼2 weeks. Crystals were cryoprotected by dehydration against increasing concentrations of KCl (1–1.5 M) prior to flash freezing in liquid nitrogen. Seleno-methionine (SeMet)-labelled protein was produced in minimal medium supplemented with SeMet, as described elsewhere (38) and optimal crystals were grown in 18% PEG 3350, 210 mM KSCN, 5% ethylene glycol and 100 mM HEPES pH 7.6 from sIHF:DNA complexes prepared at 1:1.4 ratios.
Data collection and structure determination
A complete data set of SeMet-labelled sIHF:DNA was collected at X25 beam line in NSLS (Brookhaven National Laboratory, NY, USA). Data were indexed, processed and merged using HKL2000 (39) and the structure was phased by SAD using SOLVE (40). The initial model was refined by iterative cycles of manual building in Coot and refinement in phenix.refine (41,42). The final model encompasses residues 14–105 and has all residues within the most favoured regions in the Ramachandran plot (Supplementary Table S2).
RESULTS
sIHF is expressed throughout the S. coelicolor life cycle
As a first step in investigating the role of sIHF in S. coelicolor, we overexpressed and purified it as a 6×His-tagged fusion protein and generated α-sIHF polyclonal antibodies. We followed sIHF expression throughout the life cycle of S. coelicolor (over a 4-day time course), during growth on either MS (poor carbon source) or R2YE (rich) agar media. Immunoblotting revealed that the cytoplasmic sIHF protein was present throughout development on MS agar medium, with levels gradually decreasing as development proceeded (Figure 1). On R2YE, levels were roughly equivalent at all times apart from 72 h, where there was a modest dip in expression (Figure 1). This is in contrast to mIHF and IHF, which typically accumulate later in growth (10,12,13). To ensure that we were examining equivalent amounts of protein, we separated cell-free extract samples on an SDS–polyacrylamide gel and stained the fractionated proteins with Coomassie Brilliant Blue (Supplementary Figure S1).
Figure 1.
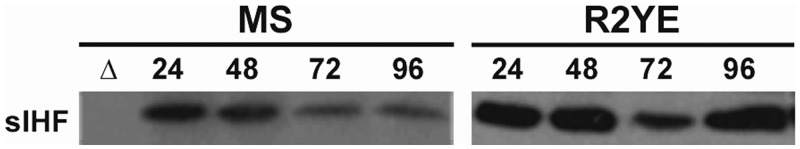
Western blot analysis of sIHF production. sIHF production is shown in wild-type S. coelicolor when grown on MS or R2YE media. Samples were taken at the indicated times (hours). The ΔsIHF-mutant strain (Δ) was used as a negative control.
Loss of sIHF impacts development and antibiotic production
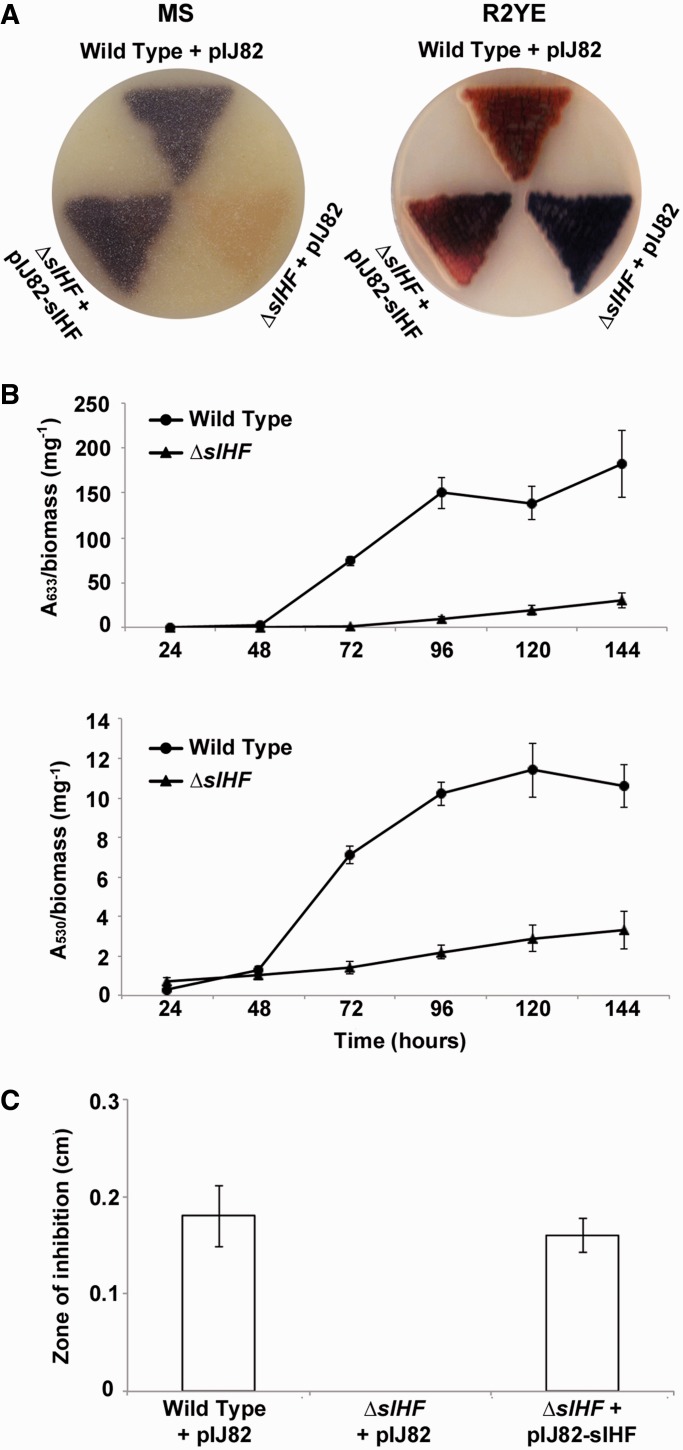
Despite the essential nature of the sIHF orthologue in Mycobacterium (mIHF), it turned out to be remarkably straightforward to delete sIHF in S. coelicolor. One of the most immediately striking phenotypes of the sIHF knockout mutant was its aberrant production of pigmented antibiotics when grown either on solid agar (Figure 2A) or in liquid culture (Figure 2B). During growth on MS agar medium, there was no obvious actinorhodin (blue) production, and little undecylprodigiosin (red) production relative to its wild-type parental strain; however, on R2YE, the mutant exhibited enhanced actinorhodin production (Figure 2A). During growth in a complex liquid medium (YEME-TSB), production of both pigmented antibiotics was significantly delayed and dramatically reduced for the ΔsIHF-mutant strain compared with the wild type (Figure 2B). We also tested CDA production using a plate-based bioassay and found that after 48 h of growth, the mutant failed to produce any CDA, while the wild-type strain produced levels that effectively inhibited growth of the indicator strain (Staphylococcus aureus; Figure 2C). The mutant phenotypes could be restored to near wild-type levels by re-introducing the wild-type sIHF gene under the control of its own promoter, on the integrating plasmid vector pIJ82 (Figure 2A and C).
