SUMMARY
Splicing and translation are highly regulated steps of gene expression. Altered expression of proteins involved in these processes can be deleterious. Therefore, the cell has many safeguards against such misregulation. We report that the oncogenic splicing factor SRSF1, which is overexpressed in many cancers, stabilizes the tumor-suppressor protein p53 by abrogating its MDM2-dependent proteasomal degradation. We show that SRSF1 is a necessary component of an MDM2/ribosomal-protein complex—separate from the ribosome—that functions in a p53-dependent ribosomal-stress checkpoint pathway. Consistent with the stabilization of p53, increased SRSF1 expression in primary human fibroblasts decreases cellular proliferation and ultimately triggers oncogene-induced senescence (OIS). These findings underscore the deleterious outcome of SRSF1 overexpression and identify a cellular defense mechanism against its aberrant function. Furthermore, they implicate the RPL5-MDM2 complex in OIS, and demonstrate a link between spliceosomal and ribosomal components—functioning independently of their canonical roles—to monitor cellular physiology and cell-cycle progression.
INTRODUCTION
The SR proteins are a phylogenetically conserved protein family involved in constitutive and alternative splicing (Long et al., 2009). SR proteins play additional key roles in the interplay between various steps in gene expression (Zhong et al., 2009). The shuttling SR protein SRSF1 is an essential, prototypical family member that functions in multiple steps of gene expression besides splicing, including chromatin remodeling, transcription, nonsense-mediated mRNA decay (NMD), mRNA export and stability, and translation (Das et al., 2007; Loomis et al.,2009; Michlewski et al., 2008; Sanford et al., 2004; Zhang et al., 2004). Furthermore, alterations in SRSF1 expression affect cell-cycle progression and cell viability (Li et al., 2005). For example, SRSF1 knockdown promotes apoptosis by altering the splicing of pro-apoptotic genes (Moore et al., 2010). SRSF1 is a proto-oncogene that is overexpressed in many different cancers, e.g., because of an increase in its gene copy number or through transcriptional activation by MYC (Anczuków et al., 2012; Das et al., 2012; Karni et al., 2007). The oncogenic properties of SRSF1 are mediated in part through altering splicing of various oncogenes and tumor suppressors (Anczuków et al., 2012; Ghigna et al., 2005; Karni et al., 2007), as well as through activation of the mTOR pathway (Karni et al., 2008; Michlewski et al., 2008).
Normal cells resist oncogenic transformation by activating an intricate anti-tumorigenic pathway, mediated by multiple cell-cycle regulators and tumor suppressors. The tumor-suppressor protein p53 (TP53) is one such critical regulator of cellular homeostasis. In response to cellular stress, multiple mitogenic and genotoxic stresses converge to induce a p53-dependent response, resulting in cell-cycle arrest, apoptosis, DNA repair, or replicative senescence (Ko et al., 1996).
Furthermore, aberrant activation of oncogenes in primary cells likewise activates p53-mediated tumor-suppressive barriers, leading to cell-cycle arrest and the onset of premature cellular senescence. This phenomenon, referred to as oncogene-induced senescence (OIS), is one of the ways cells guard themselves against oncogenic transformation (Alimonti et al., 2010; Courtois-Cox et al., 2008; Bartkova et al., 2006; Narita et al., 2006).
Considering its central role in controlling the cell-cycle, the regulation of p53 is critically important for cell viability. The major regulator of p53 is the ubiquitin ligase MDM2. Upon binding to p53, MDM2 promotes p53 nuclear export and ubiquitylation, leading to its degradation. Additionally, MDM2 binding conceals the N-terminal activation domain of p53, inhibiting its transcriptional activity (Boyd et al., 2000). Many factors regulate p53 by altering its interaction with MDM2, including a subset of ribosomal proteins (RPs) that function independently of the ribosome (Dai and Lu, 2004; Dai et al., 2004; Horn et al., 2008; Zhang et al., 2003). One such ternary complex, consisting of L5, L11, and L23, binds and sequesters MDM2, blocking its ability to ubiquitylate p53, and consequently increasing p53 protein stability and activity (Dai et al., 2004). The RP-MDM2-p53 pathway acts as a stress sensor for aberrant ribosome biogenesis and function, which can be triggered in response to various cues, such as nutrient changes (Bhat et al., 2004) and oncogenic activation, that impose a large burden on the translational machinery. For example, the RP-MDM2-p53 pathway forms an effective barrier against MYC-induced lymphomagenesis (Macias et al., 2010). Furthermore, mutations in the RP-interacting regions of MDM2 have been found in human tumors (Schlott et al., 1997).
Here we describe a novel mechanism of OIS in response to overexpression of SRSF1, through previously unidentified physical and functional connections between SRSF1 and the RP-MDM2 complex, which result in p53 activation, and consequently oncogenic-stress-induced senescence.
