Abstract
Autophagy is a highly conserved process that maintains intracellular homeostasis by degrading proteins or organelles in all eukaryotes. The effect of autophagy on fungal biology and infection of insect pathogens is unknown. Here, we report the function of MrATG8, an ortholog of yeast ATG8, in the entomopathogenic fungus Metarhizium robertsii. MrATG8 can complement an ATG8-defective yeast strain and deletion of MrATG8 impaired autophagy, conidiation and fungal infection biology in M. robertsii. Compared with the wild-type and gene-rescued mutant, Mratg8Δ is not inductive to form the infection-structure appressorium and is impaired in defense response against insect immunity. In addition, accumulation of lipid droplets (LDs) is significantly reduced in the conidia of Mratg8Δ and the pathogenicity of the mutant is drastically impaired. We also found that the cellular level of a LD-specific perilipin-like protein is significantly lowered by deletion of MrATG8 and that the carboxyl terminus beyond the predicted protease cleavage site is dispensable for MrAtg8 function. To corroborate the role of autophagy in fungal physiology, the homologous genes of yeast ATG1, ATG4 and ATG15, designated as MrATG1, MrATG4 and MrATG15, were also deleted in M. robertsii. In contrast to Mratg8Δ, these mutants could form appressoria, however, the LD accumulation and virulence were also considerably impaired in the mutant strains. Our data showed that autophagy is required in M. robertsii for fungal differentiation, lipid biogenesis and insect infection. The results advance our understanding of autophagic process in fungi and provide evidence to connect autophagy with lipid metabolism.
Keywords: Metarhizium robertsii, autophagy, appressorium, sporulation, cell differentiation, perilipin, lipid metabolism, virulence
Introduction
Autophagy is a self-eating process that maintains intracellular homeostasis by degrading proteins or organelles and is conserved in all eukaryotes.1 It plays a crucial role in establishing infections by plant pathogenic fungi such as Magnaporthe oryzae,2-5 Fusarium graminearum or Colletotrichum spp,6,7 and the mammalian pathogens like Cryptococcus neoformans, Candida albicans and Aspergillus fumigatus.8 Insect pathogenic fungi like Metarhizium robertsii, M. acridum and Beauveria bassiana are widely used in the biological control of different insect pests and are model organisms to study the mechanisms of insect-fungus interactions.9,10 Several virulence-related genes have been characterized in these fungi,11 but none associated with autophagy.
A variety of proteins in the autophagy pathways have been identified and functionally characterized.12 Thus far, 36 autophagy-related (Atg) proteins have been identified in yeast13 and 23 in the plant pathogen M. oryzae.4 Of these, Atg8 has been identified as one of the key autophagic proteins involved in multiple cellular processes including autophagy, lipid metabolism and membrane trafficking.14 The ATG8 gene encodes a highly conserved ubiquitin-like protein that is associated with the autophagosome membrane and microtubule, and is used as a marker of autophagy.13 Gene-deletion studies have revealed the involvement of ATG8 in differentiation and development in filamentous fungi.12 In M. oryzae, MoAtg8 mediates autophagic cell death and is required for fungal infection.2 Deletion of AoATG8 in Aspergillus oryzae leads to the impairment of conidiation and aerial hyphal growth.15 Disruption of ATG8 in the plant pathogen Ustilago maydis abolishes autophagosome accumulation in the vacuoles and impairs fungal growth and virulence.16 Besides ATG8, serial gene deletions of other ATG genes in M. orzyae leads to loss of virulence in 16 of 22 null mutants, due to impairment of appressorium differentiation/maturation and conidial programmed cell death.4 It is unclear whether Atg-mediated autophagy is also required for fungal differentiation and virulence in the insect pathogens.
During autophagy, Atg8 binds to phosphatidylethanolamine on the phagophore membrane and remains there through the maturation process of the autophagosome.13 This membrane binding is facilitated by the carboxyl-terminal glycine residue in Atg8.17 It remains undetermined whether the C-terminal Gly residue is essential for Atg8 function in filamentous fungi. In mammalian cells, autophagy is known to regulate lipid metabolism,18,19 although its effect on lipid breakdown or biogenesis that results in the turnover of cellular lipid droplets (LDs) is still unknown.
In this study, the ortholog of yeast ATG8 gene, designated as MrATG8 was deleted and complemented in the insect pathogenic fungus M. robertsii. We found that MrATG8 is required for conidiation, appressorium differentiation, LD accumulation and virulence in M. robertsii. In addition, the orthologous genes of yeast ATG1 (encoding a serine/threonine protein kinase), ATG4 (encoding a cysteine protease) and ATG15 (encoding a triglyceride lipase), labeled as MrATG1 (MAA_03501), MrATG4 (MAA_04475) and MrATG15 (MAA_00506),20 were also deleted in M. robertsii. These mutants showed similar defects in cellular LD storage and virulence although, in contrast to Mratg8Δ, appressorium formation was not affected. The results of this study improve the understanding of the effects of autophagy on cell differentiation, lipid storage and virulence in the insect pathogenic fungus M. robertsii.
Results
MrATG8 gene characterization and protein localization
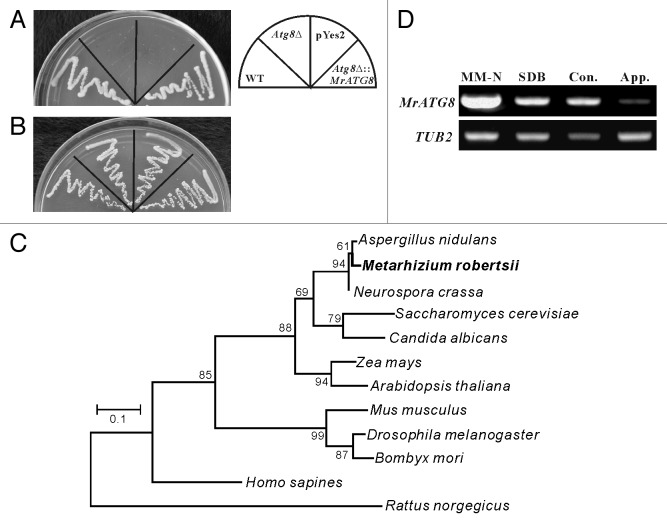
The sequence of S. cerevisiae Atg8 protein was used as query in Blast searches against the M. robertsii genome database,20 and the retrieved putative protein with 78% amino acid identity was designated as MrAtg8 (MAA_02674). To determine whether MrATG8 gene is functionally related to the yeast ATG8, the complete MrATG8 ORF was transformed into the Atg8Δ yeast strain. After nitrogen starvation for 18 d, the MrATG8-rescued yeast strain survived and grew normally similar to the wild type (WT) while the Atg8Δ mutant and control that was transformed only with the empty vector did not survive (Fig. 1A). All strains grew equally well on a nutrient-rich medium (Fig. 1B). This is not surprising, considering that MrAtg8 is highly conserved among fungi, plants and insects (Fig. 1C). Transcription of MrATG8 was also highly activated in response to nitrogen starvation that is similar to the observations in yeast and other filamentous fungi.12 In addition, the gene was also transcribed by the fungus when grown in a nutrient-rich medium, Sabouraud dextrose broth (SDB), and during conidiation and appressorium formation (Fig. 1D), which is consistent with studies in mammals that show the constitutive occurrence of autophagy.21
Figure 1. Characterization of MrATG8 gene function, evolution and expression. (A) Complementation of yeast ATG8 gene with MrATG8. In contrast to the wild-type (WT) yeast, Atg8Δ and the mutant transformed with the pYes2 vector did not survive after nitrogen starvation, however, the mutant transformed with the MrATG8 gene survived like the WT. (B) All yeast strains grew equally well on a nutrient-rich YPD medium. (C) Phylogenetic analysis indicating that the MrATG8 of M. robertsii is highly similar to the orthologs from various fungi, plants and insects. (D) RT-PCR analysis of MrATG8 expression by the fungus under various conditions. MM-N, mycelia cultured in MM-N medium for 4 h; SDB, mycelial sample cultured in SDB for 24 h; Con., conidia harvested from CM cultures; App., appressoria induced on locust hind wings. TUB2, tubulin β-2 gene, was used as a reference.
