Abstract
Phage T7 helicase unwinds double-stranded DNA (dsDNA) by encircling one strand while excluding the complementary strand from its central channel. When T7 helicase translocates on single-stranded DNA (ssDNA), it has kilobase processivity; yet, it is unable to processively unwind linear dsDNA, even 60 base-pairs long. Particularly, the GC-rich dsDNAs are unwound with lower amplitudes under single-turnover conditions. Here, we provide evidence that T7 helicase switches from ssDNA to dsDNA during DNA unwinding. The switching propensity is higher when dsDNA is GC-rich or when the 3′-overhang of forked DNA is <15 bases. Once helicase encircles dsDNA, it travels along dsDNA and dissociates from the end of linear DNA without strand separation, which explains the low unwinding amplitude of these substrates. Trapping the displaced strand with ssDNA binding protein or changing its composition to morpholino oligomer that does not interact with helicase increases the unwinding amplitude. We conclude that the displaced strand must be continuously excluded and kept away from the central channel for processive DNA unwinding. The finding that T7 helicase can switch from ssDNA to dsDNA binding mode during unwinding provides new insights into ways of limiting DNA unwinding and triggering fork regression when stalled forks need to be restarted.
INTRODUCTION
Helicases are motor proteins that use the chemical energy of nucleoside 5′-triphosphate (NTP) hydrolysis to catalyse a variety of DNA and RNA metabolic processes that involve such reactions as separating the strands of the double helical DNA, removing secondary structures in RNA, catalysing Holliday junction branch migration, or dissociating proteins bound to nucleic acids (1–7). During DNA replication, helicases unwind the double-stranded DNA (dsDNA) to create single-stranded DNA (ssDNA) templates that are copied by the DNA polymerase (8,9). Many hexameric helicases have been shown to assemble into rings around DNA (10,11). When the hexameric ring binds to only one strand of the dsDNA, it can translocate on ssDNA in an NTPase-dependent manner to unwind the dsDNA (12–15). However, when the ring binds and moves on dsDNA, it catalyses NTPase-dependent branch migration reactions (16,17). It is expected that the topological linkage of the helicase ring with the DNA confers this class of helicases a high processivity of translocation, which would be necessary for accurate replication of genomes.
Bacteriophage T7 gp4 (T7 helicase) is a hexameric ring-shaped helicase that unwinds dsDNA using the energy derived from deoxythymidine 5′-triphosphate (dTTP) hydrolysis (18,19). T7 helicase has served as a model system to understand the mechanisms of NTP-coupled translocation and DNA unwinding by the hexameric helicases (18,19). T7 helicase translocates on ssDNA in the 5′-3′ direction with a high processivity. The measured processivity indicates that T7 helicase can travel ∼75 kb length of ssDNA before dissociation (20–22). Similarly, in single molecule optical trap unwinding assays, T7 helicase unwinds kb length dsDNA (23). Hence, it is surprising that T7 helicase shows a very low unwinding processivity in ensemble strand separation assays. The single turnover strand separation assays with dsDNA of different lengths indicated that T7 helicase unwinds on an average 60 bp of dsDNA before dissociation (21,24).
In this article, we report a series of experiments carried out to understand the mechanistic basis for the poor processivity of DNA strand separation by the T7 helicase. Our results, using modified unwinding substrates and ensemble single turnover kinetic approaches, indicate that T7 helicase switches from translocating on ssDNA to dsDNA during unwinding and the probability of this reaction is higher when the dsDNA is longer and/or is GC-rich. This activity limits the amplitude of DNA unwinding by providing a path for helicase dissociation from the end of the linear DNA substrate explaining its lower processivity of DNA unwinding as compared with translocation on ssDNA. We show that the efficiency and processivity of DNA unwinding is linked to the ability of the helicase to exclude the complementary DNA strand from the central channel.
MATERIALS AND METHODS
Proteins and buffer
The M64L mutant of T7 gp4 (referred here as T7 helicase) was purified as described previously (25). The M64L mutation inactivates the second initiation codon in the open reading frame (ORF) of T7 gp4 and produces only the gp4A protein. T7 gp2.5 was purified as described (26), Escherichia coli single-stranded DNA-binding protein (SSB) was purified as described (27). Protein concentrations were determined by absorbance measurements at 280 nm in 8 M urea using the calculated extinction coefficients. Buffer A [50 mM Tris–Cl (pH 7.6), 40 mM NaCl and 10% (v/v) glycerol] was used throughout the experiments unless specified otherwise. Quenching solution consisted of 100 mM ethylenediaminetetraacetic acid (EDTA), 0.4% (v/v) sodium dodecyl sulphate and 20% glycerol.
Oligodeoxynucleotides
Oligodeoxynucleotides (Supplementary Tables S1 and S2) were purchased from Intergrated DNA Technologies (Coralville, IA). The sequences of the DNA oligos used to prepare the helicase substrates ds18, ds30, ds40, ds50, ds60, ds90 were the same as reported previously (24). The 25-mer M-oligo was purchased from Gene tools. The DNAs were purified on a 15% polyacrylamide gel/8 M urea run in Tris/Borate/EDTA (TBE) buffer, the major DNA band was excised and the DNA was electroeluted using an Elutrap apparatus (Whatman Schleicher & Schuell). The DNA concentration was determined from its absorbance at 260 nm in TBE buffer containing 8 M urea. The extinction coefficient for each DNA was calculated from those of the individual bases (M−1 cm−1): εA = 15 200, εC = 7050, εG = 12 010 and εT = 8400. The 5′-strand was end-labelled using [γ-32P] adenosine triphosphate (Amersham Pharmacia Biotech) and T4 polynucleotide kinase (New England Biolabs). The radiolabelled helicase substrate was prepared by mixing the 5′-strand with a 1.5-fold excess of the complementary non-radiolabelled 3′-strand. The DNA mixture was heated at 95°C for 5 min and allowed to slowly cool down to room temperature to form the dsDNA.