Figure 2.
Streptomyces coelicolor antibiotic production in the absence of sIHF. (A) Plasmid-containing wild type (M600) and ΔsIHF in-frame deletion strains, as well as the complemented ΔsIHF-mutant strain, were grown on MS (left) and R2YE (right) agar media for 5 and 4 days, respectively. Images were taken from the underside of each plate to effectively display antibiotic production. (B) Actinorhodin (top) and undecylprodigiosin (bottom) production by wild-type and ΔsIHF-mutant strains in complex liquid YEME-TSB medium over a 6-day time course. (C) CDA production assay by plasmid-containing wild-type and ΔsIHF-mutant strains, along with the complemented ΔsIHF-mutant strain.
We also found the ΔsIHF mutant grew more slowly than the wild type, and exhibited sporulation defects, including reduced levels of sporulation, as well as aberrant spore septum placement and consequently greater heterogeneity in spore size (Supplementary Figure S2), confirming previous observations reported by Yang et al. (23).
Aberrant chromosome compaction and segregation during sporulation of an sIHF-mutant strain
During vegetative and aerial hyphal growth, chromosomes appear diffuse, and it is only during sporulation—the reproductive phase of Streptomyces development—that the chromosomes are compacted and segregated into the future spore compartments. Given the abnormal spores resulting from the deletion of sIHF, we were interested in determining whether sIHF-mutant strains also exhibited defects in chromosome organization during sporulation.
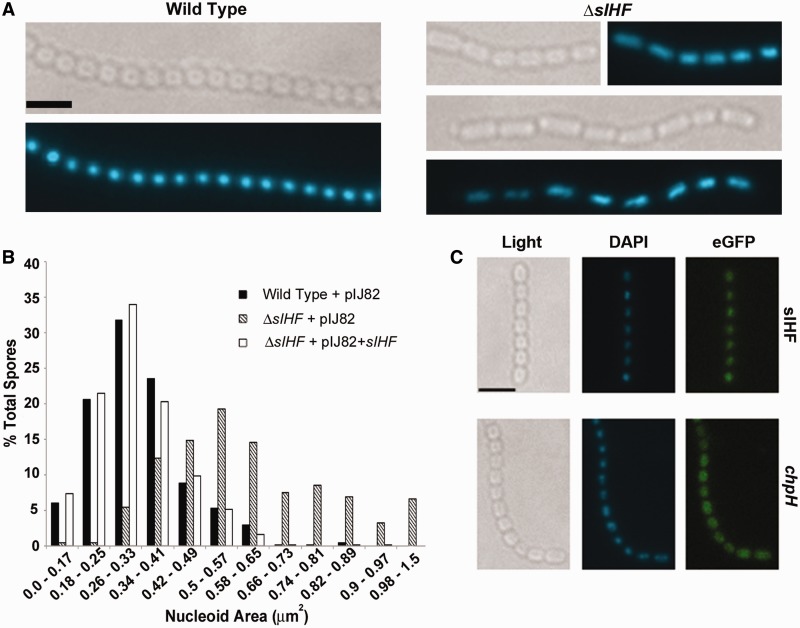
We first compared the overall DNA content of spores (n > 1000) and found that anucleate spores were far more common in the ΔsIHF-mutant strain (8.6%) than in the wild-type strain (0.3%). Introducing a wild-type copy of sIHF into the ΔsIHF mutant reduced the level of anucleate cells to ∼2%. When comparing the nucleoids of wild type and mutant spores, we also observed differences in their levels of chromosome compaction, with mutant spore DNA seeming more diffuse than those of the wild type, and in some instances adopting a ‘bilobed’ configuration (Figure 3A), although we did occasionally observe this for wild-type chromosomes as well (Figure 3C). We quantified the nucleoid area for both wild-type and ΔsIHF-mutant spores and found wild-type nucleoids were indeed more compact than those of the mutant, with the average size being 0.33 µm2, versus 0.60 µm2 for the ΔsIHF mutant (Figure 3B). Similar to what was observed for spore size, nucleoid area also showed a different distribution for the two strains, with >90% of wild-type nucleoids occupying an area of between 0.18 and 0.57 µm2, while the same proportion of mutant nucleoids exhibited a range of 0.31–0.98 µm2. Complementation with a wild-type copy of sIHF restored wild-type-like chromosome compaction in the ΔsIHF mutant (Figure 3B). We considered the possibility that the more diffuse nucleoids observed in the ΔsIHF mutant might reflect increased DNA content, but we failed to detect any significant differences in the DNA content of wild type and mutant spores (n > 1000), as determined by measuring DAPI fluorescence intensity per spore.