Similar to the pattern of Atg protein localization in yeast and Magnaporthe cells,22-24 punctate localization of MrAtg8 was observed in conidia, germlings, appressoria and hyphal bodies (fungal cells harvested from the hemolymph of infected insect) by N-terminal fusion with the red fluorescent protein (RFP) (Fig. 2A–D). In contrast, the diffused signal is present in the conidia of a strain transformed only with the RFP gene (Fig. 2E). Relative to the conidia, stronger signals were observed in the mother cell of germling and appressorium, indicating an increased occurrence of autophagy in these cells. By fluorescent staining, we also found that MrAtg8 was not localized on lipid droplets (LDs) (Fig. S1).
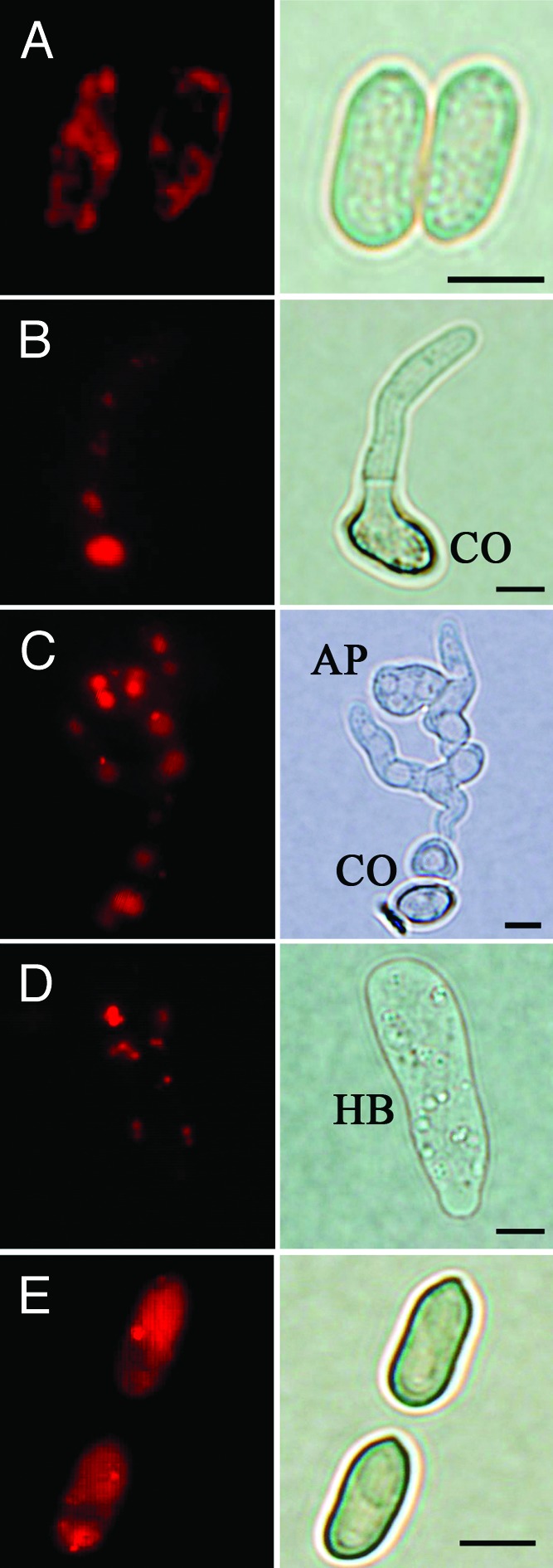
Figure 2. Cellular localization of MrAtg8 in various cell types of M. robertsii. An RFP-MrAtg8 fused construct was introduced into the wild-type strain to examine the localization of MrAtg8 in fungal cells under various developmental stages. Punctate RFP signals were detected in conidia (A), germlings (B), appressorium (C) and hyphal bodies (D). However, the strain only transformed with an RFP gene demonstrated a diffused RFP signal in conidia (E). CO, conidium; AP, appressorium. Hyphal bodies (HB) were harvested from the hemolymph of fungal infected silkworm larvae. Scale bar: 5 µm.
Effects of MrAtg8 on conidiogenesis and appressorium formation
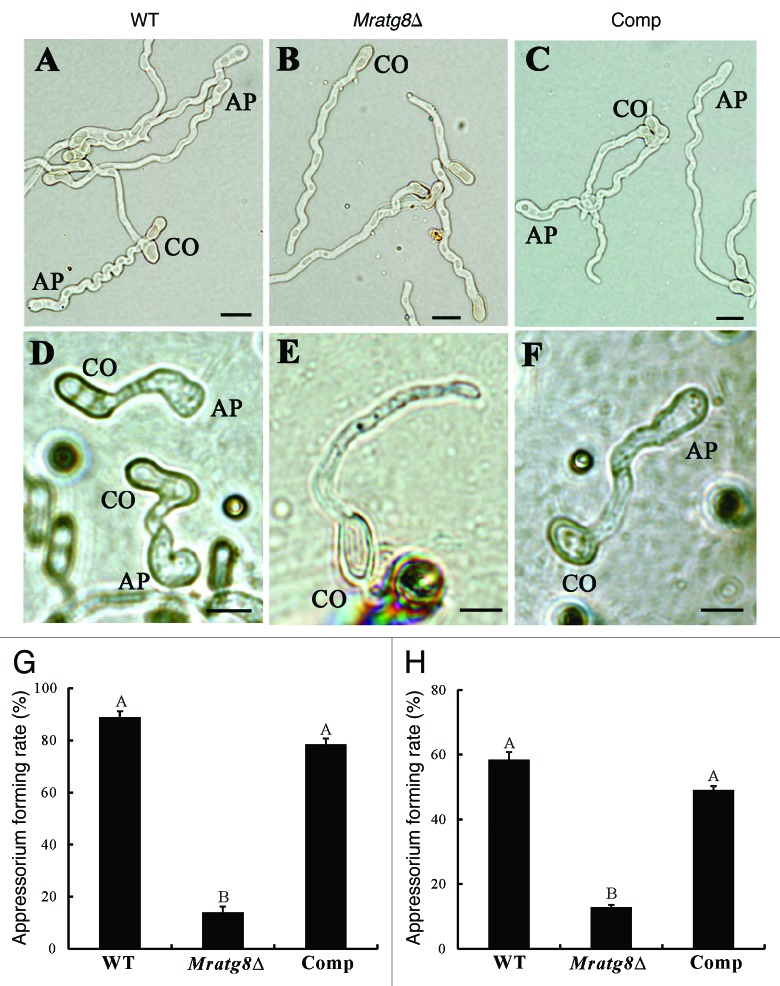
To study MrATG8 gene function, targeted gene replacement was performed to delete MrATG8 in M. robertsii via Agrobacterium tumefaciens-mediated transformation (ATMT) (Fig. S2A). Deletion of MrATG8 in drug-resistant transformants was verified by PCR (Fig. S2B) and confirmed by RT-PCR analyses (Fig. S2C). For mutant complementation, the complete MrATG8 ORF and its promoter region were amplified and transformed into Mratg8Δ and the resulting mutant was designated as Comp. When cultured on different media, we found that the WT, Mratg8Δ and Comp could sporulate on a complete medium (CM) (Fig. 3). While there was no difference between the WT and Comp on minimal medium (MM) and potato dextrose agar (PDA) that no conidiophores were produced by the null mutant on these media (Fig. 3; Fig. S3). This suggests that MrAtg8 affects fungal conidiation in a nutrient-dependent manner. To examine the effect of MrAtg8 on infection-structure differentiation, conidia of the WT, Mratg8Δ and Comp of M. robertsii were harvested from CM and used for appressorium induction either on a plastic hydrophobic surface or locust hind wings.25 Sixteen hours postinoculation, the WT and Comp produced appressoria on both surfaces while most of the conidia of Mratg8Δ germinated without germ-tube tip swelling, indicating a significant failure to form appressoria either on host or nonhost surfaces (Fig. 4A–H).