DNA unwinding kinetics
DNA unwinding kinetics was measured in a RQF3 rapid quench-flow instrument (KinTek Corp., PA) at 18°C (24). T7 helicase (100 nM), DNA substrate (2.5 nM) and dTTP (2 mM) in buffer A containing EDTA (5 mM) was loaded in one syringe of the quenched-flow apparatus. A solution containing MgCl2 (final free concentration of 2 mM) in Buffer A and trap DNA (3 µM of unlabelled strand of the same sequence as the radiolabelled strand in the experiment) was loaded in the second syringe. The indicated concentrations are final after mixing. Equal volumes of the contents of the two syringes were mixed to initiate the reaction that were continued for predetermined time intervals after which they were quenched and loaded on a 4–20% native polyacrylamide gel run in TBE buffer. The unwinding reactions with ssDNA-binding protein contained 5 µM final concentration of T7 gp2.5 or 1 µM E. coli SSB, and these were added with the MgCl2 at the start of the reaction. No DNA trap was added with the MgCl2 when ssDNA-binding protein was present. It was added instead, with the quenching solution to prevent the unwound strands from reannealing. The ds30-B·S DNA was prepared by adding a 30-fold excess of streptavidin (recombinant from Sigma Chemicals) over dsDNA and incubating the mixture for at least 15 min before adding T7 helicase (28). The fraction of ssDNA generated at t, F was calculated from Equation (1) (12).
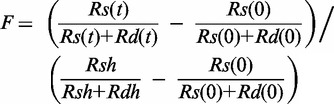 |
(1) |
where Rs(t) and Rd(t) are counts in each band corresponding to ssDNA and dsDNA, respectively, at time t. Rs(0) and Rd(0) are the corresponding counts at t = 0. Rsh and Rdh are counts of a negative control to measure the extent of maximum unwinding in which the sample was heated at 95°C for 5 min.
Four-way junction resolution/unwinding kinetics were measured in a RQF3 rapid quench-flow instrument (KinTek Corp., PA) at 18°C. T7 helicase (100 nM), DNA substrate (2.5 nM), and dTTP (2 mM) were mixed in buffer A containing EDTA (5 mM) and loaded in one syringe of the quench-flow apparatus. A solution containing MgCl2 (final free concentration of 2 mM) in Buffer A and trap DNA (3 µM of unlabelled 5′-strand) was loaded in the second syringe. Equal volumes of the contents of the two syringes were mixed to initiate the reaction that were continued for 0.1–120 s after which they were quenched with sodium dodecyl sulphate–EDTA solution and loaded on a 10% native polyacrylamide gel run in TBE buffer.
Kinetic analysis
Kinetic analysis of the unwinding time courses was performed using MATLAB software with Optimization toolbox (The MathWorks, Inc., Natick, MA). A step-wise unwinding model of helicase action was assumed (29,30). Best fits were obtained assuming unwinding by two populations of helicase species with identical step size s, but different stepping rates (k1 and k2). The stepping kinetics was described by Equations (2)–(4).
| (2) |
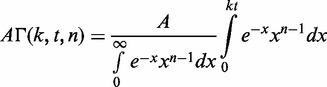 |
(3) |
where F is a fraction of unwound DNA substrate molecules, A1 and A2 are the amplitudes of unwinding and t is reaction time. The number of steps, n, taken by the helicase to unwind the substrate was calculated as
| (4) |
where L is the number of base pairs in the DNA substrate, Lm is the length of the shortest DNA duplex that can stay together under the experimental conditions, and it was fixed to 12 bp, based on previous studies (24). The processivity per step (P) was determined from the plot of amplitude (A) versus dsDNA length (l = L−Lm) using Equation (5) as described (29).
| (5) |
A0 is the maximum amplitude that provides a measure of the fraction of helicase-DNA complex initially present in the reaction.
RESULTS
T7 helicase translocates on dsDNA in the absence of the 3′-overhang in the forked DNA
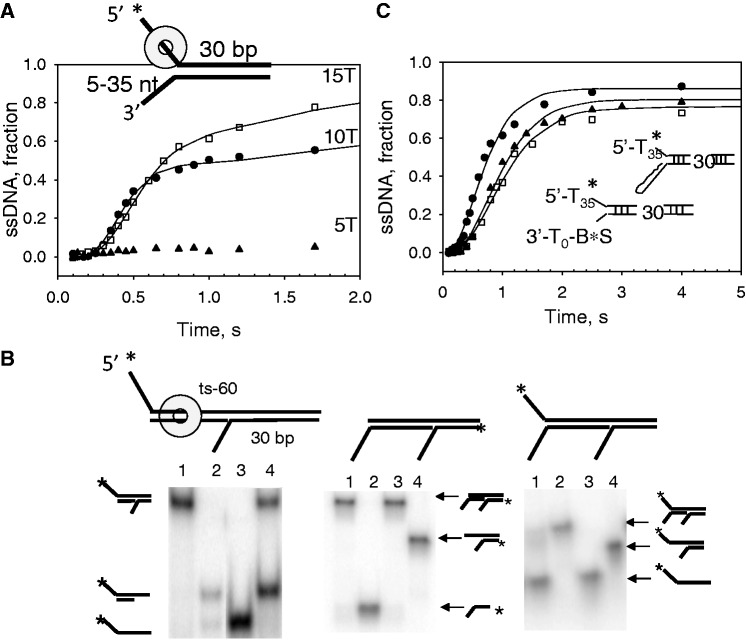
T7 helicase requires two ssDNA overhangs flanking the dsDNA to separate the dsDNA strands (12,13,31). The 5′-overhang is required for helicase loading and translocation, but the role of the 3′-overhang is less clear. Studies of related ring-shaped helicases indicate that the ssDNA overhang in the displaced strand may interact externally with the helicase to increase the DNA-binding affinity and/or serve as a steric block to prevent the helicase from surrounding the dsDNA (14,15,32,33). Consistent with these observations, we find that the length of the 3′-overhang affects the yield of ssDNA products observed in our single turnover unwinding assays (Figure 1A).