Figure 3.
sIHF localization within spore compartments relative to chromosomal DNA. (A) Light and DAPI fluorescence images of wild-type (left panels) and ΔsIHF-mutant (right panels) strains. Each image set depicts the same spore chain. The scale bar represents 2.6 µm. (B) Comparison of nucleoid areas for wild type, mutant and complemented strains, with calculated areas rounded to the nearest 0.01 µm2. 500–600 spores were examined for each strain. (C) Light and fluorescence microscopy (DAPI and GFP) images of wild-type spores expressing an sIHF-eGFP translational fusion (top panels) or PchpH-eGFP transcriptional fusion (bottom panels). The DAPI panel shows chromosomal DNA localization, while the eGFP panel shows the localization of eGFP when fused to either sIHF or the chpH promoter. Scale bar represents 3.2 µm.
Given the effect that sIHF deletion had on chromosome architecture in sporulating cells, we also probed the intracellular localization of sIHF using an sIHF–eGFP fusion. This construct effectively complemented the mutant phenotype (restoring antibiotic production on MS agar and wild-type spore size to the mutant; Supplementary Figure S3), indicating that the fusion protein was functional. Using fluorescence microscopy, we found sIHF–eGFP co-localized with the condensed nucleoid during sporulation (Figure 3C). To ensure that the pattern we observed reflected nucleoid association and not simply cytoplasmic distribution, we compared the fluorescence pattern observed for the sIHF–eGFP-containing strain, with that of wild-type spores expressing eGFP under the control of the chpH promoter, which is expressed at high levels during sporulation (43). In this instance, the eGFP pattern was more diffuse than that of sIHF–eGFP and occupied the entire spore compartment (Figure 3C). Considering the nucleoid localization of sIHF, together with the sporulation and chromosome segregation/compaction defects observed for the sIHF-mutant strain, this suggests that sIHF may play a key role in coupling changes in chromosome architecture with appropriate cell division during Streptomyces sporulation.
sIHF interacts non-specifically with double-stranded DNA
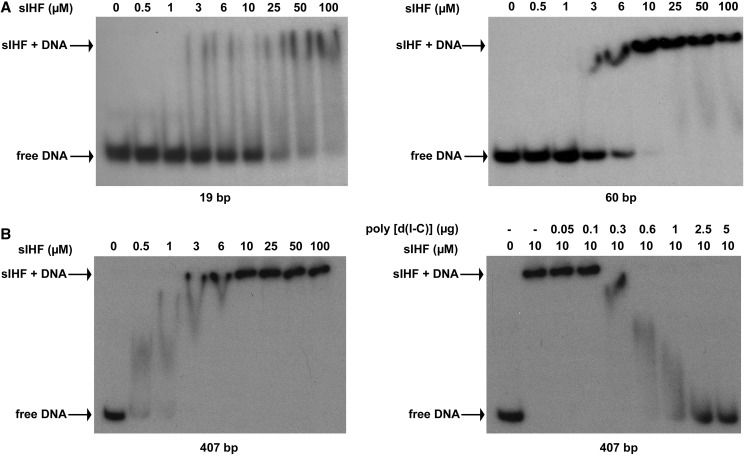
Recent studies have suggested a role for sIHF as a transcriptional regulator (23,44), and previous work on the mycobacterial orthologue mIHF showed it to associate indiscriminately with double-stranded DNA (9). We set out to systematically assess the DNA-binding specificity of sIHF using EMSAs. We started by incubating purified protein with a short DNA duplex (19 bp) and found the probe was shifted in a concentration-dependent manner, although a clearly defined shift was not observed (Figure 4A). To explore the length dependence of sIHF binding, we next tested two double-stranded DNA probes that were approximately two and three times the length of the 19 bp probe (43 and 60 bp, respectively); these probes corresponded to the coding sequence of a gene (SCO4676) whose product has no obvious role in the development or antibiotic production of S. coelicolor under the conditions examined here (Hindra and Elliot,M.A., unpublished data). In each case, a complete shift of all labelled probe was observed with 10 µM sIHF, giving a discrete complex at the top of the gel (Figure 4A and Supplementary Figure S4). When we extended the length of the labelled probe to 407 bp, sIHF binding was further enhanced: >50% of the probe was shifted in the presence of 0.5 µM sIHF, a complete shift was observed with 3 µM sIHF and 10 µM sIHF led to a discrete shifted complex that was unaffected by further protein addition (Figure 4B). These results imply that effective complex formation between sIHF and DNA is length dependent, and that sIHF binding may be cooperative.
Figure 4.
DNA binding by sIHF. (A) EMSAs showing sIHF binding to probes of increasing length. Increasing concentrations of sIHF were added to the indicated DNA probes. Panels represent the binding of sIHF to a 19-bp DNA probe (left) and a 60-bp DNA probe (right). (B) EMSAs showing the binding affinity of sIHF to a 407-bp DNA probe. The left panel shows sIHF–DNA complex formation in the presence of increasing concentrations of sIHF. The right panel shows the effect of increasing poly(dI-dC) concentrations on sIHF–probe complexes.
We next probed the binding specificity of sIHF, conducting competition assays using increasing concentrations of poly(dI-dC), together with the labelled 60 (Supplementary Figure S4) and 407 bp probes (Figure 4B). We found that as little as 0.3 µg of poly(dI-dC) was sufficient to eliminate all binding to the 60 bp probe, while 2.5 µg of poly(dI-dC) abolished binding to the 407 bp probe, confirming that there was little specificity associated with sIHF–DNA binding. This is notable, when considering that 1 µg of poly(dI-dC) (three times the concentration needed to eliminate all binding to the 60 bp probe) is frequently used as an internal competitor in DNA-binding reactions (e.g. [37,45]). Interestingly, the amount of poly(dI-dC) required to effectively compete for sIHF binding was proportional to the length of the labelled probe: the 407 bp probe is approximately seven times longer than the 60 bp probe, and required eight times the amount of poly(dI-dC) for full competition. These results all support a model in which sIHF binds DNA without any obvious sequence specificity, in a concentration and length-dependent manner.