RESULTS
SRSF1 is a Component of the RPL5-MDM2 Complex
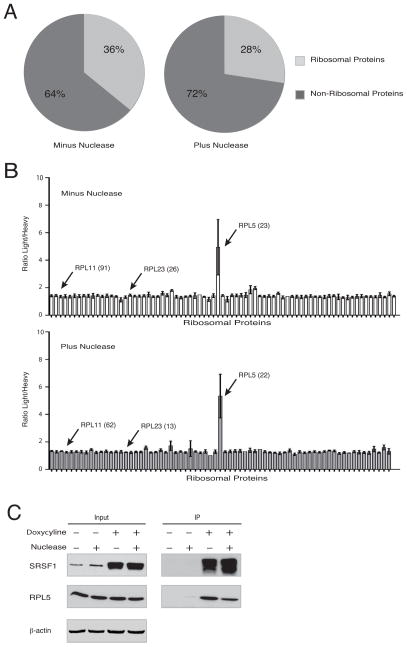
To identify and study how SRSF1 protein–protein interactions regulate the multiple cellular processes in which SRSF1 is involved, we performed immunoprecipitation (IP), followed by mass spectrometry (MS) of T7-tagged SRSF1 from doxycycline-inducible HeLa cells. Approximately 1/3 of all SRSF1-interacting partners identified were components of the ribosome (Figure 1A). As SRSF1 is an RNA-binding protein, we also performed IP-MS following extensive nuclease digestion, to distinguish protein-protein interactions from interactions mediated by RNA. Very few of the SRSF1/RP interactions were lost with nuclease treatment, indicating that most of these interactions are independent of RNA (Figure 1A and Figure S1A).
Figure 1. SRSF1 Interacts Specifically with RPL5.
(A) Percentage of detected SRSF1 interactions that are ribosomal (light gray). T7-SRSF1 was immunoprecipitated from lysates of doxycycline-induced HeLa cells (HeLa TT7-SRSF1) with or without nuclease treatment, and interacting proteins were identified by MudPIT MS.
(B) I-DIRT ratios of light/heavy peptides are shown for all RPs co-immunoprecipitated with T7-SRSF1, with or without nuclease treatment. Bars show average value (+/− s.d.) for each protein, based on individual peptides identified, and total number of peptides for the indicated proteins are shown in parentheses.
(C) HeLa TT7-SRSF1 cells were left untreated or induced with doxycycline for 36 h. Lysates were immunoprecipitated with T7 monoclonal antibody, with or without nuclease treatment. Whole-cell lysates (Input) and IPs were analyzed by immunoblotting with the indicated antibodies.
However, a major difficulty when assessing interactions with RPs is that they are generally considered common contaminants in MS experiments, due to their small size, positive charge, and high abundance (Trinkle-Mulcahy et al., 2008). On the other hand, as SRSF1 has roles in translation regulation (Michlewski et al., 2008; Sanford et al., 2004; Sun et al., 2010), these interactions cannot be ignored. To determine which, if any, SRSF1/RP interactions are biologically relevant and take place intracellularly, as opposed to those established post-lysis or during the IP, we performed a quantitative Isotopic Differentiation of Interactions as Random or Targeted (I-DIRT) IP-MS experiment (Tackett et al., 2005) (outlined in Figure S1B) using the inducible T7-tagged SRSF1 cells. In short, I-DIRT is a quantitative form of mass spectrometry in which, by mixing differentially labeled control and experimental lysates at equal concentrations prior to the IP, one can determine which peptides found to interact with the target (T7-SRSF1) come from the control cells, and which come from the experimental cells. A Light/Heavy ratio of around 1 indicates that, though the proteins were identified to interact with overexpressed T7-SRSF1, this interaction was not formed in the cell, but instead formed post-lysis; whereas a Light/Heavy ratio greater than 3 indicates that the majority of the peptides stem from the experimental T7-SRSF1-overexpressing cells, and thus the interaction is indeed physiological.
Strikingly, of the 74 and 69 RPs identified to interact with T7-SRSF1 without and with nuclease treatment, respectively, only RPL5 had a light/heavy ratio greater than 3-fold over background (Figure 1B and Table S1). This indicates that the T7-SRSF1–RPL5 interaction formed predominantly intracellularly, and that only RPL5 specifically interacts with SRSF1; in contrast, the additional RPs identified formed interactions with T7-SRSF1 post-lysis, and thus interacted non-specifically. Furthermore, the SRSF1–RPL5 interaction persisted after nuclease treatment, suggesting that this interaction is not mediated through RNA (Figures 1B and 1C). Though SRSF1 co-sediments with monosomes and polysomes in cytoplasmic extracts (Sanford et al., 2004), no other RPs were identified by I-DIRT as specifically interacting with SRSF1 in the absence of nuclease treatment. To verify that the ribosome is not involved in the interaction of SRSF1 and RPL5, as well as to identify where in the cell this interaction takes place, we immunoprecipitated endogenous SRSF1 from nuclear and cytoplasmic fractions of HeLa cells. We detected no interaction between endogenous SRSF1 and RPL5 in the cytoplasmic fraction, but endogenous SRSF1 efficiently co-IPd with RPL5 in the nuclear fraction (Figure S1C). Taken together, these data indicate that SRSF1 does not interact directly with the intact ribosome, and that the SRSF1-RPL5 interaction occurs in the nucleus, independently of the ribosome or the large subunit.