Figure 1.
DNA unwinding analysis of fork DNAs with modified 3′-overhangs. The single turnover unwinding kinetics was measured at 18°C as described in the Experimental Procedures. (A) shows the unwinding kinetics of ds30-5T (filled triangle), ds30-10T (filled circle) and ds30-15T (open square) fit to Equations (2)–(4) (Experimental Procedures) to obtain the average unwinding rate (ku = k × s) and amplitude (A) for ds30-10T, ku = 33 bp/s (A = 0.68) and ds30-15T, ku = 26 bp/s (A = 0.91). (B) Native polyacrylamide gel image shows the unwinding products of tandem substrate under single turnover reaction conditions. Left panel, lanes 1–3 are standards, and lane 4 shows products after 2 min of reaction. Middle panel, lane 1 shows reaction after 2 min, and lanes 2–4 are standards. Right panel, lane 1 shows reaction after 2 min, and lanes 2–4 are standards. (C) The unwinding kinetics of ds30-15T (filled circle) in comparison with that of the ds30-B·S (filled triangle) and ds30-hp (open square) DNAs. The average unwinding rates for ds30-B·S, ku = 19 bp/s (A = 0.8), ds30-hp, ku = 18 bp/s (A = 0.78). Asterisk represents the position of radiolabelling.
Figure 1A shows the single-turnover unwinding kinetics of the forked substrate with 30 bp (33% GC bp) dsDNA, a fixed length dT35 5′-overhang and 5-nt, 10-nt or 15-nt 3′-ssDNA overhang (Supplementary Table S1). In these and the rest of the all-or-none single-turnover unwinding assays in this article, T7 helicase (100 nM) was loaded on radiolabelled forked substrates (2.5 nM) in the presence of 2 mM dTTP (30). The unwinding reaction was initiated by addition of Mg(II) and trap DNA, which traps free helicase present at the onset of the reaction and any that dissociates during the reaction or on completion of the reaction (34). The trap DNA in all the experiments was the non-radiolabelled 5′-strand of the fork DNA; hence, it also binds to the 3′-ssDNA strand generated by the helicase action to prevent reannealing of the unwound strands. Under these conditions, the unwinding amplitude measures the fraction of helicase molecules that complete the unwinding reaction after a single DNA binding event. We have also used dT-100 as the trap DNA and obtained similar amplitudes and rates of unwinding (17). T7 helicase does not unwind the forked substrate when the 3′-overhang is 5 nt in length, but as the length of the 3′-overhang is increased, the unwinding amplitude increases with 10-nt and 15-nt overhangs unwound with 60 and 80% amplitudes, respectively. The results indicate that the 3′-overhang needs to be ∼15 nt in length for optimal unwinding of the dsDNA (12).
To investigate why the short 3′-overhang substrate was not unwound, we used tandem DNA substrates with 30-bp proximal and 30-bp distal dsDNA regions (33–40% GC content) (Figure 1B). Each of the three tandem substrates contained a 15-nt 3′-overhang in front of the distal dsDNA region but was missing one or the other overhang in front of the proximal dsDNA. The proximal dsDNA was not unwound when it was missing the 3′-overhang, but the distal dsDNA was still unwound efficiently (Figure 1B, left panel, lane 4). None of the dsDNA regions were unwound when the proximal dsDNA was missing the 5′-overhang (Figure 1B, middle panel, lane 1), which indicates that T7 helicase does not load internally on the dsDNA to unwind the distal dsDNA. Both dsDNA regions were unwound when the proximal duplex contained 5′ and 3′ overhangs (Figure 1B, right panel, lane 1). Later in the text, we present more detailed unwinding kinetic studies of these tandem DNAs. The results thus far indicate that the 3′-overhang is required for DNA unwinding, which is consistent with the strand exclusion model. Moreover, our results indicate that in the absence of the 3′-overhang, T7 helicase does not dissociate from the DNA, but it moves over the dsDNA to unwind the distal dsDNA. These findings are in agreement with similar behaviours reported for other ring-shaped helicases (14,15,35). Our studies were carried out under single turnover conditions in the presence of a protein trap, and therefore indicate that the helicase can switch between ssDNA and dsDNA regions without dissociating into solution.
To determine whether the 3′-overhang in the forked DNA is required only for steric exclusion or also for interactions with the helicase, we prepared DNA substrate that contained a 10-bp DNA hairpin (ds30-hp) or a biotin-streptavidin complex (ds30-BS) instead of the 3′-overhang (DNA sequences, Supplemental Table S1). T7 helicase efficiently unwinds both of these 3′-strand modified DNAs (Figure 1C). The unwinding amplitude (∼80%) of the hairpin or the BS-modified substrate was similar to that of the substrate with the 15-nt ssDNA 3′-overhang. These results indicate that the interactions with the 3′ ssDNA overhang are not necessary for DNA unwinding. Nevertheless, the rate of DNA unwinding of the duplex or BS modified DNA was 1.5–2-fold slower than the fork DNA with 15-nt 3′-overhang DNA. The slower rate may be due to inhibition of helicase movement by the bulky moieties, or that the 3′-overhang interactions somehow increase the efficiency of DNA unwinding.