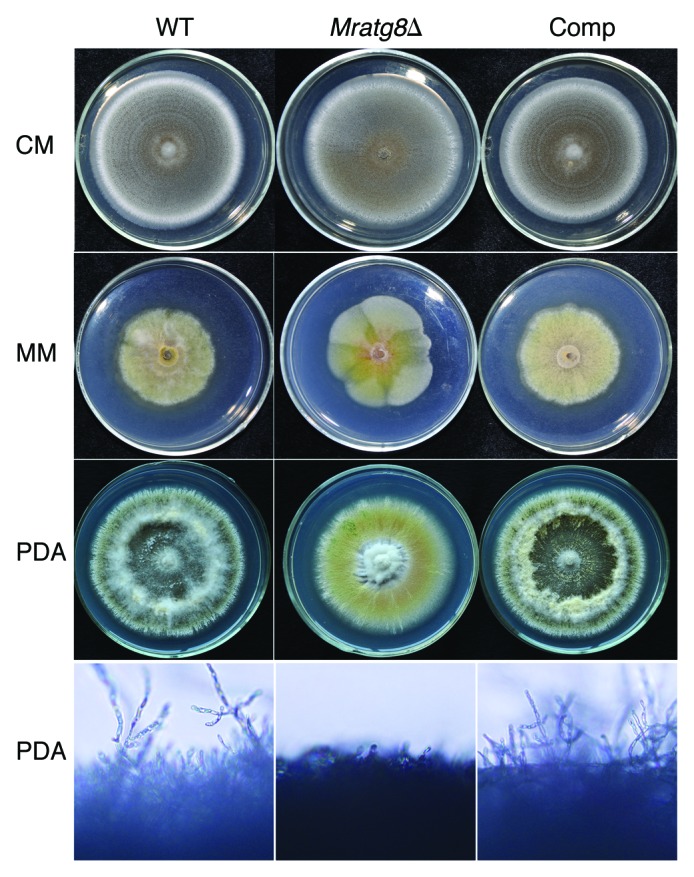
Figure 3. Phenotypic characterization on different media. The wild-type (WT), Mratg8Δ and complemented mutant (Comp, Mratg8Δ complemented with the MrATG8 gene) strains sporulated uniformly on a complete medium (CM) while in contrast to the WT and Comp, Mratg8Δ failed to produce conidia either on a nutrient-poor minimum medium (MM) or a nutrient-rich potato dextrose agar (PDA), indicating the loss of ability to form conidiophores.
Figure 4. Appressorium induction. Conidia of the wild-type (WT), Mratg8Δ and Comp mutant harvested after 20 d on CM plates were subjected to appressorial induction for 16 h either on a hydrophobic surface (A–C; sterile plastic Petri dishes in six cm diameter containing 3 ml of 0.0125% yeast extract) or on locust hind wings (D–F). In contrast to the WT (A and D) and Comp (C and F), appressorium production by Mratg8Δ (B and E) were significantly impaired on the surfaces of either plastic Petri dishes (G) or locust hind wings (H). The same letter labeled within (G or H) means that no statistical difference was detected between the samples at a level of p < 0.01. AP, appressorium. CO, conidium. Scale bar: 5 μm.
Deletion of MrATG8 leads to impaired formation of autophagic bodies and lipid droplets
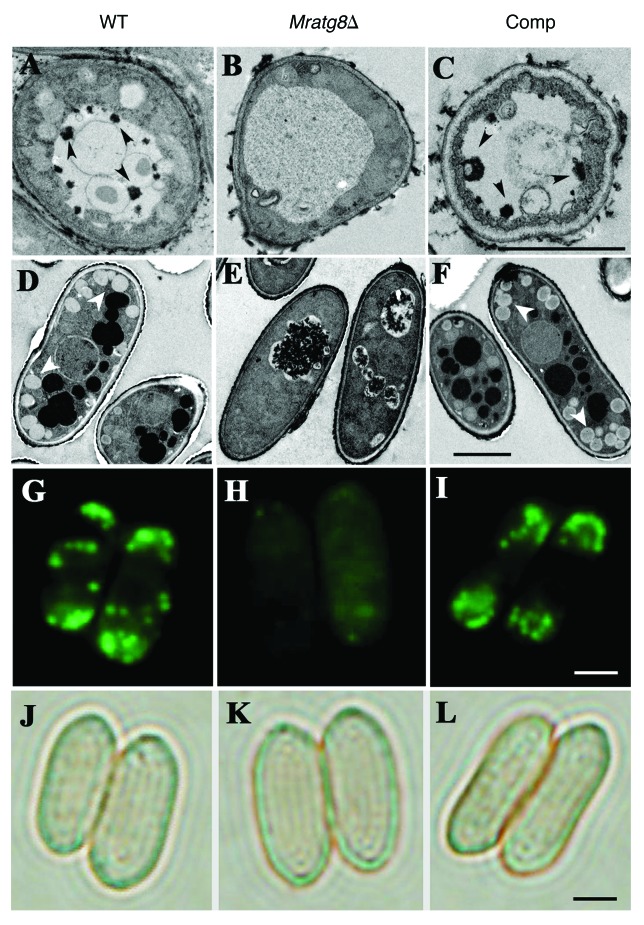
Using transmission electron microscopy (TEM), autophagic bodies were not visible in the vacuoles of Mratg8Δ following nitrogen starvation while they were observed in the Comp cells that was similar to the WT (Fig. 5A–C). Our TEM analysis also found that, the accumulation of LDs in conidia was also significantly impaired in Mratg8Δ which is in contrast to the WT and Comp (Fig. 5D–F). This was confirmed by fluorescent staining using a LD-specific dye Bodipy (Fig. 5G–L). Thus, consistent with the studies in mammalian cells,18,19 the association of autophagy with lipid metabolism is also observed in M. robertsii.
Figure 5.MrATG8 effects on the accumulation of autophagosome and lipid droplets (LDs). After growth in an MM-N medium for 4 h, vacuolar autophagic bodies (arrows) were evident in the WT (A) and Comp (C) cells but not in the gene-deletion mutant (B). Conidia in the wild type (WT), Mratg8Δ and Comp mutant harvested after 20 d on CM plates were used for TEM analysis. In contrast to the WT (D) and Comp (F), accumulation of LDs (arrows) was significantly reduced in Mratg8Δ (E). This was confirmed by Bodipy fluorescent dye staining that shows the lack or presence of a few LDs in Mratg8Δ conidia (comparing H to G and I). (J–L) show the corresponding bright-field images of (G–I), respectively. Scale bar: 2 μm.
Lipid storage is controlled by LD-specific proteins like perilipin.26 We previously identified a mammalian perilipin-like protein Mpl1 that regulates LD storage, appressorium turgor pressure and virulence in Metarhizium.27 To investigate whether the impaired LD accumulation in Mratg8Δ is connected to MPL1 gene transcription and protein expression, we performed RT-PCR and western blot analyses. Interestingly, the transcription of MPL1 in Mratg8Δ was not significantly different when compared with the WT and Comp (Fig. 6A). However, consistent with the reduced numbers of LDs in Mratg8Δ cells (Fig. 5E and H), Mpl1protein level was also lower than in the WT and Comp cells (Fig. 6B). This suggests that impaired lipid storage in Mratg8Δ is concomitantly associated with reduced Mpl1 protein levels.
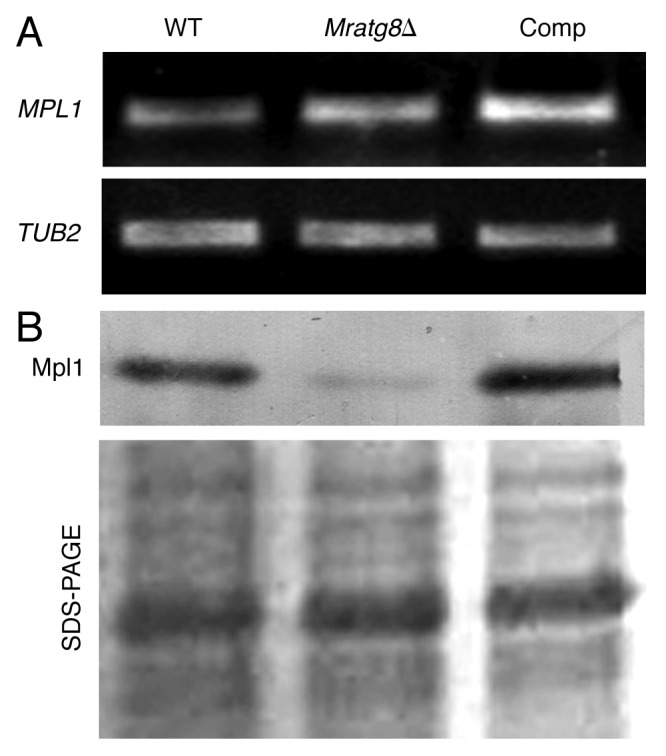
Figure 6. Transcription and protein expression of lipid droplet specific gene. RT-PCR analysis of the Metarhizium LD specific gene, MPL1. No obvious difference in gene transcription was observed among the WT, Mratg8Δ and Comp strains (A). Western blot analysis indicated that, in contrast to the WT and Comp, cellular accumulation of Mpl1 protein was significantly reduced in Mratg8Δ (B).