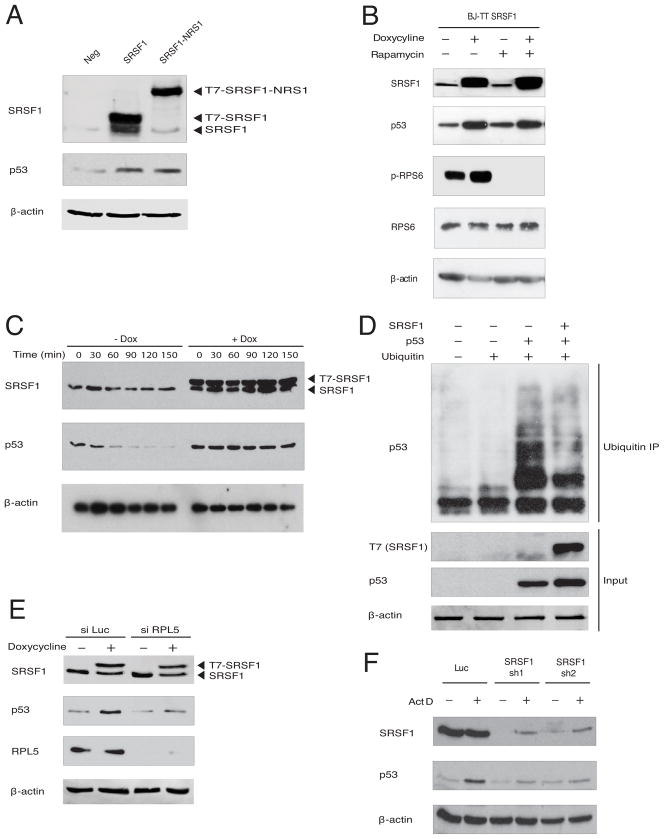
As RPL5 has functions separate from its roles in ribosome function and biogenesis, we used co-IP of endogenous SRSF1 to examine whether SRSF1 is also a component of the RPL5/11/23-MDM2 complex. Consistent with the I-DIRT data, we detected an interaction between endogenous SRSF1 and RPL5; however, we did not detect interactions between endogenous SRSF1 and RPL11 or RPL23 (though overexpressed T7-SRSF1 did co-IP with RPL11 in HeLa cells, data not shown). In addition, we found that SRSF1 interacts with the E3 ubiquitin ligase MDM2 (Figure 2A and Figure S2A). As variations in the canonical RPL5/11/23-MDM2 complex have been previously reported (Horn and Vousden, 2008), our data suggest that SRSF1 is part of an SRSF1-RPL5-MDM2 complex distinct from the canonical complex.
Figure 2. SRSF1 is a Component of an RP-MDM2 Complex and Induces p53 Protein Expression.
(A) BJ fibroblasts were either left untreated or treated with 5 nM actinomycin D for 8 h, followed by 50 μM of proteasomal inhibitor MG132 for 8 h. Lysates were immunoprecipitated with either control IgG or AK96 monoclonal antibody against SRSF1. Whole-cell lysates (Input) and IPs were analyzed with the indicated antibodies.
(B) BJ TT7-SRSF1 fibroblasts or empty-vector control (BJ TT7) were treated with doxycycline for 36 h and 5 nM actinomycin D for 8 h and analyzed by immunoblotting, as indicated. Values represent fold change in protein levels, relative to a loading control. Representative western blots are shown. Data are means +/− s.d. (n=3), *P<0.05, **P<0.01, *** P<0.001.
(C) Total RNA from BJ TT7-SRSF1 cells was amplified by radioactive RT-PCR and analyzed by native PAGE. Values represent fold change in mRNA levels relative to β-actin. Data are means +/− s.d. (n=3), *P<0.05, **P<0.01, *** P<0.001.
See also Figure S2.
SRSF1 Stabilizes p53 through RPL5 and is Necessary for Ribosomal Stress-Induced p53 Activation
Induction of ribosomal stress, such as by treatment with 5 nM actinomycin D, a concentration that specifically inhibits RNA polymerase I, thereby perturbing ribosome biogenesis, has been shown to trigger the formation of the RP-MDM2 complex (Dai and Lu, 2004; Dai et al., 2004; Horn et al., 2008; Zhang et al., 2003). Interestingly, we observed that the interaction of endogenous SRSF1 with RPL5 and MDM2 was strengthened upon actinomycin D treatment (Figure 2A and Figure S2A), highlighting a physiological role for this complex in response to ribosomal stress. We did not observe any change in the sub-cellular localization of SRSF1 upon actinomycin D treatment (Figure S2B).