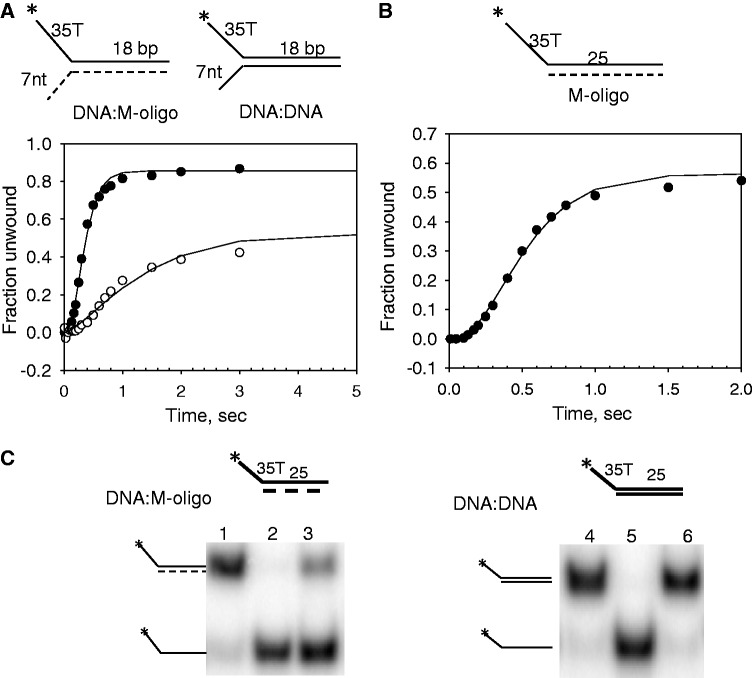
T7 helicase unwinds a morpholino–DNA hybrid substrate without requiring 3′-overhang
To investigate whether the interactions of the 3′-overhang and the displaced 3′-strand with the helicase are required for dsDNA unwinding, a modified unwinding substrate was prepared in which the 3′-strand was replaced with a morpholino oligo (M-oligo). The morpholino nucleic acid has standard bases to pair in the same manner as DNA, but the bases are bound to morpholine rings instead of deoxyribose rings and linked through phosphorodiamidate groups that lack negative charges as in the phosphates of DNA. The M-oligo is water soluble, and the hybrid duplex (M-oligo/DNA) is as stable as or more stable than the DNA duplex (36,37). Several experiments showed that the M-oligo does not interact with T7 helicase. In one experiment, the binding of a radiolabeled DNA-oligo to T7 helicase was measured in the presence of increasing amounts of the M-oligo of the same length and sequence. Even a 10-fold excess of the M-oligo did not inhibit the binding of the radiolabeled DNA-oligo to the T7 helicase (Supplementary Figure S1A). Similarly, the M-oligo did not stimulate the dTTPase activity of T7 helicase (Supplementary Figure S1B), and when added at a high concentration, the M-oligo did not inhibit the DNA-oligo stimulated dTTPase activity (Supplementary Figure S1C). These results indicate that the neutral backbone M-oligo does not interact with the T7 helicase.
An 18-bp M-oligo:DNA hybrid with 30% GC content was prepared with a 35-nt ssDNA 5′-overhang and a 7-nt M-oligo 3′-overhang. A DNA:DNA substrate of the same sequence was made as a control (Figure 2A). Interestingly, T7 helicase unwinds the M-oligo:DNA with ∼90% amplitude and 2.5-fold faster rate than the DNA:DNA substrate (Figure 2A). When a 25-bp M-oligo:DNA substrate was made without any 3′-overhang, it was also unwound efficiently, which was unexpected, as an analogous DNA substrate was not unwound at all (Figure 2B and C). The results indicate that keeping the 3′-overhang or the displaced strand away from the helicase or removing any interactions with the displaced strand in fact increases unwinding efficiency.
Figure 2.
Unwinding kinetics of 3′-morpholino/5′-DNA forked substrate by T7 helicase. (A) Single-turnover unwinding kinetics of ds18-DNA:DNA (open circle) and ds18-DNA:M-oligo (filled circle) with 7 nt 3′-tail fit to average unwinding rates ku = 5 bp/s (A = 0.56) and ku = 35 bp/s (A = 0.9), respectively. (B) The ds25-DNA:M-oligo without any 3′-tail was unwound with single-turnover rate, ku = 26 bp/s (A = 0.52). (C) The native polyacrylamide gel image shows unwinding of the ds25-DNA:M-oligo (left panel) and ds25-DNA:DNA (right panel) under single-turnover conditions. Lanes 1 and 4 show the control unreacted starting DNA. Lanes 2 and 5 show the control radiolabelled 5′-strand. Lanes 3 and 6 show the reaction products after 1 min of unwinding.
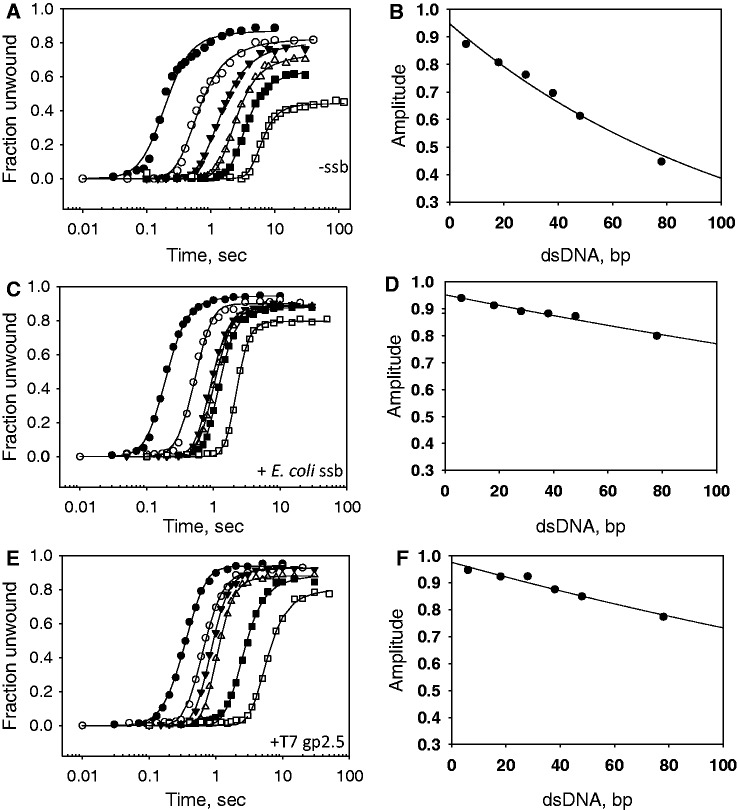
Single-strand DNA-binding proteins increase the unwinding processivity
The aforementioned results indicate that if the displaced strand is kept away from the helicase, it increases the unwinding efficiency of T7 helicase. Therefore, we investigated whether trapping the displaced ssDNA strand with ssDNA-binding protein, such as E. coli SSB or T7 gp2.5, will improve the processivity. It is important to pre-bind T7 helicase to the forked substrates in the presence of dTTP and then add ssDNA-binding protein with Mg(II) at the start of the reaction, otherwise it is observed that SSB competes with the helicase and inhibits unwinding at high concentrations (38). The unwinding kinetics were measured with forks of different lengths from 18 to 90 bp (30–35% GC bp) in the absence and in the presence of ssDNA-binding proteins (Figure 3A, C and E). The series of unwinding kinetics were fit to the stepping model [Equations (2)–(4)] to obtain the unwinding rates and amplitudes. The amplitudes were plotted as a function of the dsDNA lengths and fit to Equation (5) to obtain the unwinding processivity (24,29,30). In the absence of the ssDNA-binding protein, the single-turnover unwinding amplitudes decreased steeply, as the length of the dsDNA increased from 18 bp to 90 bp (Figure 3A and B). The processivity value of 0.9911 (±0.0007) indicates that T7 helicase unwinds on an average 60–110 bp before dissociation, consistent with previous measurements (24).