The carboxyl-terminal glycine residue in MrAtg8 is essential for its function
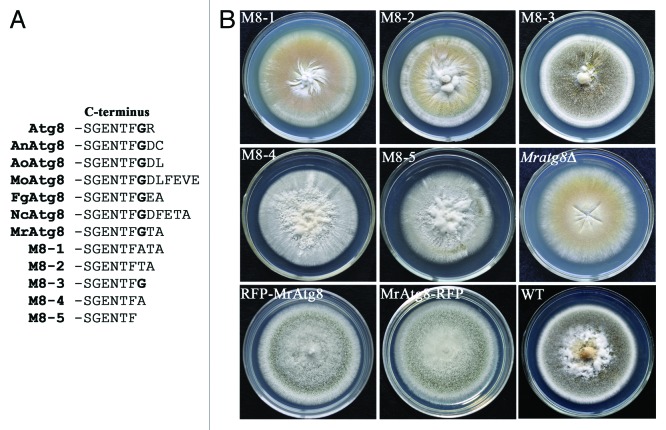
In yeast, the C-terminal glycine residue of Atg8, which is exposed by Atg4 protease cleavage, is essential for membrane binding, and therefore the autophagosome formation and cytoplasm-to-vacuole targeting pathway.17 This conserved Gly residue (G116) is also present in MrAtg8 followed by Thr and Ala residues instead of an Arg residue in yeast Atg8 (Fig. 7A). To study the roles of MrAtg8 C-terminal residues G116, T117 and A188, five variants were generated by designing PCR primers to introduce mutations (Table S1) in the MrATG8 gene. The acquired mutants were designated as M8-1 (G116 replaced with A116), M8-2 (deletion of G116), M8-3 (deletions of T117 and A118), M8-4 (deletions of G116 and T117) and M8-5 (deletions of G116, T117 and A118) (Fig. 7A). Growth tests on PDA medium showed that only the growth of M8-3 was similar to the WT (Fig. 7B). M8-3 could also form appressoria (a differentiation rate at 82.22%) on a hydrophobic surface similarly to the WT (88.4%) (Fig. S4) and kill insects in a similar manner like the WT (Table 1), indicating that G116 residue is crucial to maintain the complete function of MrAtg8. In addition, we found that fusion of the RFP protein either on the N or C terminus of MrAtg8 had no apparent alteration in conidiation of transformants in comparison to the WT (Fig. 7B).
Figure 7. Mutagenesis of MrAtg8 C-terminal residues. (A) Alignment of MrAtg8 with the orthologs of yeast (Atg8, NP_009475), A. nidulans (AnAtg8, AN5131), A. oryzae (AoAtg8, Q2UBH5), M. oryzae (MoAtg8, MGG_01062), F. graminearum (FgAtg8, FGSG_10740) and Neurospora crassa (NcAtg8, NCU01545) showing the conservation of C-terminal residues and the mutation strategies of MrAtg8 to generate M8-1 to M8-5. (B) Phenotyping of the corresponding transformants on PDA medium. RFP-Atg8 and Atg8-RFP cultures are mutants transformed with the MrAtg8 N- and C-terminal fused RFP, respectively.
Table 1. Comparison of virulence between the wild type and mutants assayed on silkworm larvae.
| Strains | LT50 (days) | Significance* |
|---|---|---|
| WT |
4.0 ± 0.39 |
- |
|
Mratg1Δ |
5.0 ± 0.62 |
χ2 = 12.77; p = 0 |
|
Mratg4Δ |
4.6 ± 0.34 |
χ2 = 4.86; p = 0.028 |
|
Mratg8Δ |
NC |
χ2 = 63.12; p = 0 |
|
Mratg15Δ |
4.3 ± 0.16 |
χ2 = 8.51; p = 0.004 |
| Comp |
4.5 ± 0.21 |
χ2 = 0.38; p = 0.54 |
| M8-3 | 4.0 ± 0.15 | χ2 = 0.04; p = 0.85 |
Each mutant was compared with the wild type (WT), respectively and a Log-Rank test was conducted to examine the significance of difference between treatments. Comp, Mratg8Δ gene-rescued strain; M8-3, Mratg8Δ mutant complemented with mutagenized MrATG8 gene by deletions of protein C-terminal T117 and A118 residues. NC, not calculated due to mortality < 50%.
Mratg8 is essential for full virulence of M. robertsii
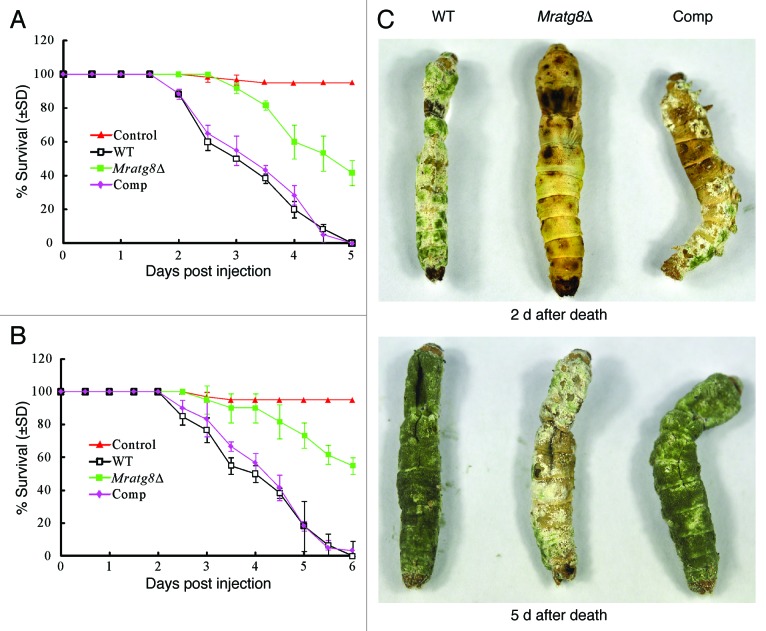
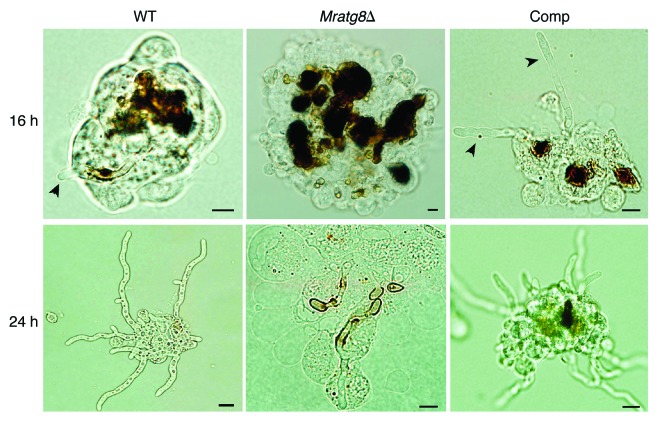
We performed both injection and immersion assays to examine the effects of MrATG8 deletion on fungal virulence against the silkworm larvae. The values of median lethal time (LT50) were estimated and compared among the WT, Mratg8Δ and gene rescued mutant Comp. Injection of the WT and Comp spores killed the insects in five days but Mratg8Δ only caused 41% mortality (Fig. 8A). Thus, the difference between the WT (LT50 = 3.0 ± 0.30) and Mratg8Δ (LT50 = 5.0 ± 0.35) (χ2 = 47.42, p = 0), and between Mratg8Δ and Comp (LT50 = 3.5 ± 0.27) (χ2 = 42.42, p = 0) was significant although the difference between the WT and Comp was not significant (χ2 = 0.22, p = 0.64). After topical infection, WT and Comp killed insects in six days but Mratg8Δ failed to cause even 50% mortality (Fig. 8B). The difference was thus significant between the WT and Mratg8Δ (χ2 = 63.1, p = 0) and between the mutant and Comp (χ2 = 54.5, p = 0) (Table 1). We found that deletion of MrATG8 delayed fungal development and mycosis on insect cadaver, although conidia were produced on insect cuticles (Fig. 8C). Microscopic observations indicated that the conidia of Mratg8Δ were encapsulated and heavily melanized by insect hemocytes 16 h postinjection, while both the WT and Comp could escape from the hemocyte nodules up to 24 h postinjection and produce branched mycelia (Fig. 9). These observations indicate that deletion of MrATG8 impaired fungal defense-response against host immunity thereby lowering virulence.