As the major role of the RPL-MDM2 complex is to sequester MDM2 and stabilize p53 (Dai and Lu, 2004; Dai et al., 2004; Horn et al., 2008; Zhang et al., 2003), we investigated whether overexpression of SRSF1 affects p53 expression in non-transformed human BJ fibroblasts. We generated doxycycline-inducible BJ cells overexpressing SRSF1, or with empty vector as a control. Induction of SRSF1, but not the control, led to increased levels of p53 protein, similar to those seen by treatment with 5 nM actinomycin D (Figure 2B).
This increase in p53 was not due to a direct interaction between SRSF1 and p53 (Figure 2A and Figures S2A and S2C), nor to changes in transcription, splicing, or mRNA stability of the major p53 isoform, as p53 mRNA levels did not change upon overexpression of SRSF1 (Figure 2C). Furthermore, overexpression of SRSF1 and/or actinomycin D treatment did not lead to activation of p14/ARF (Figure 2C), an upstream activator of p53, whereas both mRNA and protein levels of the p53 target gene p21 increased when p53 was upregulated by either means (Figure 2C and Figure S2D).
Though our interaction studies suggest that SRSF1 does not interact with the ribosome, considering the involvement of SRSF1 in translation, we analyzed the effects on p53 expression in BJ cells of nuclear-retained SRSF1 (SRSF1-NRS) (Cazalla et al., 2002) (Figure S5A), a chimeric protein unable to regulate translation (Sanford et al. 2004). Stable overexpression of wild-type SRSF1 or SRSF1-NRS increased p53 expression to comparable levels (Figure 3A). Furthermore, SRSF1 has been shown to activate the mTOR pathway (Karni et al., 2008; Michlewski et al., 2008), which in turn regulates p53 mRNA translation (Astle et al., 2012). However, we saw no reduction in the ability of SRSF1 to induce p53 when cells were pretreated with the mTOR inhibitor rapamycin (Figure 3B). We conclude that SRSF1 increases p53 protein expression and activity, independently of direct or indirect effects on p53 transcription, splicing, mRNA stability, or translation.
Figure 3. SRSF1 Blocks the Ubiquitylation of p53 and is a Necessary Component of the RPL5-MDM2 Complex.
(A) BJ cells were transduced with empty vector, T7-SRSF1, or T7-NRS-SRSF1. Post puromycin selection, whole-cell lysates were analyzed by immunoblotting with the indicated antibodies.
(B) BJ TT7-SRSF1 cells were treated with or without doxycycline for 36 h, followed by rapamycin (200 nM) for 8 h. Lysates were analyzed by immunoblotting with the indicated antibodies.
(C) BJ TT7-SRSF1 cells were treated with or without doxycycline for 36 h, followed by cycloheximide (10 μg/mL) for the indicated times. Lysates were analyzed by immunoblotting.
(D) H1299 cells lacking endogenous p53 were transfected with His-Ub, Flag-p53, and/or T7-SRSF1 plasmids, or the corresponding empty vectors. Cells were lysed under denaturing conditions and incubated with nickel-agarose beads for 3 h. Input and nickel-bound proteins were analyzed by immunoblotting with the indicated antibodies.
(E) BJ TT7-SRSF1 cells were transfected with luciferase or a pool of RPL5 siRNA for 36 h, followed by doxycycline induction for 36 h, as indicated, and analyzed by immunoblotting.
(F) U2OS cells were transduced with luciferase or SRSF1 shRNA, selected with puromycin, followed by actinomycin D (5 nM) treatment for 8 h, as indicated, and analyzed by immunoblotting.
See also Figure S3.
To measure the effect of SRSF1 on p53 protein stability, we performed a cycloheximide-chase experiment in BJ fibroblasts, and monitored p53 levels with and without SRSF1 induction (Figure 3C). Wild-type p53 had a half-life of ~15 min in the control cells. Overexpression of SRSF1 greatly increased the stability of p53, to the extent that no appreciable decline in p53 levels was seen over a 150-min time course, suggesting that SRSF1, like additional components of the RPL-MDM2 complex, inhibits degradation of p53.
To directly determine whether SRSF1 influences p53 ubiquitylation, we measured the ubiquitin status of p53, with and without SRSF1 overexpression in H1299 cells, which lack endogenous p53 (Dai et al., 2004). Transient overexpression of T7-SRSF1 with Flag-p53 and His-Ub decreased the levels of ubiquitylated p53, while increasing the steady-state levels of p53 (Figure 3D). Thus, the effect of SRSF1 on p53 expression occurs at the level of protein stability, with SRSF1 blocking the ubiquitylation and proteasome-mediated degradation of p53.