sIHF specificity in binding to DNA with distinct conformations
Having determined that sIHF was required for normal chromosome segregation and compaction through our genetic analyses, and having shown that it was a promiscuous binding protein in vitro, we were interested in trying to understand how its DNA-binding activity impacted chromosome dynamics in the cell. Many nucleoid-associated proteins bind DNA without any defined sequence specificity, but preferentially associate with defined DNA structures. For example, in E. coli, H-NS binds curved DNA with highest affinity (46,47), while HU has strong affinity for distorted (e.g. gapped) DNA configurations (48). We therefore set out to determine whether sIHF had increased affinity for DNA having different structures, or whether it influenced DNA structure upon binding. We began by comparing the affinity of sIHF for linear DNA versus curved and gapped sequences, but found no reproducible difference in sIHF affinity for any of these different DNA conformations (data not shown). We also compared the relative affinity of sIHF for linear versus supercoiled DNA, and again, we observed equivalent binding to both DNA forms (data not shown). We went on to investigate the ability of sIHF to associate with single-stranded DNA, as this has been shown to be a substrate for several nucleoid-associated proteins in other bacteria, including the SMC protein in Bacillus subtilis (49). We tested binding using labelled probes corresponding to either strand of the 60 bp probe tested above. These single-stranded sequences did not have any significant hairpin potential or extended self-complementarity. While we observed concentration-dependent interactions with the single-stranded probes, complete shifting of the probe was never observed, even in the presence of 100 µM sIHF (Supplementary Figure S4). This is in stark contrast to the equivalent double-stranded sequence, where 10 µM sIHF was sufficient to shift all probe (Figure 4A), indicating that sIHF has a strong preference for double-stranded DNA, relative to single stranded. To determine whether sIHF influenced DNA structure upon binding, as has been observed for nucleoid-associated proteins such as IHF (50,51), we tested the ability of sIHF to bend DNA upon binding using FRET analysis, but did not detect any obvious changes in fluorescence when comparing samples incubated in the presence or absence of sIHF (data not shown). Collectively, these results suggest that, apart from preferentially binding double-stranded DNA, sIHF does not discriminate between different DNA configurations, at least in vitro.
sIHF enhances topoisomerase activity
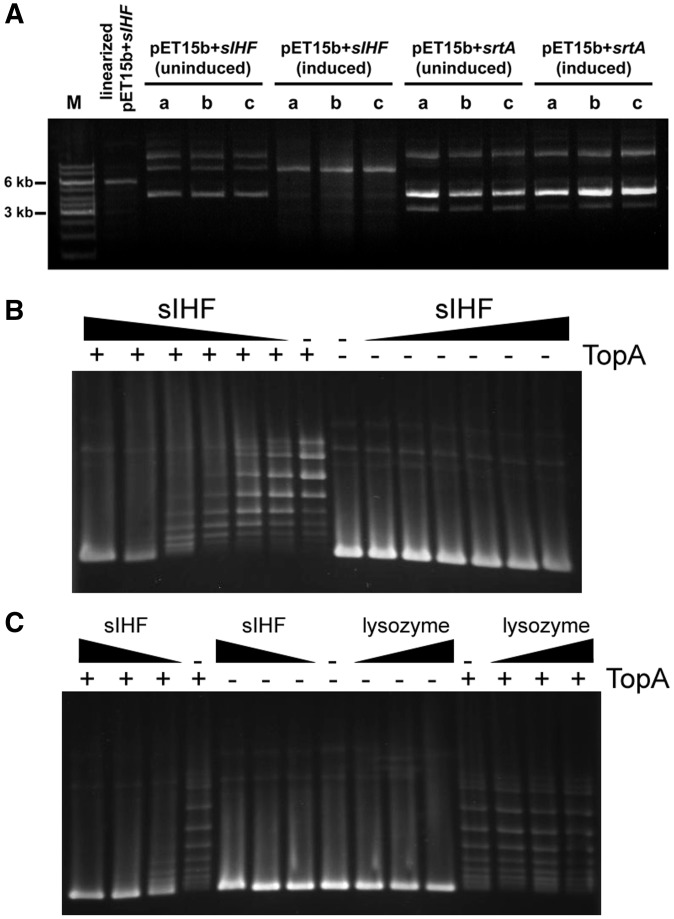
In an attempt to further probe the means by which sIHF impacts chromosome organization, we wanted to assess its effect on enzymes that influence DNA supercoiling and chromosome compaction. As a first step, we took advantage of our sIHF overexpression construct, and introduced this construct into E. coli, and compared the topology of plasmids isolated following sIHF overexpression/induction relative with those isolated from an uninduced strain. We consistently observed different plasmid topologies for the sIHF overexpressing strain relative to its uninduced control (Figure 5A). To ensure that the addition of IPTG did not impact plasmid conformation, we also examined plasmids isolated from strains expressing an unrelated protein (SrtA) from the same plasmid, and we did not see any differences in plasmid configuration in the induced versus uninduced strain (Figure 5A). We confirmed in each case, that expression of sIHF and SrtA had been effectively induced, and that these proteins were present at much higher levels than in the uninduced cultures (Supplementary Figure S5).
Figure 5.
sIHF alters plasmid DNA supercoiling. (A) pET15b containing either sIHF (left) or srtA (right), isolated from E. coli. IPTG was added to three independent cultures (a, b and c) to induce overexpression of sIHF and srtA, while three others were grown without induction prior to plasmid harvest. For size comparison, a marker (M) was run alongside linearized (NdeI digested) pET15b + sIHF. (B) The minimal amount of sIHF able to affect TopA activity was assayed by incubating supercoiled pUC19 plasmid DNA with serially diluted sIHF (1:540 to 1:17 [pUC19:sIHF] molar excess) in the presence/absence of TopA. (C) Supercoiled pUC19 plasmid DNA was incubated alone, or together with increasing concentrations of either sIHF or lysozyme (at 1:135, 1:270 and 1:540 [DNA:protein] molar excess), in the presence/absence of S. coelicolor TopA (8 µg/ml) as indicated.