Figure 3.
Unwinding kinetics in the presence and absence of ssDNA-binding proteins. The single-turnover unwinding kinetics was measured for ds18 (Filled circle), ds30 (○), ds40 (filled inverted triangle), ds50 (open inverted triangle), ds60 (filled square) and ds90 (open square) at 18°C in the absence of ssDNA-binding protein (A), in the presence of E. coli SSB (C) or in the presence of T7 gp2.5 (E). Each curve was fit to Equations (2)–(4) to obtain the unwinding rates. The average unwinding rate is 19 ± 7 bp/s in the absence of ssDNA-binding protein, 32 ± 5 bp/s in the presence of E. coli SSB and 23 ± 8 bp/s in the presence of T7 gp2.5. The amplitude from the individual fits is plotted against the corrected dsDNA length (L-Lm) and fit to Equation (5) to obtain processivity of 0.9911 (±0.0007) in the absence of ssDNA-binding proteins (B), 0.9979 (±0.0002) in the presence of E. coli SSB (D) and 0.9971 (±0.0003) in the presence of T7 gp2.5 (F).
In the presence of E. coli SSB (Figure 3C and D) and T7 gp2.5 (Figure 3E and F), the unwinding amplitudes were higher and decreased only slightly with increasing dsDNA lengths. The processivity value of 0.9979 (±0.0002) in the presence of E. coli SSB and 0.9971 (±0.0003) in the presence of T7 gp2.5 indicates that T7 helicase unwinds on an average 500 bp before dissociation. The rate of DNA unwinding increased ∼30% in the presence of E. coli SSB and remained unchanged in the presence of T7 gp2.5. These results indicate that ssDNA-binding proteins increase the processivity of T7 helicase. This increase in processivity could be due to specific interactions of T7 gp2.5 with T7 helicase; however, no such interactions have been observed between T7 helicase and E. coli SSB (39). We therefore propose that the increase in processivity of DNA unwinding by ssDNA-binding proteins is due to trapping the displaced strand and preventing its direct interactions with the helicase.
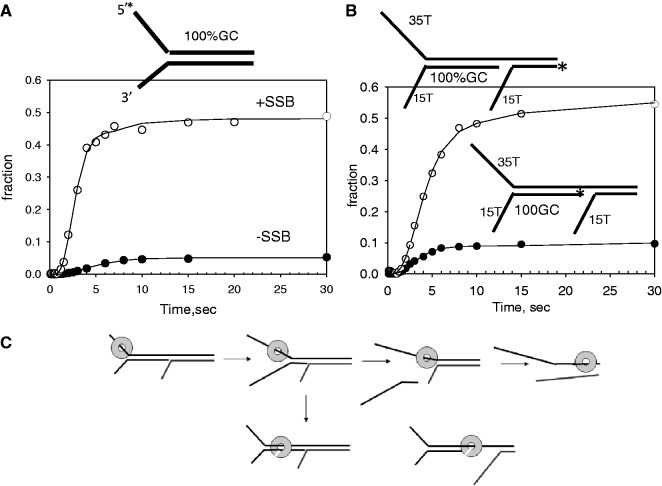
Switching from ssDNA to dsDNA accounts for the lower amplitudes of unwinding GC-rich dsDNA
The 30 bp dsDNA substrate with 100% GC-content is unwound very inefficiently, with <10% of the dsDNA unwound under single turnover conditions. Interestingly, in the presence of E. coli SSB, the extent of unwinding increases to 50% (Figure 4A). To understand the basis of the low unwinding amplitude of the GC-rich dsDNA, we prepared a tandem, forked unwinding substrate with a proximal dsDNA region that was 100% GC and a distal dsDNA that was 50% GC-rich. As the tandem substrate contains both 5′-overhang and 3′-overhang (see schematic in Figure 4B), we expect that T7 helicase will unwind both the proximal and distal dsDNA regions, as observed earlier in the text (Figure 1B). However, if the helicase dissociates during unwinding the GC-rich proximal dsDNA region, then the unwinding yield of both the proximal and distal dsDNA will be equally low. On the other hand, if the helicase does not dissociate but switches from ssDNA to dsDNA at any point during unwinding, then the helicase can travel over the dsDNA without unwinding the proximal DNA, and if it switches back to ssDNA, it can unwind the distal dsDNA (Figure 4C). In this case, the unwinding yield of the distal DNA will be higher than the proximal DNA. Our results show that the unwinding yield of the proximal 100% GC dsDNA was 10%, but the unwinding yield of the distal dsDNA was ∼50% (Figure 4B). These results indicate that when the dsDNA is GC-rich, instead of dissociating, the helicase goes over the dsDNA without unwinding it.