We next determined whether this increased stability of p53 upon SRSF1 overexpression is dependent on the interaction of SRSF1 with RPL5. siRNA-mediated knockdown of RPL5 in BJ cells severely abrogated the effect of SRSF1 overexpression on p53, whereas a luciferase siRNA had no effect (Figure 3E), consistent with the inability of SRSF1 to interact with MDM2 upon RPL5 knockdown (Figure S3A). Furthermore, upon knockdown of SRSF1, actinomycin D or 5-fluorouracil (Sun et al., 2007) treatment no longer induced p53 protein accumulation to the same levels as in the control, and inhibited the RPL5-MDM2 interaction (Figures 3F, S3B, and S3C), suggesting that SRSF1 is necessary for upregulation of p53 by ribosomal stress. Knockdown of SRSF1 alone did not affect p53 protein levels, as compared to the luciferase control (Figure 3F). Importantly, DNA-damage-induced p53 activation was not affected by knockdown of SRSF1 (Figure S3D), indicating that SRSF1 functions specifically through the ribosomal-stress pathway. Taken together, our data indicate that SRSF1 is dependent on its interaction with RPL5 to increase the stability of p53, and that SRSF1 is a necessary component of the RPL-MDM2 complex that stabilizes p53 in response to ribosomal perturbation.
SRSF1 Overexpression Induces Senescence in Primary Fibroblasts
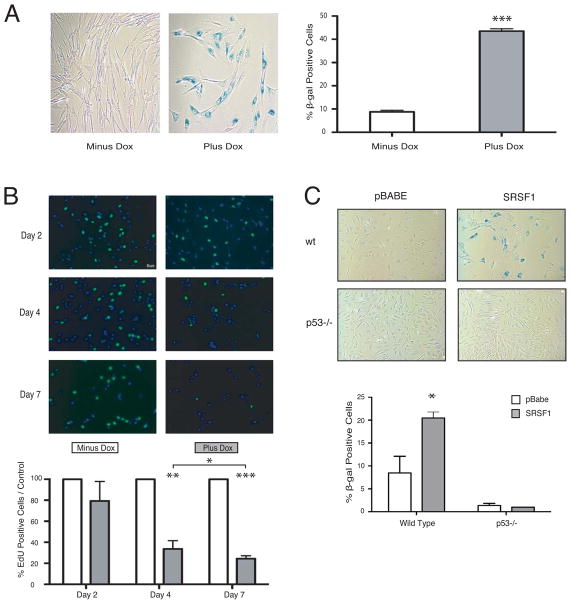
As SRSF1 is a proto-oncogene whose overexpression is sufficient to transform immortal rodent fibroblasts (Karni et al., 2007), this role of SRSF1 in stabilizing p53 may seem counterintuitive. However, overexpression of other potent oncogenes, such as H-RasV12, in primary cells likewise induces p53 activity, leading to a state of premature cellular senescence termed oncogene-induced senescence, or OIS (Chicas et al., 2010; Serrano et al., 1997). In this way, p53 forms an essential barrier against cellular transformation upon oncogenic stress (Courtois-Cox et al., 2008). We therefore characterized cell morphology and senescence-associated β-galactosidase (SA-β-gal) accumulation to determine whether modest overexpression of SRSF1 in non-transformed BJ fibroblasts induces senescence. Prolonged overexpression of SRSF1 resulted in enlarged and flattened cells with an increased number of intracellular vesicles—phenotypes typical of senescence (Chicas et al., 2010; Serrano et al., 1997) (Figure 4A, left panel and Figure S4A). In addition, SRSF1 overexpression resulted in a 5-fold increase in the number of SA-β-gal-stained cells, compared to uninduced control cells (Figure 4A, right panel).
Figure 4. Overexpression of SRSF1 Leads to Senescence of Primary Fibroblasts through p53 Induction.
(A) BJ TT7-SRSF1 cells were induced with doxycycline for 7 d, fixed, stained with X-gal, and observed at 20× magnification (left panels). 200 cells were counted for each condition (right panel); n=6. Means +/− s.d. are shown; ***P = 0.0003.
(B) BJ TT7-SRSF1 cells were induced with doxycycline for 2, 4, or 7 d, incubated with 10 μM EdU, and observed at 20× magnification (top panels). 100 cells were counted for each condition (lower panel); n=6. Means +/− s.d. are shown; *P = 0.05, **P = 0.005, ***P = 0.0008.
(C) Wild-type or p53-null MEFs transduced with control or T7-SRSF1-expressing retroviruses were fixed and stained with X-gal (top panels). 200 cells were counted for each condition (lower panels); n=3. Means +/− s.d. are shown; *P = 0.05.
See also Figure S4.