These results suggested that sIHF might influence DNA supercoiling. To further investigate this possibility, we conducted an in vitro assay aimed at assessing the effect of sIHF on the activity of TopA—the sole type I topoisomerase encoded by S. coelicolor. Using purified TopA, we followed its activity in the presence and absence of sIHF, using supercoiled E. coli plasmid DNA as a substrate. We found sIHF had a profound effect on plasmid topology, appearing to counteract the relaxation activity of TopA (Figure 5B). Importantly, sIHF alone did not affect plasmid supercoiling and relaxation, nor was the sIHF concentration used for these reactions sufficient to shift the plasmid DNA (Figure 5B and C). To confirm that the observed effects were specific to sIHF, we repeated the experiment, substituting lysozyme (another small protein) for sIHF, and did not observe any change in plasmid topology (Figure 5C). These findings imply that sIHF makes a significant contribution to DNA conformation and topology in a topoisomerase-dependent manner.
Structure of sIHF bound to DNA
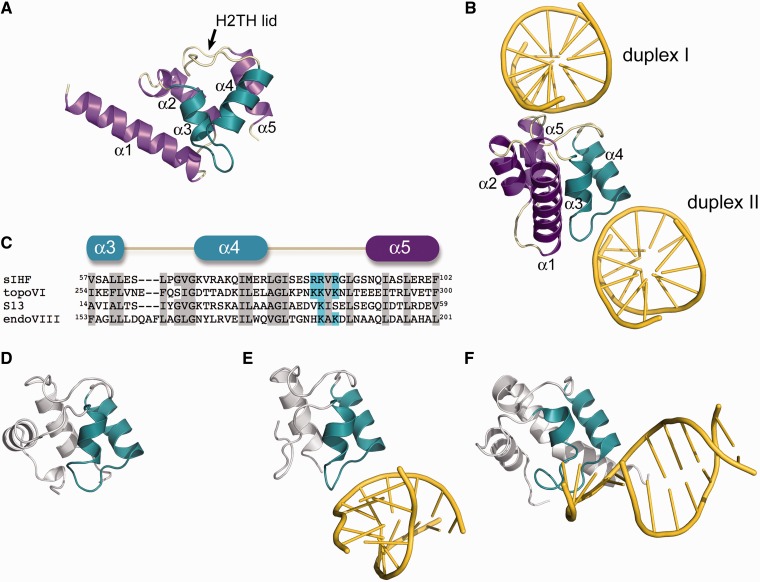
The in vitro and in vivo behaviour of sIHF determined to this point were not consistent with that of any known nucleoid-associated protein, and while the activity of the sIHF orthologue in the mycobacteria (mIHF) in promoting phage DNA integration mirrored that of IHF in E. coli, we suspected that neither sIHF nor mIHF were structural or functional homologues of IHF. We therefore sought to determine the crystal structure of sIHF in complex with the 19-bp DNA duplex used for our initial EMSA experiments (Figure 4A). Crystals of sIHF bound to DNA diffracted to 2.7 Å resolution and, of the 107 amino acids of sIHF, we were able to model residues 14–103 in the experimental electron density maps (Supplementary Table S2). The sIHF structure is composed of a long and protruding N-terminal α-helix (α1) followed by four shorter α-helices that define the core of the protein (Figure 6A). The arrangement of the helical core resembles that of a small domain found in the structures of type IIb topoisomerases (specifically topoisomerase VI), the endonuclease VIII family of base excision repair enzymes and the ribosomal protein S13 (52–55) (Figure 6D–F). This domain includes a characteristic helix-two turns-helix (H2TH) motif, spanning α3 to α4 in sIHF (Figure 6C). While the specific function of the H2TH domain in these proteins is unknown, the structures of the small S13 ribosome subunit and endonuclease VIII bound to DNA reveal that the two turns connecting the helices of the motif may enhance binding to nucleic acids in an unspecific manner (Figure 6E and F).
Figure 6.
Structure of sIHF. (A) Ribbon diagram of sIHF with helices shown in purple, loops in cream and the H2TH motif coloured in teal. (B) Interaction of sIHF with neighbouring DNA molecules (shown in yellow). The view is rotated 90° with respect to the view in panel (A) and the two neighbouring DNA duplexes are labelled for reference. (C) Sequence alignment of the hydrophobic core of sIHF, the H2TH domain of topoisomerase VI, the N-terminal domain of the ribosomal protein S13 and the H2TH domain of endonuclease VIII. Conserved hydrophobic residues are shadowed in grey and conserved charged residues in the H2TH lid are highlighted in cyan. (D) Ribbon diagram of the H2TH domain of topoisomerase VI (residues Lys230-Phe306, PDB ID 1MU5). (E) Ribbon diagram of residues Ala1-Phe62 from the ribosomal protein S13 (PDB ID 2GY9). (F) Ribbon diagram of the H2TH domain of endonuclease VIII (residues Pro132-Gln214, PDB ID 1K3W).
sIHF binds DNA as a monomer, and although a 19-bp DNA duplex was used to assemble the sIHF–DNA complex, only eight nucleotides fit in the asymmetric unit. Adjacent DNA molecules stack end-to-end, forming a pseudo-continuous duplex that runs parallel to one axis of the unit cell. Given its lack of binding specificity, sIHF has the potential to associate with any sequence within these long duplexes, and it appears that the crystal captured an average distribution of sIHF over the entire length of the duplex DNA. Accordingly, the phosphate backbone of the DNA duplex was clearly defined in the experimental electron density maps (Figure 7), but the nucleotide sequence of the DNA duplex bound by sIHF could not be assigned, reinforcing the idea that the 8 bp in the asymmetric unit represent an average of different bases. Since we used a GC-rich sequence to prepare the duplex DNA, we modelled the base moieties as guanines or cytosines based on the relative size of the electron density.