Figure 8. Insect bioassays. (A) Survival of silkworm larvae injected with 10 µl of 5 × 106 conidia/ml suspensions (control insects were injected with 10 µl sterilized water). Median lethal time (LT50) was estimated as 3.0 ± 0.30 d for the wild type (WT), 5.0 ± 0.35 d for Mratg8Δ and 3.5 ± 0.27 d for Comp, respectively. (B) Survival of silkworm larvae following immersion in 2 × 107 conidia/ml suspensions for 30 sec (control insects were treated with sterilized water). LT50 values were estimated as 4.0 ± 0.39 d for the WT and 4.5 ± 0.21 d for Comp, respectively. Mratg8Δ caused only 45% mortality. (C) Development and conidiation differences among the WT, Mratg8Δ and Comp on insect cadavers. Insects newly killed by the fungi were kept moisturized for different times for mycosis, which showed that, in contrast to the WT and Comp, Mratg8Δ demonstrated defective sporulation on insect cadavers.
Figure 9. Fungal development in insect hemocoel. Relative to the WT and Comp, Mratg8Δ conidia were heavily encapsulated and melanized by insect hemocytes 16 h after injection. Arrows indicate the escaped hyphal tubes. Twenty-four hours postinjection, mycelia of the WT and Comp branched and escaped from the hemocyte nodules but not in the gene-deletion mutant. Scale bar: 10 µm.
Characterization of various autophagy-related genes
Similar to the plant pathogen M. orzyae (Table S2), 23 yeast-like ATG genes were identified in M. robertsii by whole-genome survey.20 To determine whether other autophagy-related genes are required for fungal development and pathogenicity, the orthologous genes of yeast ATG1 (MAA_03501, designated as MrATG1, 46% identity), ATG4 (MAA_04475, MrATG4, 40% identity) and ATG15 (MAA_00506, MrATG15, 38% identity) were deleted in M. robertsii, respectively. TEM analysis showed that, similar to Mratg8Δ, all three null mutants had defects in LD accumulation in conidia (Fig. 10A). While all three mutants could similarly produce conidia on CM medium only the WT and Mratg15Δ could sporulate on PDA medium and Mratg1Δ and Mratg4Δ lost their abilities to sporulate. For gene-complemented mutants, their sporulation abilities on PDA were restored (Fig. S5). Most significantly, we found that the deletion of these three genes did not impair appressoria formation by the mutants when compared with the WT (Fig. 10A; Fig. S6). Consistent with the observation in Mratg8Δ, our western blot analysis revealed that the LD surface protein Mpl1 was downregulated in these three mutants when compared with the WT, particularly in Mratg4Δ (Fig. 10B). In contrast to Mratg8Δ, topical infection of Mratg1Δ, Mratg4Δ and Mratg15Δ caused 58%, 82% and 91% insect mortality, respectively. A significant difference (p < 0.05) was observed between each mutant and the WT in the time taken to kill insect hosts (Table 1).
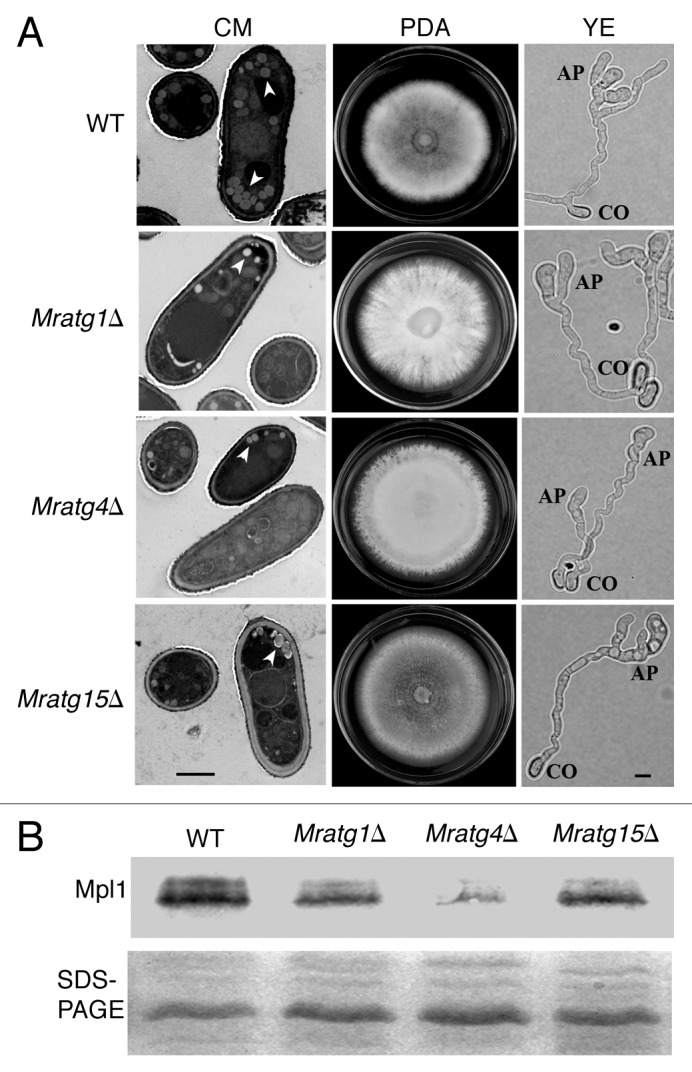
Figure 10. Phenotypic characterization of the various autophagy gene deletion mutants and western blot analysis. (A) Compared with the wild type (WT), accumulation of lipid droplets (arrows) was reduced in the conidia of Mratg1Δ, Mratg4Δ and Mratg15Δ. Mratg1Δ and Mratg4Δ lost abilities to sporulate on the potato dextrose agar (PDA). Like the WT, all three mutants formed appressoria on the hydrophobic plastic surface in yeast extract (YE) medium. CO, conidium; AP, appressoirum; scale bar: 2 μm. (B) western blot analysis showing the reduced level of Mpl1 protein in the conidia of Mratg1Δ, Mratg4Δ and Mratg15Δ in comparison to the WT.
Discussion
The most primordial function of autophagy is to recycle proteins and organelles within cells as an adaptation to nutrient deprivation.1,13 Current understanding of autophagy has extended its function to cell differentiation, polarity, lipid metabolism, immunization and aging.28,29 Autophagy has been studied in a few species of filamentous fungi, especially the plant pathogen M. oryzae.12 Deletion of various autophagy-related genes revealed that autophagy is required for fungal conidiation, infection structure formation and virulence.4,12 In this report, we conducted a comprehensive study of autophagy-related genes in an insect pathogenic fungus M. robertsii by deleting MrATG1, MrATG4, MrATG8 and MrATG15 genes. Disruption of any of these four genes reduced conidial LD accumulation and impaired fungal virulence against silkworm larvae but only Mratg8Δ lost the ability to form appressoria. Most significantly, we found that the LD-specific perilipin-like protein, Mpl1, was downregulated in the mutants specifically in the conidia of Mratg4Δ and Mratg8Δ consistent with impaired LD accumulation. We also found that the localization of MrAtg8 was punctate in the various cell types.