Figure 4.
Unwinding kinetics of GC-rich forked and tandem substrate. (A) Single turnover unwinding kinetics of the 30 bp 100% GC dsDNA with and without E. coli SSB. The average unwinding rate was ku = 8 bp/s (A = 0.046) in the absence of SSB and ku = 10 bp/s (A = 0.44) in the presence of E. coli SSB. (B) The proximal 100% GC-rich dsDNA region of the tandem forked DNA substrate was unwound with ku = 6 bp/s (A = 0.13) and the distal 50% GC rich dsDNA region was unwound with ku = 5.2 bp/s (A = 0.46). (C) The schematic representation shows the normal pathway of unwinding both dsDNA regions of the tandem dsDNA substrate, and the pathway where the helicase encircles and does not unwind the initial GC-rich dsDNA but unwinds the distal dsDNA region.
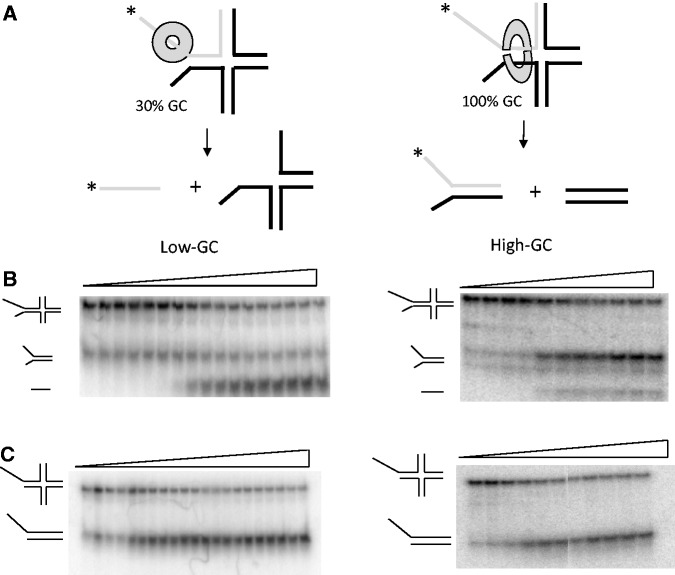
The tendency to switch from traveling on ssDNA to dsDNA is higher when the dsDNA is GC-rich, as shown by the following experiments. Here, instead of using the unwinding of the distal DNA as a signal for helicase traveling over dsDNA, we used branch migration as the signal. Two types of homologous Holliday junction substrates with a forked arm were prepared (Supplementary Table S2). The dsDNA flanking the fork arm in one substrate was 100% GC and in another 30% GC-rich. T7 helicase binds to the 5′-overhang of the fork and owing to the presence of the 3′-overhang, it is expected to unwind the dsDNA to generate a ssDNA product. If T7 helicase goes over the dsDNA at any point and translocates along the dsDNA arm, then it is expected to resolve the four-way junction rather than unwind the DNA strands (17) (Figure 5A). Under single turnover conditions in the presence of a trap, T7 helicase unwinds the 30% GC substrate with a high efficiency producing ssDNA product, but it preferentially resolves the 100% GC substrate (Figure 5B). Control experiments with both the low-GC and the high-GC substrates lacking the 3′-overhang resulted in the resolution of the four-way junction, with no strand separation (Figure 5C). These experiments indicate that the propensity to travel over the dsDNA is higher when the dsDNA is GC rich and more stable.
Figure 5.
Unwinding and branch migration kinetics on GC-rich forked four-way junction. (A) Schematic representation of the unwinding of forked four-way junction substrates by T7 helicase. The Low-GC substrate (left panel) is preferentially unwound by T7 helicase, whereas the high-GC substrates (right panel) is preferentially resolved. (B) Single-turnover unwinding kinetics (0.1–120 s) of forked four-way junction substrates with a low-GC loading arm (left panel) or a high-GC loading arm (right panel). The low-GC substrate gets unwound, whereas the high-GC substrate gets resolved. (C) Single-turnover unwinding kinetics (0.1–120 s) of control four-way junction substrates lacking the 3′-overhang get resolved irrespective of the GC-content.
Our results show that T7 helicase does not dissociate from the DNA when it is unwinding GC-rich dsDNA, but instead it switches between encircling ssDNA and dsDNA. Such events during unwinding provide a path for helicase sliding along dsDNA and dissociating from the end of linear substrates without strand separation, explaining the lower amplitude of unwinding. The experiments also show that the helicase not only switches its DNA-binding mode from ssDNA to dsDNA but also switches its DNA motor activity from ssDNA to dsDNA.
DISCUSSION
Ring-shaped helicases encircle the DNA they travel along and therefore are expected to display a high translocation processivity. Our previous studies have shown that T7 helicase travels along ssDNA with a high processivity but unwinds linear dsDNA with an unusually low processivity (21,24). These results indicate that although T7 helicase rarely dissociates when it is moving along ssDNA, it dissociates more often when unwinding linear dsDNA. The experiments in the present study show that T7 helicase actually does not dissociate from the ss–ds junction but instead travels over the dsDNA to dissociate from the end of the linear substrate without unwinding it. It is known that T7 helicase can bind and move on dsDNA in a dTTPase-dependent manner (17), and this reaction of the ring-shaped helicase is prevented during DNA unwinding by providing an overhang in the displaced strand (14,15). T7 helicase requires a 3′-overhang to catalyse unwinding (12,13,31), and studies in this article confirm that the overhang needs to be at least 10-nt long and optimally 15-nt long. The overhang may serve as a steric block or have specific interactions with the helicase (32,33). As the 3′-overhang can be replaced by a duplex structure or a biotin-streptavidin complex, we argue that the 3′-overhang acts as a steric block, rather than having a role where interactions with the 3′-overhang assists unwinding. Consistent with this, when the displaced strand was replaced with a morpholino oligo, which has no affinity for T7 helicase, we observed better DNA strand separation even in the absence of a 3′-overhang. Similarly, trapping the displaced strand by ssDNA-binding proteins, such as E. coli SSB or T7 gp2.5, increased the unwinding processivity of the T7 helicase. Thus, keeping the displaced strand away from the helicase increases the unwinding efficiency. This hypothesis also explains the high unwinding processivity of T7 helicase in single molecule optical tweezers assays, where the unwound DNA strands are tethered and splayed under force, which provides efficient displaced strand exclusion (23).