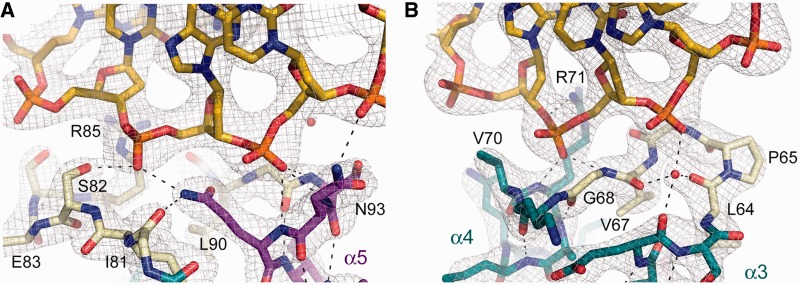
Figure 7.
sIHF–DNA interaction. (A) Detail of the interaction between the H2TH lid and duplex I. The refined model is shown as colour-coded sticks and the 2Fo-Fc electron density map contoured at 1.0 σ. Hydrogen bonds are shown as dashed lines. (B) Detail of the interaction between the H2TH motif and duplex II. The refined model is shown as colour-coded sticks and the 2Fo-Fc electron density map contoured at 1.0 σ.
sIHF contacts two of these pseudo-continuous DNA duplexes via distinct regions (Figures 6B and 7). The protein associates with duplex I through the 11-residue ‘lid’ that connects helices α4 and α5. This lid appears to have a dual function, serving to both conceal the hydrophobic core of the protein (Figure 6A) and provide a flat surface rich in positively charged residues (Figure 6C) that cradles the phosphate backbone of one of the DNA strands of duplex I. One of the duplex phosphate moieties further stabilizes this interaction by neutralizing the dipole moment of helix α5 (Figure 7A). Interestingly, this lid adopts a nearly identical conformation in the structures of topoisomerase VI and ribosomal protein S13 (Figure 6D–E). In the monomer and dimer structures of topoisomerase VI, the lid is exposed to the solvent and is more accessible than the H2TH motif in sIHF; however, it is not possible to infer a role for this feature in DNA binding by topoisomerase VI, as DNA is not present in that structure (52).
The interface between sIHF and duplex II is less extensive than that of sIHF and duplex I, involving only the second turn of the H2TH motif and one phosphate moiety neutralizing the dipole moment of helix α4 (Figure 7B). However, this interface loosely resembles the interaction of both S13 with RNA and the H2TH motif of endonuclease VIII with DNA (Figure 6B, E and F). The H2TH motifs of S13 and endonuclease VIII are, however, embedded within larger structures that make extensive contacts with DNA or RNA molecules, and hence for these proteins, the DNA-binding role of the H2TH domain is predicted to be largely peripheral (54,56,57), with other protein domains ensuring proper DNA binding.
DISCUSSION
We have shown here that sIHF makes important contributions to Streptomyces development and secondary metabolism, but is dispensable for growth and viability. Loss of sIHF leads to defects in chromosome compaction, chromosome segregation, antibiotic production and spore septum placement. Collectively, our data strongly support a role for sIHF as a nucleoid-associated protein governing chromosome dynamics in S. coelicolor. This function is most evident during sporulation, which is the only time in the Streptomyces life cycle that chromosome compaction and segregation occur. While a number of classical nucleoid-associated proteins in S. coelicolor impact chromosome behaviour during sporulation, ΔsIHF mutants exhibit distinct phenotypic characteristics when compared with these other mutant strains. For example, the increased nucleoid size observed for the ΔsIHF mutant is a property shared with smc (58), hupS (59) and dpsA (60) mutants in S. coelicolor. These all encode classical nucleoid-associated proteins: SMC proteins play a pivotal role in the structural maintenance of chromosomes (61); HupS is one of two HU-like proteins encoded by S. coelicolor (59); and Dps-like proteins protect DNA from stressful conditions, in part through their ability to compact the chromosome (62). In a hupS mutant, chromosome condensation defects are coupled with reduced spore pigmentation and compromised spore dormancy, but the spores themselves are of uniform shape and size and contain appropriately segregated chromosomes (59), unlike those of the ΔsIHF mutant. Mutations in smc do not affect spore shape but lead to increased anucleate spores (58,63), equivalent to that observed here for the ΔsIHF mutants (∼7–8%). The ‘bilobed’ chromosomal architecture occasionally observed for the ΔsIHF mutant was not shared with either smc or hupS mutants, but has been seen in scpA and scpB mutants (63), where these genes encode two accessory proteins required for SMC function (64). Finally, dpsA mutants exhibit variation in spore size and defects in chromosome condensation, like ΔsIHF mutants, but have wild-type levels of anucleate spores (60). It will be interesting to see whether there is any functional redundancy shared by sIHF and these diverse nucleoid-associated proteins.
Like many well-characterized nucleoid-associated proteins in E. coli, sIHF is a small, positively charged protein. It is, however, structurally unrelated to not only the archetypal IHF, HU and H-NS proteins but also to all known nucleoid-associated proteins. The sIHF structure has revealed that it binds DNA as a monomer, and that DNA binding encompasses 8 bp. It appears to have no obvious DNA-binding specificity, based on both our DNA-binding assays and the sIHF–DNA crystal structure. We cannot, however, exclude the possibility that higher affinity sIHF-binding sequences exist until a comprehensive binding screen is undertaken. For instance, IHF can associate with non-specific DNA sequences, although it recognizes a preferred sequence with ∼1000-fold greater affinity (65). In binding DNA, sIHF interacts primarily with the phosphate backbone, but several positively charged residues interact with DNA through the minor groove—a feature shared with many architectural proteins including IHF- and HU-like proteins. sIHF does, however, lack an analogous proline residue found in every member of the HU/IHF family and, therefore, unlike these proteins, its binding does not lead to significant bending of the DNA (50). Indeed, the overall architecture and DNA-binding mechanism of sIHF are unrelated to other nucleoid-associated proteins, confirming that sIHF is not a homologue of IHF, and that it represents a new category of nucleoid-associated protein. The observation that sIHF has two potential DNA contact points suggests that it may have the capacity to bridge different DNA molecules, or stabilize (but not introduce) DNA bends. This differs from the DNA-bridging capabilities of H-NS and Lsr2, which bind single sites and bring disparately oriented DNA segments together through oligomerization.