In yeast cells, Atg8 is cleaved by the cysteine protease Atg4 to remove its C-terminal residues and expose Gly116 for Atg8 conjugation with phosphatidylethanolamine (PE) which results in targeting to autophagosomal membranes.30 Although Atg8 is highly conserved in eukaryotes, the C-terminal residues after Gly116 differ in their composition and length in different organisms (Fig. 7A). Our experiments with protein mutations revealed that the amino acids following Gly116 are dispensable for MrAtg8 function. Interestingly, we found that the mutation of MrAtg8 that deleted all residues beyond Gly116 still rescued Mratg8Δ phenotype, i.e., mutant M8-3 (Fig. 7; Fig. S4). This mutation did not require the function of protease MrAtg4. Thus, it is surprising that the Atg8 orthologs have not evolved with Gly as the last residue. Consistent with the successful cleavage of human microtubule-associated protein 1 light chain 3 (LC3, an ortholog of yeast Atg8) C terminus tagged with FLAG or His (6),31 MrAtg8-RFP could rescue Mratg8Δ phenotype (Fig. 7B), suggesting that the residue length of MrAtg8 C terminus does not prevent the protease cleavage. Mutagenesis of LC3 with Ala substituted for G120 (G120A) does not result in cleavage of the mutant protein,31 emphasizing the importance of the C-terminal Gly residue. Similar results were obtained with the mutated MrAtg8 (G116A) strain failing to rescue the Mratg8Δ phenotype (mutant M8-1, Fig. 7). The nonsporulation phenotypes of M8-2, M8-4 and M8-5 mutants confirmed that the C-terminal Gly is crucial for MrAtg8 function, including conjugation with PE.
Genes affecting the virulence of Metarhizium have been functionally identified from those mediating conidial adhesion, controlling cuticle penetration and invading host immunities.9,32 Similar to plant pathogenic fungi,12,33 which require autophagy for infections, deletions of autophagy genes in M. robertsii also led to reduced pathogenicity. However, while the Moatg8 null- mutant of M. oryzae formed appressoria,2,4 Mratg8Δ lost the ability to produce an infection structure. Taken together with the reduced level of LD in conidia, the significantly decreased virulence of Mratg8Δ might be due to the impaired ability of the fungus to penetrate insect cuticle during topical infection. To some extent, it is similar to the reduced virulence of M. robertsii with deletion of the perilipin-like protein gene, MPL1, that leads to decreased turgor pressure within the appressoria.27 However, in our injection assays that bypass insect cuticle Mratg8Δ showed a significantly reduced virulence while Mpl1Δ killed insects like the WT strain.27 This could be due to the inability of Mratg8Δ to counteract insect defense responses (Fig. 9).
In yeast, Atg8 is processed by the cysteine protease Atg4 to expose the C-terminal glycine residue for membrane targeting.34 MoAtg4 of M. orzyae interacts with MoAtg8 and deletion of MoATG4 leads to a significant reduction in conidiation, delay in appressorium formation and a lower turgor pressure.24 Similarly, Mratg4Δ lost the ability to sporulate on PDA medium and also showed a significantly reduced virulence when compared with the WT strain. Consistent with reduced virulence in M. orzyae with MoATG1deletion3,4 and F. graminearum with FgATG15 (FGSG_02519) deletion,6 Mratg1Δ and Mratg15Δ of M. robertsii were also slower in killing insects when compared with the WT strain. This could be, in part, due to the reduced turgor pressure of appressoria caused by impaired LD accumulation in conidia. It is interesting to note that Atg15 extends yeast life span by mediating intravacuolar membrane disintegration.35 The role of MrAtg15 in M. robertsii remains to be determined.
Although autophagy is highly conserved in eukaryotic cells, differences have been observed between yeasts and mammalian cells.36 When compared with yeasts,13 our genome survey indicated that 11 of 34 yeast ATG orthologs (ATG10, ATG14, ATG19, ATG23, ATG25, ATG28, ATG30, ATG31, ATG32, ATG33 and ATG34) are not present in the genomes of M. robertsii and M. oryzae (Table S2), implying that different mechanisms are employed by the yeasts and filamentous fungi for degradation of proteins or organelles. In particular, ATG32 and ATG33 essential for the specific autophagic elimination of mitochondria (mitophagy),37 are not present in M. robertsii or M. oryzae. Interestingly, the ortholog of yeast ATG11, a mitophagy receptor,38 is present in the genomes of M. robertsii (MAA_04312, 20% identity) and M. oryzae (MGG_04486, 22% identity). Three of five ATGs, ATG25, ATG28 and ATG30, are also absent in Metarhizium or Magnaporthe (Table S2). The protein products of these genes are required for vacuolar degradation of peroxisomes, i.e., the pexophagy.39 Yeast ATG14 encodes the autophagy-specific subunit of phosphatidylinositol-3-kinase complex.40 The ortholog is also absent in filamentous fungi (Table S2), suggesting an alternative strategy in filamentous fungi to form the proper kinase complex to phosphorylate phosphatidylinositol, an essential process in autophagy.
We also found a variety of phenotypic differences in different organisms with disruptions of ATG gene orthologs. For example, Mratg8Δ of M. robertsii could not produce appressoria but Moatg8Δ of M. oryzae could form appressoria on hydrophobic surface.2,4 Interestingly, both Mratg8Δ and Moatg8Δ showed a significant reduction in the ability to infect insect and rice hosts, respectively, while the ability of F. graminearum Fgatg8Δ to infect barley and wheat remained like the WT.41 Similar to Aoatg4Δ and Aoatg15Δ of A. orzyae,42 Mratg4Δ and Mratg15Δ lost the ability to form conidia. In contrast, Moatg4Δ and Moatg15 of M. oryzae can produce conidia but exhibit significantly reduced abilities of conidgiogenesis.4,24 There are also differences between fungal and mammalian autophagy gene-deletion mutants. For example, in contrast to the increased lipid contents in mammalian cells followed by deletions of autophagy genes,18,21 reduced LD accumulation was evident in the four null mutants of M. robertsii. Compared with the WT cells, fewer LD were also observed in the conidia of Moatg1Δ of M. oryzae3 and Fgatg15Δ of F. graminearum.6 Studies in different organisms indicate that autophagy has evolved multiple times. For example, autophagy in mammals is much more ancient than previously anticipated and in yeast the autophagic pathway appears to be specialized.36 The evolution of autophagy in filamentous fungi remains to be determined. Nevertheless, our study indicated that a comprehensive understanding of the mechanisms and effects of autophagy by using various organisms as research models is resourceful.
Given that autophagy regulates lipid metabolism it is reasonable to assume a connection between lipolysis and autophagy,18,19,21 however, the exact mechanism(s) of this connection is still unknown. In mammalian cells two distinct functions have been suggested for autophagy in LD homeostasis; one is the regulation of LD breakdown in hepatocytes and the second is the control of LD formation in adipocytes associated with adipose differentiation.21 Because of this, inhibition or genetic knockdown of autophagy markedly increases lipid content in hepatocytes and LDs destined for degradation could be visualized within autophagic vacuoles.18 Deletion of autophagy gene in the protozoan parasite Leishmania major markedly increases cellular content of phospholipids PE and phosphatidylcholine.43 In contrast, knockdown of autophagy genes in adipocytes block LD formation due to lack of LD biogenesis.19 Before this study, fewer LDs have been observed in the conidia of Moatg1Δ and FgAtg15Δ of the plant pathogens M. oryzae and F. graminearum, respectively.3,6 Thus, to a certain extent fungal spores are similar to animal adipocytes where autophagy controls LD biogenesis rather than breakdown. According to the current theory, LD biogenesis involves neutral lipid accumulation within the membrane bilayer of endoplasmic reticulum followed by budding off and enwrapping by a phospholipid monolayer containing specific proteins including perilipin.44 Except for the perilipin-like protein Mpl1, mammalian LD associated proteins like the adipose differentiation-related protein (PLIN2/ADRP, also called perilipin-2) and tail-interacting protein of 47 kDa (PLIN3/TIP47, also called perilipin-3) are not present in the genomes of Metarhizium20 and other filamentous fungi (our own blast searches). Thus, perilipin-like protein is largely responsible for the regulation of lipid storage and metabolism in filamentous fungi. In mammalian cells, perilipin (PLIN1), PLIN2 or PLIN3 protein, if not bound to LDs, will be covalently ubiquitinated and targeted for proteasomal degradation.26 In this respect, since the transcription of MPL1 is not affected in Mratg8Δ (Fig. 6A), the reduction of Mpl1 protein in mutant conidia could be presumably due to proteasomal degradation when there is not enough LDs for Mpl1 to bind. MrAtg8 is not localized on LDs (Fig. S1), implying the absence of protein-protein interaction between MrAtg8 and Mpl1. It remains to be established whether the deletion of autophagy gene affects neutral lipid accumulation or the formation phospholipid monolayer. On the other hand, we cannot preclude that autophagy is involved in LD degradation in fungi. Indeed, it has been observed in F. graminearum that deletions of autophagy genes block LD degradation in mycelia upon nutrient deprivation.6,41
Collectively, our data indicated that autophagy plays an important role in the differentiation and virulence of an insect pathogenic fungus, M. robertsii and the fungus represents a genetically tractable new system to study the mechanisms of autophagy and how that relates to other physiological processes, including lipid metabolism.