We find that the helicase’s tendency to switch binding modes from ssDNA to dsDNA at the forked junction is high when the dsDNA is GC-rich. The GC-rich dsDNAs are unwound at slower rates as compared with AT-rich dsDNA (22,40). The slower rate of unwinding GC-rich dsDNA may allow the helicase more time to engage the displaced strand and surround the dsDNA. Alternatively, the GC base pairs reannealing at the unwinding junction may allow T7 helicase to surround the dsDNA. Either way, excluding the displaced strand from binding to T7 helicase by addition of the SSBs increases the unwinding amplitude, which implies that SSB binding to the displaced strand prevents the helicase from surrounding the dsDNA. Similarly, changing the composition of the displaced strand to a morpholino oligo that does not interact with the helicase prevents the helicase from binding the duplex, either because the helicase does not engage the morpholino oligo or the helicase cannot encircle the DNA:M-oligo hybrid duplex. The results indicate that for T7 helicase to surround the dsDNA, it needs to have interactions with the sugar-phosphate backbone of the displaced strand. After encircling the dsDNA, T7 helicase moves on dsDNA and when provided with a four-way DNA junction in its path, the helicase is able to resolve the junction. Similarly, when provided with a tandem fork, it is able to switch from dsDNA-binding mode back to ssDNA and unwind the downstream dsDNA in the tandem substrate. Thus, T7 helicase can toggle between ssDNA and dsDNA binding, and this switch occurs without the helicase physically dissociating from the DNA substrate.
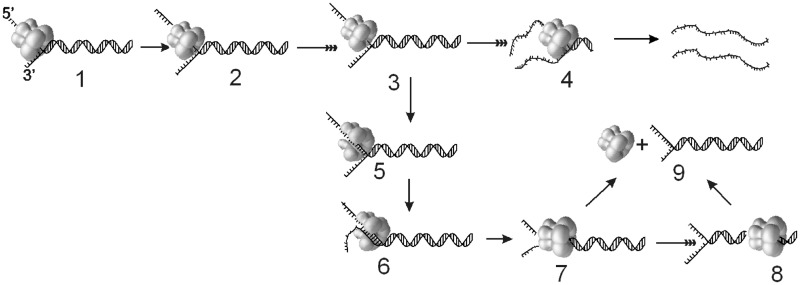
We propose the following mechanism to explain the ability of T7 helicase to switch between its DNA-binding modes (Figure 6). This mechanism is consistent with the structural studies of ring-shaped helicases and also explains many of the peculiar behaviours of helicases reported in the literature, as discussed later in the text. T7 helicase unwinds DNA by moving along one strand of the DNA (the 5′-strand or the loading strand) and excluding the complementary strand (the 3′-strand or the displaced strand) from the central channel (strand exclusion model). As the subunit–subunit interactions within the hexamer ring are not extensive (41–43), it is possible that the ring undergoes spontaneous opening and closing transitions during its movement along the DNA. Transient ring opening does not appear to completely dissociate the helicase from DNA, as evident from the very slow rate of dissociation of the translocating helicase from ssDNA (21,44). However, if the nascent displaced strand is in physical proximity of the helicase, it enters the central channel of the open ring, anneals with the loading strand and consequently, the helicase ring ends up surrounding the dsDNA. Once bound to dsDNA, the helicase ring moves along dsDNA and dissociates from the end of the linear DNA without unwinding it. This phenomenon explains the unexpected low processivity of DNA unwinding with linear substrates. To accommodate the dsDNA in the central channel, the helicase ring may adopt the lockwasher conformation, first observed in Rho helicase and recently in DnaB helicase (45,46). Unlike Rho, where the lockwasher conformation has an open gap, the DnaB lockwasher structure is bridged by a linker helix, which will prevent the displaced strand from entering the central channel. In the DnaB class of helicases, which includes T7 helicase, the subunit–subunit interactions involve packing of a linker helix against the neighboring subunit. To surround the dsDNA, the linker helix in DnaB and T7 helicase must disengage from the neighbouring subunit. Plasticity in the helicase ring has also been suggested for the viral helicase, SV40 large T antigen, which is able to bypass bulky adducts on the tracking strand (47). Transient ring opening and the tendency to surround dsDNA during unwinding may also explain peculiar behaviour of helicases, such as premature rezipping of DNA during DNA unwinding and repetitive unwinding observed in single molecule experiments (48–51).
Figure 6.
Model that explains the low processivity of DNA unwinding by T7 helicase. T7 helicase initiates unwinding from the 5′-ssDNA overhang. The upper highlighted pathway (species 1–4) shows a series of intermediates that result in the complete unwinding of the dsDNA. The branched pathway (3–9) shows that the occasional ring opening reaction (3→4→5) can result in T7 helicase encircling the displaced strand and the dsDNA (5→6→7). The helicase ring bound to the dsDNA can dissociate in solution (7→9) or move along the dsDNA (7→8) and fall off from the end of the linear DNA without unwinding the DNA strands (8→9).