As senescence occurs with a gradual decline in cell proliferation (Chicas et al., 2010; Serrano et al., 1997), we used EdU (a BrdU analog) incorporation to measure cell proliferation in the presence of overexpressed SRSF1 over a seven-day period. Within four days of SRSF1 overexpression, cell proliferation was drastically reduced to only 20% of control cells (Figure 4B). Senescence-associated heterochromatic foci (SAHF), an additional hallmark of senescent cells (Narita et al., 2006; Chicas et al., 2010), were also observed in SRSF1-overexpressing but not in control cells (Figure S4A). However, unlike other senescence-inducing oncogenes, such as Ras, Mos, and Cdc 6 (Di Mocco et al., 2006), SRSF1 overexpression did not trigger an early phase of hyper-proliferation (Figure 4B), nor did it cause detectable DNA damage, as Western blotting and immunofluorescence showed no increase in γ-H2AX levels or CHK1 phosphorylation (Figure S4B, left and right panels, respectively). Furthermore, modest overexpression of SRSF1 in p53-null MEFs (Figure S4C) failed to trigger premature cellular senescence (Figure 4C), indicating that the ability of SRSF1 to activate the tumor-protective senescence response is dependent on an intact p53 pathway. This is once again in contrast to RasV12-induced senescence, for which p53 has been reported to be dispensable for OIS in primary human fibroblasts (Serrano et al., 1997). Based on these results, we predict that tumors driven by SRSF1 overexpression would be compromised in their p53 tumor-suppressive pathway, with mutations, loss or silencing of p53 itself, or any of its regulators or critical downstream target genes. To test this hypothesis, we analyzed public microarray data from different cancer types to ascertain the TP53 expression status of SRSF1-overexpressing tumors (Figure S4D). We found significant anti-correlation between SRSF1 and TP53 expression in cancers of the kidney (282 tumor samples), colon (293 tumor samples), and breast (352 tumor samples), with SRSF1-overexpressing tumors having a tendency to downregulate their TP53 expression.
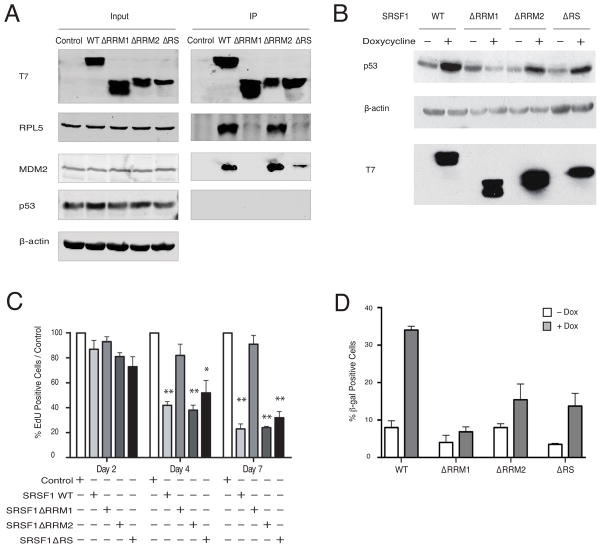
RRM1 is Required for Interaction with the RPL5-MDM2 Complex, p53 Induction, and OIS
To further dissect the role of the SRSF1-RP-MDM2 complex in regulating p53 and inducing premature senescence, we examined the interaction of several SRSF1 domain mutants (Figure S5A) with the other components of the complex. Whereas the ΔRRM2 mutant was indistinguishable from wild-type SRSF1, the ΔRRM1 mutant was almost completely defective in interacting with RPL5 or MDM2 (Figure 5A). Interestingly, the ΔRS mutant, which is defective in nuclear-cytoplasmic shuttling, and primarily accumulates in the cytoplasm (Cáceres et al., 1998), still co-immunoprecipitated with MDM2—though the interaction was much weaker than that for wild-type SRSF1 or ΔRRM2—but it did not interact with RPL5 (Figure 5A). Similarly, whereas SRSF1-NRS was able to interact with RPL5 and MDM2, SRSF1-AAA, another mutant of SRSF1 that accumulates in the cytoplasm (Sinha et al., 2010), failed to interact with RPL5, yet weakly interacted with MDM2 and very marginally induced p53 protein (Figures S5B and S5C). We were unable to observe induction of p53 protein in HeLa cells, in which p53 protein is rapidly degraded due to the expression of HPV16-E6 (May et al., 1991), unless we massively overexpressd SRSF1. Therefore, we examined the effects of these various mutants on induction of p53 and premature cellular senescence in BJ cells. Whereas ΔRRM1 was unable to induce p53, both ΔRRM2 and ΔRS overexpression led to a partial increase in p53 protein (Figure 5B), though in both cases the degree of p53 induction was lower than observed with wild-type SRSF1. Furthermore, only ΔRRM1 overexpression failed to slow down cell proliferation, as assayed by EdU labeling (Figure 5C and Figure S5D). Unlike the wild-type protein, none of the deletion mutants was able to induce premature cellular senescence (Figure 5D), despite the ability of ΔRS and ΔRRM2 to both induce p53 expression and growth arrest to some extent; we attribute this to a threshold effect for p53 induction to elicit senescence. These data suggest that SRSF1 interacts with the RPL-MDM2 complex through RRM1, and through this interaction SRSF1 leads to senescence in primary fibroblasts.