Materials and Methods
Fungal strains and growth conditions
The wild-type strain and mutants of M. robertsii ARSEF 2575 (previously named as M. anisopliae) were routinely cultured on potato dextrose agar (PDA, Difco, 213200) at 25°C. For characterization of growth, fungi were additionally cultured on a minimum medium (MM)27 and a complete medium (CM).24 For liquid incubation, fungi were grown in Sabouraud dextrose broth (SDB, Difco, 238220) and MM-N (MM without the nitrogen source and agar). Yeast strains SEY6210 (WT, MATα, ura3 leu2 his3 trp1 lys2 suc2) and the ATG8 deletion mutant (KVY5, apg8Δ-HIS3)22 were included in this study for gene complementation assay. These were cultured in YPD (1% yeast extract, 2% peptone and 2% glucose) and SD-N (2% glucose and 0.17% yeast nitrogen base with neither amino acids nor ammonium sulfate).
Yeast complementation
For functional complementation of yeast Atg8Δ gene, the full-length cDNA of MrATG8 was amplified from a cDNA library with primers A8XbU and A8XbD (Table S1) and cloned into the XbaI site of the yeast expression vector pYES2 (Invitrogen, V825-20) under the control of the GAL1 promoter. The resultant vector pYES-MrATG8 was then transformed into the yeast KVY5 strain using a small-scale yeast transformation protocol.27 The uracil prototrophic transformants were selected and verified by PCR. The complementation effect of Metarhizium MrATG8 gene was tested in a starvation challenge as described previously.3 Briefly, the yeast cells (SEY6210, KVY5 and MrATG8 transformants) were streaked on YPD plates and cultured for two days at 30°C and then moved to SD-N medium for 18 d at 30°C.
Gene deletions
Targeted gene deletion of MrATG8 gene was performed by homologous recombination as we described before.45 Briefly, the 5′ and 3′ flanking sequences of MrATG8 gene were amplified using genomic DNA as a template with primer pairs ATG8UF and ATG8UR, ATG8DF and ATG8DR (Table S1), respectively. The products were digested with BamHI and SpeI, respectively, and then inserted into the corresponding sites of the binary vector pDHt-bar (conferring resistance against ammonium glufosinate) to produce the binary vector pBarATG8 for Agrobacterium-mediated fungal transformation (ATMT). For mutant complementation, the full-length ORF plus the promoter region of MrATG8 was amplified with the primers A8XbU and A8XbD (Table S1) and the purified product cloned into the binary vector pDHt-ben-gpdA (conferring resistance against benomyl) to produce pBenATG8. To investigate the effects of other autophagy genes in Metarhizium, the orthologs of yeast ATG1 (designated as MrATG1, MAA_03501), ATG4 (MrATG4, MAA_04475) and ATG15 (MrATG15, MAA_00506) were additionally deleted in M. robertsii for functional characterizations. Gene complementation was conducted for these mutants by amplification of individual ORF and transformation of the mutant using the benomyl selection marker gene BEN as described above.
RT–PCR and western blot analyses
Total RNA was extracted using a TRIzol Reagent (Invitrogen, 10296-028) from mycelia grown in MM-N medium and SDB, conidia from CM plates and appressoria differentiated on locust hind wings after 24 h of culture. RT-PCR was performed using the primers ATG8F and ATG8R as described previously.46 The tubulin β-2 gene, TUB2, (MAA_02081) was amplified using primers TUBF and TUBR (Table S1) and served as an internal positive control. To detect the expression of the mammalian perilipin-like Mpl1,27 which is a lipid droplet specific protein, the predicted antigenic peptide RADSLGDKTLDRIDERFPIVKKPTSC was synthesized, conjugated with keyhole limpet hemycyanin and used to raise polyclonal antibodies in New Zealand white rabbits. The conidia of the WT and the various mutants were harvested from the CM plates, washed three times in PBS buffer and homogenized in PBS with silica beads (1 mm in diameter, BioSpec Products Inc., 11079110z) using FastPrep (MP Biomedicals, 116004500). Equal amounts of proteins (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12% gel), transferred to polyvinylidene fluoride (PVDF) membranes and probed with the antibodies as previously described.45
MrATG8 gene point-mutations
C-terminal Gly residue is essential for Atg8-mediated autophagy.17 To verify the importance of Gly116 and other terminal amino acid(s) in MrAtg8, five variants of MrAtg8 were created by PCR using different primers (Table S1). Based on the sequence of MrATG8 gene, primer pairs A8XbU/A8XbD1, A8XbU/A8XbD2, A8XbU/A8XbD3, A8XbU/A8XbD4 and A8XbU/A8XbD5 were designed, to introduce the following mutations in MrATG8 respectively; substitution of Gly116 to Ala116, deletion of Gly116, deletions of Thr117 and Ala118, deletions of Gly116 and Thr117, and deletions of Gly116, Thr117 and Ala118 (Fig. 7A). Amplicons were inserted individually into the XbaI site of plasmid pDHt-ben-gpdA to generate pBenAtg81, pBenAtg82, pBenAtg83, pBenAtg84 and pBenAtg85, for transformation of Mratg8Δ by ATMT. The resulting transformed strains were designated as M8-1, M8-2, M8-3, M8-4 and M8-5, respectively.
MrATG8 gene fusions
Primers RfpXbF and RfpXbR (Table S1) were used to amplify the ORF of the red fluorescent protein (RFP) gene using plasmid pMT-mRFP1 (Fungal Genetic Stock Center) as the template. The amplified RFP fragment was then ligated into the XbaI site of the vector pDHt-ben-gpdA to create pBenRFP for direct transformation of the WT strain. In addition, to generate the RFP-MrATG8 and MrATG8-RFP fusion constructs, fusion PCRs were performed with corresponding primers. In the first PCR, primer pairs RfpXbF-RfA81 and RfA82-A8XbD were used to amplify the RFP gene without stop codon from the plasmid pBenRFP and MrATG8 from a cDNA library, respectively. The second PCR was performed with the primers RfpXbF and A8XbD using the PCR products from above as templates and the resulting fusion product was cloned into the XbaI restriction site of vector pDHt-ben-gpdA to produce pRFP-ATG8. The same strategy was employed to generate pATG8-RFP by using the primer pairs A8XbU-A8Rf1 and A8Rf2-RfpXbR in the first PCR that amplified MrATG8 without the stop codon and the primers A8XbU and RfpXbR in the second PCR amplification. The resulting fusion product was cloned into the XbaI site of pDHt-ben-gpdA to generate pATG8-RFP. The orientation of each construct was verified by additional PCRs and sequencing. All these constructs were transformed into the Mratg8Δ mutant strain by ATMT. To localize the distribution of MrAtg8 protein, pRFP-ATG8 was also introduced into the wild-type strain of M. robertsii.
Fluorescent staining and microscopy
Fluorescence microscopy was performed on an Olympus BX51 microscope (Olympus, BX51-33P). For lipid-droplet staining, conidia harvested from CM plates were stained with the Bodipy dye (Invitrogen, D3922) as described previously.27 Images of cells were captured with an Olympus DP71 CCD camera using the Image-Pro® Express software.
Transmission electron microscopy (TEM) analysis
For TEM observations, fungal conidia collected from 14-d-old CM plates were cultured in SDB liquid medium at 25°C and 200 rpm for 24 h. The mycelia were harvested, washed with sterilized distilled water for three times, and transferred into a MM-N liquid medium containing 4 mM phenylmethylsulfonyl fluoride.3 After incubation at 25°C and 200 rpm for 4 h, fungal cultures were collected and fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) at 4°C overnight. The samples were then rinsed three times with phosphate buffer and fixed overnight in 1% osmium tetroxide in 0.1 M cacodylate buffer (pH 7.0) at 4°C. After rinsing three times with phosphate buffer, the samples were dehydrated in a gradient ethanol series, infiltrated with a graded series of epoxy resin in epoxy propane, and then embedded in Epon 812 resin. The ultrathin sections were stained in 2% uranium acetate followed by lead citrate and visualized under a transmission electron microscope (Hitachi, H-7650) operating at 80 kV.