Is there a role for this ssDNA–dsDNA switching activity of the helicase during DNA replication? Replicative helicases act in conjunction with the DNA polymerase and ssDNA-binding proteins to catalyse DNA replication (52–54). During normal replication, the DNA polymerase provides efficient strand exclusion by moving with the helicase and continuously binding to the displaced strand near the unwinding junction. Hence, the unwinding rate of T7 helicase is optimal in the presence of T7 DNA polymerase (54). However, when replication stops, e.g. owing to DNA polymerase stalling at a roadblock in the leading strand, the helicase activity is decoupled from DNA synthesis, but it continues to unwind dsDNA and thus create free displaced ssDNA (55,56). Under these conditions, the displaced ssDNA may interact externally and promote the helicase to surround the dsDNA. The switching from ssDNA to dsDNA would be useful in limiting helicase’s strand separation activity when replication forks stall. Another intriguing possibility is that switching of the helicase from ssDNA- to dsDNA-binding mode and spontaneous reannealing of the newly unwound DNA strands could be the trigger for replication fork regression. Future experiments with the replisome will test these ideas. Thus, our finding that the ring-shaped replicative helicase can switch between ssDNA and dsDNA provides the basis to understand the mechanisms to overcome replication blocks by facilitating reactions that are needed for repair and restart of the replication fork.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figure 1 and Supplementary Methods.
FUNDING
National Institutes of Health (NIH) [GM55310 to S.S.P.]; Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [2010-0022332 to Y.J.J.]. Funding for open access charge: NIH [GM55310].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Patel laboratory for critical discussions. All experiments were carried out by Y.J.J. and V.R.. S.S.P. supervised the project, and Y.J.J., V.R. and S.S.P. wrote the article. They thank Divya Nandakumar for fitting the unwinding kinetics data to the stepping model.
REFERENCES
- 1.Matson SW, Bean DW, George JW. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 2.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 3.Levin MK, Patel SS. In: Molecular Motors. Schliwa M, editor. Weinheim Germany: Wiley-VCH Verlag GmbH; 2003. pp. 179–198. [Google Scholar]
- 4.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 5.Patel SS, Donmez I. Mechanisms of helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 6.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 7.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 8.von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 9.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 10.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 12.Ahnert P, Patel SS. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 1997;272:32267–32273. doi: 10.1074/jbc.272.51.32267. [DOI] [PubMed] [Google Scholar]
- 13.Hacker KJ, Johnson KA. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry. 1997;36:14080–14087. doi: 10.1021/bi971644v. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan DL, Davey MJ, O'Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 16.Bujalowski W. Expanding the physiological role of the hexameric DnaB helicase. Trends Biochem. Sci. 2003;28:116–118. doi: 10.1016/S0968-0004(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 17.Rasnik I, Jeong YJ, McKinney SA, Rajagopal V, Patel SS, Ha T. Branch migration enzyme as a Brownian ratchet. EMBO J. 2008;27:1727–1735. doi: 10.1038/emboj.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donmez I, Patel SS. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006;34:4216–4224. doi: 10.1093/nar/gkl508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 20.Tabor S, Richardson CC. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc. Natl Acad. Sci. USA. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DE, Narayan M, Patel SS. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J Mol. Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 22.Donmez I, Rajagopal V, Jeong YJ, Patel SS. Nucleic acid unwinding by hepatitis C virus and bacteriophage t7 helicases is sensitive to base pair stability. J. Biol. Chem. 2007;282:21116–21123. doi: 10.1074/jbc.M702136200. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl Acad. Sci. USA. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel SS, Rosenberg AH, Studier FW, Johnson KA. Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J. Biol. Chem. 1992;267:15013–15021. [PubMed] [Google Scholar]
- 26.Kim YT, Tabor S, Churchich JE, Richardson CC. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J. Biol. Chem. 1992;267:15032–15040. [PubMed] [Google Scholar]
- 27.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 28.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 29.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 30.Pandey M, Levin MK, Patel SS. Experimental and computational analysis of DNA unwinding and polymerization kinetics. Methods Mol. Biol. 2010;587:57–83. doi: 10.1007/978-1-60327-355-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matson SW, Tabor S, Richardson CC. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 32.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3' arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DNAB helicase. J. Mol. Biol. 2004;343:101–114. doi: 10.1016/j.jmb.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 33.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem. 2007;282:34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 34.Picha KM, Patel SS. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- 35.Shin JH, Jiang Y, Grabowski B, Hurwitz J, Kelman Z. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J. Biol. Chem. 2003;278:49053–49062. doi: 10.1074/jbc.M308599200. [DOI] [PubMed] [Google Scholar]
- 36.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 37.Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–572. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J. Biol. Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 39.Nakai H, Richardson CC. The effect of the T7 and Escherichia coli DNA-binding proteins at the replication fork of bacteriophage T7. J. Biol. Chem. 1988;263:9831–9839. [PubMed] [Google Scholar]
- 40.Donmez I, Patel SS. Coupling of DNA unwinding to nucleotide hydrolysis in a ring-shaped helicase. EMBO J. 2008;27:1718–1726. doi: 10.1038/emboj.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 42.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 43.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage t7. Mol. Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 44.Matson SW, Richardson CC. Nucleotide-dependent binding of the gene 4 protein of bacteriophage T7 to single-stranded DNA. J. Biol. Chem. 1985;260:2281–2287. [PubMed] [Google Scholar]
- 45.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 46.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yardimci H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, Walter JC. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492:205–209. doi: 10.1038/nature11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun B, Johnson DS, Patel G, Smith BY, Pandey M, Patel SS, Wang MD. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–135. doi: 10.1038/nature10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl Acad. Sci. USAmerica. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 51.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong F, Weitzel SE, von Hippel PH. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc. Natl Acad. Sci. USA. 1996;93:14456–14461. doi: 10.1073/pnas.93.25.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McInerney P, O'Donnell M. Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J. Biol. Chem. 2007;282:25903–25916. doi: 10.1074/jbc.M703777200. [DOI] [PubMed] [Google Scholar]
- 56.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.