Notably, other proteins with domains bearing structural similarity to the sIHF H2TH domain (endonuclease VIII, ribosomal protein S13 and topoisomerase VI) are all nucleic acid-binding proteins; however, for those whose structures have been solved in complex with either DNA (exonuclease VIII) or RNA (S13), their H2TH domains appear to contribute only peripherally to nucleic acid association (54,56,57). It is therefore possible that sIHF acts in conjunction with other proteins, perhaps through its extended N-terminal helix (residues 14–35 in the structure), as this helix does not contribute to DNA association, and yet is highly conserved. An intriguing interaction candidate would be the single type I topoisomerase (TopA) encoded by S. coelicolor and other actinobacteria, given that sIHF impacts DNA topology in a TopA-dependent manner in in vitro topoisomerase assays, and that the H2TH domain is also found in topoisomerase VI. It is worth noting, however, that topoisomerase VI is a type II topoisomerase (cleaves both DNA strands and changes the linking number by 2), while the S. coelicolor TopA is a type I enzyme (cleaves a single DNA strand and changes the linking number by 1), and thus these enzymes would act via different mechanisms.
IHF in E. coli functions to promote phage DNA integration, nucleoid compaction through its ability to bend DNA and gene regulation through its DNA-binding capabilities. While mIHF (the sIHF orthologue in mycobacteria) is required for phage integrase activity, it is unclear as to whether sIHF will have an equivalent role, as we found plasmid vectors bearing ϕC31-(pSET152/pIJ82) integration elements were readily incorporated into the chromosome of an ΔsIHF mutant. Interestingly, phage integrase activity often proceeds via a topoisomerase I-like mechanism (66), and as discussed above, we have shown here that sIHF has both functional and structural connections with topoisomerase enzymes. While TopA has not previously been implicated in chromosome dynamics during Streptomyces sporulation, investigations in other organisms have connected topoisomerase I with both plasmid and chromosome condensation/segregation (67–69). The striking effect of sIHF on TopA activity in vitro could provide a reasonable explanation for the reduced chromosome compaction observed for an sIHF mutant during sporulation. sIHF appears to promote supercoiling in a topoisomerase-dependent manner; if this accurately reflects its role in vivo, loss of sIHF would be expected to correlate with a more diffuse chromosome architecture.
In E. coli, many nucleoid-associated proteins also function as global transcription regulators. For example, IHF influences the expression of anywhere between 150 and 500 genes, while HU affects the expression (both positively and negatively) of >900 genes (70,71). Previous work (44) has suggested that sIHF may directly regulate antibiotic production in S. coelicolor by binding upstream of two pathway-specific regulatory genes: redD (which encodes a direct activator of undecylprodigiosin production) and actII-orf4 (which encodes the direct activator of actinorhodin production). Our work supports an important role for sIHF in antibiotic biosynthesis, as an sIHF mutant was significantly impaired in its antibiotic production under the conditions we tested. We are not confident, however, that this effect is a direct one, as both our work, and work by Yang et al. (23), seem to suggest that sIHF can bind any DNA fragment ≥19 bp in vitro. Irrespective of direct or indirect sIHF association with antibiotic biosynthetic genes, it is obvious that the effect of sIHF on antibiotic production is not straightforward. Yang et al. (23) had previously reported enhanced actinorhodin and undecylprodigiosin production by their sIHF mutant under all growth conditions examined, including rich (glucose-containing) agar medium, and minimal agar medium supplemented with different carbon sources (glucose, chitin) and various amino acids. Consistent with these observations, we found actinorhodin production by the sIHF mutant was enhanced during growth on rich agar medium. We did, however, observe reduced antibiotic (actinorhodin and CDA) levels for the sIHF mutant relative to that of the wild-type strain, during growth on other solid media (soy flour with mannitol [MS] agar; nutrient agar), suggesting that the effect of sIHF on antibiotic production may be carbon source dependent. We also observed differences during liquid culture, where growth in a rich (glucose-based) liquid medium resulted in markedly reduced levels of both actinorhodin and undecylprodigiosin by the sIHF mutant, compared with its wild-type parent. Taken together, these results suggest that sIHF-mediated antibiotic effects are differentially responsive to media composition and growth conditions.
The diverse, but reproducible, phenotypic characteristics associated with sIHF mutation range from defects in antibiotic production, to aberrant sporulation, chromosome segregation and chromosome compaction. This suggests that sIHF nucleoid association and DNA binding likely impact gene expression on a global level, and this will be an exciting avenue to pursue in the future.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited with the Protein Data Bank (accession code 4ITQ).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
McMaster University, a grant from the Canadian Institutes of Health Research [MOP-67189 to A.G.]; Canada Research Chairs (CRC) program (to M.A.E.) and a graduate student award from NSERC (to J.P.S.). Funding for open access charges: McMaster University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank David Capstick and Hindra for critical reading of the article and for helpful suggestions.