Figure 5. RRM1 of SRSF1 is Required for Interaction with the RPL-MDM2 Complex, p53 Induction, and OIS.
(A) HeLa cells were transfected with wild-type SRSF1 and domain-deletion mutants. Lysates were immunoprecipitated with T7 monoclonal antibody, with nuclease treatment. Whole-cell lysates (Input) and IPs were analyzed by immunoblotting with the indicated antibodies.
(B) BJ TT7-SRSF1, BJ TT7-SRSF1-ΔRRM1, BJ TT7-SRSF1-ΔRRM2 and BJ TT7-SRSF1-ΔRS cells were treated with doxycycline for 36 h and analyzed by immunoblotting, as indicated.
(C) BJ TT7-SRSF1, BJ TT7-SRSF1-ΔRRM1, BJ TT7-SRSF1-ΔRRM2 and BJ TT7-SRSF1-ΔRS cells were induced with doxycycline for 2, 4, or 7 d, and incubated with 10 μM EdU. One hundred cells were counted for each condition; n=3. Means +/− s.d. are shown, *P = 0.05, **P = 0.005.
(D) BJ TT7-SRSF1, BJ TT7-SRSF1-ΔRRM1, BJ TT7-SRSF1-ΔRRM2 and BJ TT7-SRSF1-ΔRS cells were treated with doxycycline for 7 d, fixed, and stained with X-gal. 200 cells were counted for each condition; n=2. Ranges are shown.
See also Figure S5.
DISCUSSION
We have identified a new role of SRSF1 in regulating p53 protein stability and cell viability, summarized in our proposed model (Figure 6). We found that endogenous SRSF1 is an essential component of an RPL-MDM2 complex; through these interactions, it increases the cellular pool of active p53. Furthermore, p53 induction resulting from overexpression of SRSF1 in primary human fibroblasts leads to OIS through a pathway not previously implicated in this oncogenic-stress response.
Figure 6. A model for SRSF1’s Role in the Ribosomal-stress Pathway and Oncogene-Induced Senescence.
We have identified SRSF1 as a critical component of the RP-MDM2 complex, which is formed in response to induction of ribosomal stress. Sequestration of the E3 ligase MDM2 in this complex results in decreased ubiquitylation and increased stability of the tumor-supressor p53 protein. Moreover, we have identified and characterized an anti-tumorigenic response that primary cells mount in response to overexpression of the SRSF1 oncoprotein, which triggers the formation of a nuclear ternary SRSF1-RPL5-MDM2 complex, leading to activation of the p53-mediated tumor-suppressive pathway and OIS.
Using quantitative MS, we found that SRSF1 interacts specifically with RPL5, whereas the remaining RP interactions are non-specific. This interaction occurs independently of the ribosome and of rRNA or mRNA, as nuclease treatment did not disrupt SRSF1-RPL5 binding. Although SRSF1 co-sediments with actively translating ribosomes, and influences translation when bound to exonic splicing enhancers in mature mRNA (Sanford et al., 2004; Michlewski et al., 2008), the main cellular pool of SRSF1 does not appear to interact directly or stably with intact ribosomes.
In addition to RPL5, we identified the E3 ligase MDM2 as an SRSF1-interacting protein. Previous reports described a complex of RPL5/11/23-MDM2 (Dai and Lou, 2004; Dai et al., 2004; Horn et al., 2008; Zhang et al., 2003); however this interaction was not observed by I-DIRT or by co-IP with endogenous SRSF1 (though RPL11 was detected by co-IP when T7-tagged SRSF1 was overexpressed). The apparent lack of bona fide interactions with L11 and L23 suggests that SRSF1 functions in a separate complex from other, previously described RPL-MDM2 complexes. It is possible that SRSF1 competes with L11 or L23 for binding to RPL5 and/or MDM2, therefore displacing components of the complex. Whether particular complexes are formed in response to distinct stress signals, and whether these complexes activate distinct cellular responses in response to induction of ribosomal stress will be important questions to pursue.
SRSF1 expression within cells is tightly controlled (Sun et al., 2010). It is auto-regulated, reflecting the fact that changes in SRSF1 expression can be deleterious to cells. Indeed, even a modest 2-fold overexpression of SRSF1 in immortal murine NIH3T3 fibroblasts, which have a compromised p53 pathway, has been shown to transform them, by promoting proliferation and inhibiting apoptosis (Karni et al., 2007). Here we show that a powerful tumor-suppressive barrier has apparently evolved to guard against overexpression of SRSF1, further emphasizing SRSF1’s potential as a potent oncoprotein. Although it remains unclear how oncogenic pathways can overcome the autoregulation of SRSF1 expression, we show here that enforced overexpression of SRSF1 in normal human primary fibroblasts with intact p53 activates the p53 tumor-suppressive pathway. Consequently, the cells are forced into a state of premature senescence, which protects the host against transformation.