Appressorium induction and insect bioassays
Appressoria of the WT and mutant strains of Metarhizium was induced either on locust hind wings or on sterile plastic Petri dishes (6 cm in diameters, Fisher Scientific, 14-793-07) containing 3 ml of 0.0125% yeast extract.25 The induction rates of appressorium differentiation were quantified by examining different microscopic fields of at least 300 spores for each strain. The differences between different strains were subject to a t-test analysis. To investigate the effect of MrATG8 mutation on fungal virulence, insect bioassays were conducted against the newly emerged fifth instar silkworm, Bombyx mori.45 Conidia of the WT, Mratg8Δ and gene complementary mutants harvested from CM plates were applied topically by immersing the larvae for 30 sec in an aqueous suspension containing 1 × 107 conidia/ml. Each treatment had three replicates with 20 insects each and the experiments were repeated twice. Mortality was recorded every 12 h. Median lethal time (LT50) was calculated by Kaplan-Meier analysis with the program SPSS (Ver. 13.0). Additional insects were injected and bled at different times to observe fungal development within insect hemocoel.32
Supplementary Material
Acknowledgments
We are grateful to Dr. Xiaohong Liu for providing the yeast strains and Mr. Xiaoyan Gao for helps with the transmission electron microscopy analysis. This work was supported by the National Natural Science Foundation of China (Grant nos.: 31225023 and 30970034), the Ministry of Science and Technology of China (Grant no.: 2009CB118904) and the Chinese Academy of Sciences (Grant no. KSCX2-EW-N-06).
Glossary
Abbreviations:
- Atg
autophagy-related
- LD
lipid droplet
- ATMT
Agrobacterium tumefaciens-mediated transformation
- TEM
transmission electron microscopy
- PLIN1
perilipin
- PLIN2/ADRP
perilipin 2/adipose differentiation-related protein
- PLIN3/TIP47
perilipin3/tail-interacting protein of 47 kDa
- MPL1
Metarhizium perilipin-like protein 1
- TUB2
tubulin beta-2
- BEN
benomyl-resistance protein
- RFP
red fluorescent protein
- WT
wild type
- PDA
potato dextrose agar
- MM
minimum medium
- CM
complete medium
- SDB
Sabouraud dextrose broth
- YPD
yeast extract peptone dextrose
- PCR
polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PVDF
polyvinylidene fluoride
- LT50
median lethal time
- AP
appressorium
- CO
conidium
- ORF
open reading frame
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/23575
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23575
References
- 1.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 2.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–3. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 3.Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell. 2007;6:997–1005. doi: 10.1128/EC.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci U S A. 2009;106:15967–72. doi: 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu F, Liu XH, Zhuang FL, Zhu J, Lin FC. Analyzing autophagy in Magnaporthe oryzae. Autophagy. 2011;7:525–30. doi: 10.4161/auto.7.5.15020. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen LN, Bormann J, Le GT, Stärkel C, Olsson S, Nosanchuk JD, et al. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet Biol. 2011;48:217–24. doi: 10.1016/j.fgb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Takano Y, Asakura M, Sakai Y. Atg26-mediated pexophagy and fungal phytopathogenicity. Autophagy. 2009;5:1041–2. doi: 10.4161/auto.5.7.9316. [DOI] [PubMed] [Google Scholar]
- 8.Palmer GE, Askew DS, Williamson PR. The diverse roles of autophagy in medically important fungi. Autophagy. 2008;4:982–8. doi: 10.4161/auto.7075. [DOI] [PubMed] [Google Scholar]
- 9.St. Leger RJ, Wang CS, Fang WG. New perspectives on insect pathogens. Fungal Biol Rev. 2011;25:84–8. doi: 10.1016/j.fbr.2011.04.005. [DOI] [Google Scholar]
- 10.Xiao GH, Ying SH, Zheng P, Wang ZL, Zhang S, Xie XQ, et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St Leger RJ, Wang CS. Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl Microbiol Biotechnol. 2010;85:901–7. doi: 10.1007/s00253-009-2306-z. [DOI] [PubMed] [Google Scholar]
- 12.Pollack JK, Harris SD, Marten MR. Autophagy in filamentous fungi. Fungal Genet Biol. 2009;46:1–8. doi: 10.1016/j.fgb.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 15.Kikuma T, Ohneda M, Arioka M, Kitamoto K. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot Cell. 2006;5:1328–36. doi: 10.1128/EC.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadal M, Gold SE. The autophagy genes ATG8 and ATG1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol Plant Pathol. 2010;11:463–78. doi: 10.1111/j.1364-3703.2010.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–39. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q, Jin K, Ying SH, Zhang Y, Xiao GH, Shang YF, et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7:e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong H, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrinol Metab. 2011;22:234–40. doi: 10.1016/j.tem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong B, Liu XH, Lu JP, Zhang FS, Gao HM, Wang HK, et al. MgAtg9 trafficking in Magnaporthe oryzae. Autophagy. 2009;5:946–53. doi: 10.4161/auto.5.7.9161. [DOI] [PubMed] [Google Scholar]
- 24.Liu TB, Liu XH, Lu JP, Zhang L, Min H, Lin FC. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy. 2010;6:74–85. doi: 10.4161/auto.6.1.10438. [DOI] [PubMed] [Google Scholar]
- 25.Wang CS, St Leger RJ. Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot Cell. 2005;4:937–47. doi: 10.1128/EC.4.5.937-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–40. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang CS, St Leger RJ. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J Biol Chem. 2007;282:21110–5. doi: 10.1074/jbc.M609592200. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimoto K, Takano Y, Sakai Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010;584:1350–8. doi: 10.1016/j.febslet.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–46. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 30.Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta. 2012;1824:44–50. doi: 10.1016/j.bbapap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–10. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Kang Q, Lu Y, Bai L, Wang CS. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci U S A. 2012;109:1287–92. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2012;8:e1002514. doi: 10.1371/journal.ppat.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 35.Tang F, Watkins JW, Bermudez M, Gray R, Gaban A, Portie K, et al. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy. 2008;4:874–86. doi: 10.4161/auto.6556. [DOI] [PubMed] [Google Scholar]
- 36.King JS. Autophagy across the eukaryotes: Is S. cerevisiae the odd one out? Autophagy. 2012;8:1159–62. doi: 10.4161/auto.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanki T, Klionsky DJ, Okamoto K. Mitochondria autophagy in yeast. Antioxid Redox Signal. 2011;14:1989–2001. doi: 10.1089/ars.2010.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manjithaya R, Nazarko TY, Farré JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–73. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol. 2011;2011:713435. doi: 10.1155/2011/713435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Josefsen L, Droce A, Sondergaard TE, Sørensen JL, Bormann J, Schäfer W, et al. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum. Autophagy. 2012;8:326–37. doi: 10.4161/auto.18705. [DOI] [PubMed] [Google Scholar]
- 42.Kikuma T, Kitamoto K. Analysis of autophagy in Aspergillus oryzae by disruption of Aoatg13, Aoatg4, and Aoatg15 genes. FEMS Microbiol Lett. 2011;316:61–9. doi: 10.1111/j.1574-6968.2010.02192.x. [DOI] [PubMed] [Google Scholar]
- 43.Williams RA, Smith TK, Cull B, Mottram JC, Coombs GH. ATG5 is essential for ATG8-dependent autophagy and mitochondrial homeostasis in Leishmania major. PLoS Pathog. 2012;8:e1002695. doi: 10.1371/journal.ppat.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robenek H, Buers I, Hofnagel O, Robenek MJ, Troyer D, Severs NJ. Compartmentalization of proteins in lipid droplet biogenesis. Biochim Biophys Acta. 2009;1791:408–18. doi: 10.1016/j.bbalip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Wang CS, St Leger RJ. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc Natl Acad Sci U S A. 2006;103:6647–52. doi: 10.1073/pnas.0601951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan Z, Shang Y, Gao Q, Zheng P, Wang CS. A phosphoketolase Mpk1 of bacterial origin is adaptively required for full virulence in the insect-pathogenic fungus Metarhizium anisopliae. Environ Microbiol. 2009;11:2351–60. doi: 10.1111/j.1462-2920.2009.01961.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.