REFERENCES
- 1.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 2.Browning DF, Grainger DC, Busby SJW. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 5.Guo F, Adhya S. Spiral structure of Escherichia coli HUαβ provides foundation for DNA supercoiling. Proc. Natl Acad. Sci. USA. 2007;104:4309–4314. doi: 10.1073/pnas.0611686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Gordon BRG, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon BRG, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedulla ML, Lee MH, Lever DC, Hatfull GF. A novel host factor for integration of mycobacteriophage L5. Proc. Natl Acad. Sci. USA. 1996;93:15411–15416. doi: 10.1073/pnas.93.26.15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedulla ML, Hatfull GF. Characterization of the mIHF gene of Mycobacterium smegmatis. J. Bacteriol. 1998;180:5473–5477. doi: 10.1128/jb.180.20.5473-5477.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 12.Bushman W, Thompson JF, Vargas L, Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditto MD, Roberts D, Weisberg RA. Growth phase variation of integration host factor level in Escherichia coli. J. Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MH, Hatfull GF. Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J. Bacteriol. 1993;175:6836–6841. doi: 10.1128/jb.175.21.6836-6841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 16.Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harries DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 17.Cerdeño-Tárraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, Besra GS, Churcher C, James KD, De Zoysa A, et al. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 2003;31:6516–6523. doi: 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thanky NR, Young DB, Robertson BD. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis. 2007;87:231–236. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Flärdh K, Richards DM, Hempel AM, Howard M, Buttner MJ. Regulation of apical growth and hyphal branching in Streptomyces. Curr. Opin. Microbiol. 2012;15:737–743. doi: 10.1016/j.mib.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 21.Hopwood DA. Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford, NY: Oxford University Press; 2007. [Google Scholar]
- 22.Champness WC, Chater KF. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp. In: Piggot P, Moran CP, Youngman P, editors. Regulation of Bacterial Differentiation. Washington, DC: American Society for Microbiology; 1994. pp. 61–94. [Google Scholar]
- 23.Yang YH, Song E, Willemse J, Park SH, Kim WS, Kim EJ, Lee BR, Kim JN, van Wezel GP, Kim BG. A novel function of Streptomyces integration host factor (sIHF) in the control of antibiotic production and sporulation in Streptomyces coelicolor. Antonie Van Leeuwenhoek. 2012;101:479–492. doi: 10.1007/s10482-011-9657-z. [DOI] [PubMed] [Google Scholar]
- 24.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 25.Chakraburtty R, Bibb MJ. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J. Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen GR, Bibb MJ. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 30.Sun J, Kelemen GH, Fernández-Abalos JM, Bibb MJ. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- 31.Duong A, Capstick DS, Di Berardo C, Findlay KC, Hesketh A, Hong HJ, Elliot MA. Aerial development in Streptomyces coelicolor requires sortase activity. Mol. Microbiol. 2012;83:992–1005. doi: 10.1111/j.1365-2958.2012.07983.x. [DOI] [PubMed] [Google Scholar]
- 32.Redenbach M, Kieser HM, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood DA. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 33.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:33–42. [Google Scholar]
- 34.Haiser HJ, Yousef MR, Elliot MA. Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J. Bacteriol. 2009;191:6501–6512. doi: 10.1128/JB.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenzie NL, Nodwell JR. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J. Bacteriol. 2007;189:5284–5292. doi: 10.1128/JB.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang SG, Jin W, Bibb M, Lee KJ. Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3(2) grown in continuous culture. FEMS Microbiol. Lett. 1998;168:221–226. doi: 10.1111/j.1574-6968.1998.tb13277.x. [DOI] [PubMed] [Google Scholar]
- 37.Hindra, Pak P, Elliot MA. Regulation of a novel gene cluster involved in secondary metabolite production in Streptomyces coelicolor. J. Bacteriol. 2010;192:4973–4982. doi: 10.1128/JB.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta. Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonine PV, Gross-Kunstleve RW, Echols M, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SS, Yang YH, Song E, Kim EJ, Kim WS, Sohng JK, Lee HC, Liou KK, Kim BG. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2) J. Ind. Microbiol. Biotechnol. 2009;36:1073–1083. doi: 10.1007/s10295-009-0591-2. [DOI] [PubMed] [Google Scholar]
- 45.Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3 (2) Mol. Microbiol. 2001;40:257–269. doi: 10.1046/j.1365-2958.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- 46.Owen-Hughes TA, Pavitt GD, Santos DS, Sidebotham JM, Hulton CS, Hinton JCD, Higgins CF. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 47.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 48.Swinger KK, Rice PA. Structure-based analysis of HU-DNA binding. J. Mol. Biol. 2007;365:1005–1016. doi: 10.1016/j.jmb.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirano M, Hirano T. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 51.Lorenz M, Hillisch A, Goodman SD, Diekmann S. Global structure similarities of intact and nicked DNA complexed with IHF measured in solution by fluorescence resonance energy transfer. Nucleic Acids Res. 1999;27:4619–4625. doi: 10.1093/nar/27.23.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000;19:3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brodersen DE, Clemons WM, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 56.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 57.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 58.Kois A, Swiatek M, Jakimowicz D, Zakrzewska-Czerwińska J. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J. Bacteriol. 2009;191:310–319. doi: 10.1128/JB.00513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salerno P, Larsson J, Bucca G, Laing E, Smith CP, Flärdh K. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J. Bacteriol. 2009;191:6489–6500. doi: 10.1128/JB.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, Del Sol R. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 2009;73:1186–1202. doi: 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- 61.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 62.Nair S, Finkel SE. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004;186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dedrick RM, Wildschutte H, McCormick JR. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J. Bacteriol. 2009;191:320–332. doi: 10.1128/JB.00858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- 65.Wang S, Cosstick R, Gardner JF, Gumport RI. The specific binding of Escherichia coli integration host factor involves both major and minor grooves of DNA. Biochemistry. 1995;34:13082–13090. doi: 10.1021/bi00040a020. [DOI] [PubMed] [Google Scholar]
- 66.Kikuchi Y, Nash HA. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc. Natl Acad. Sci. USA. 1979;76:3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Q, Pongpech P, DiGate RJ. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc. Natl Acad. Sci. USA. 2001;98:9766–9771. doi: 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Minden JS, Marians KJ. Escherichia coli topoisomerase I can segregate replicating pBR322 daughter DNA molecules in vitro. J. Biol. Chem. 1986;261:11906–11917. [PubMed] [Google Scholar]
- 69.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 70.Gama-Castro S, Jiménez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muñiz-Rascado L, Martínez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee ASN, Luscombe NM. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012;40:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.