Interestingly, premature senescence induced by SRSF1 overexpression in normal cells is distinct from the classical Ras-induced senescence (Serrano et al., 1997), which is primarily a DNA-damage response induced by hyper-proliferation and is dependent on p16 activation (Bartkova et al., 2006; Chicas et al., 2010; Di Mocco et al., 2006). In contrast, SRSF1-induced senescence does not result in an early hyper-proliferative phase, DNA damage, or activation of p14/ARF, and it is likely due to aberrant activation of the ribosomal-stress pathway, involving distinct regulators and mediators. Thus, the differences in the senescence phenotypes may stem from the distinct underlying mechanisms of induction. Furthermore, we show that SRSF1 is directly involved in mounting the anti-tumorigenic response to its own overexpression, through initiating the formation of an SRSF1-RPL5-MDM2 complex, and thereby stabilizing p53. Hence, it appears that SRSF1 autoregulates not only its expression but also its function, so as to allow the organism to resist oncogenic transformation.
Aberrant activation of the PI3K/AKT signaling pathway likewise induces premature cellular senescence in the absence of hyper-proliferation or DNA damage (Alimonti et al., 2010; Astle et al., 2011). However, AKT-induced senescence is dependent on mTOR, a protein kinase that controls cell growth (Astle et al., 2011). SRSF1 is also an activator of the mTOR pathway, but it bypasses upstream PI3K/AKT signaling (Karni et al. 2008). Considering the critical role of mTOR in regulation of ribosome biogenesis (Mayer et al., 2006), these studies further emphasize the emerging concept of the central role that the ribosome plays in regulating cellular homeostasis and oncogenesis. Additionally, this is the first time that ribosomal stress has been implicated in OIS; however, whether this is a common mechanism of tumor protection caused by ribosomal perturbation, or is unique to the SRSF1 proto-oncogene will need further investigation.
EXPERIMENTAL PROCEDURES
Plasmids
pCG-T7-SRSF1, pCG-T7-SRSF1-ΔRRM1, pCG-T7-SRSF1-ΔRRM2, pCG-T7-SRSF1-ΔRS, pCG-T7-SRSF1-NRS, and pCG-T7-SRSF1-AAA were described previously (Cáceres et al., 1998; Cazalla et al., 2002; Sinha et al., 2010). pMSCV-TT-IRES-Puro was modified from the TMP vector (Dickins et al., 2005) and was a gift from Scott Lowe (MSKCC). pMSCV-TT-T7SRSF1 constructs were generated by subcloning the SRSF1 cDNAs from the pCG-T7-SRSF1 plasmids into pMSCV-TT-IRES-Puro after amplification with Xho1-forward and EcoRl-reverse primers. pCDNA3.1-p53 and pCDNA3.1-HA-Ub were purchased from Addgene. For primer sequences, see Supplemental Methods.
Cell Culture, Virus and Stable Cell Line Generation
BJ, HeLa, U2OS, and NCI-H1299 cell culture conditions were as recommended by ATCC. Viruses were produced as described (Karni et al., 2007). To generate stable doxycycline-inducible cell lines, Tet-on Advanced HeLa, BJ, and U2OS cells (Clontech) were infected with pMSCV-TT-T7SRSF1 constructs or pMSCV-TTIRES-Puro, and selected with puromycin (Sigma). For inducible expression of SRSF1, doxycycline was added at 0.01 to 1.0 μg/mL for 36 h. For I-DIRT (Tackett et al., 2005), HeLa cells were passaged for six doublings in DMEM (Thermo) without L-lysine and L-arginine, supplemented with either L-lysine-2HCl and L-arginine-HCl for light media, or 13C6 L-lysine-2HCl and 13C6 15N4 L-arginine-HCl (Thermo) for heavy media, to a final concentration of 0.1 mg/mL each.
Cell Lysis and Protein Analysis
For protein analysis of whole-cell lysates, cells were lysed in RIPA buffer plus Protease Inhibitor Cocktail EDTA-free (Roche). For whole-cell lysate preparation followed by IP, cells were first lysed in NP-40 Lysis Buffer (0.05–0.5 % (v/v) NP-40, 100–500 mM NaCl, 50 mM Tris, pH 7.4, 1 mM DTT) plus protease inhibitor cocktail. Lysates were sequentially passed through a syringe with a 20G, 22G, and 26G needle. Nuclear/cytoplasmic fractionation was adapted from (Allemand et al., 2005). Where indicated, nuclease was added (1 U/mL of RNase A (Ambion), 40 U/mL RNase T1 (Ambion), 500 U/mL Benzonase Nuclease (EMD) plus 2 mM MgCl2) and incubated on ice for 30 min. Lysates were cleared by high-speed centrifugation at 13,000 g for 15 min at 4 °C.
Immunoblotting
Lysates were separated by SDS-PAGE, probed with the indicated antibodies, and when appropriate, quantified using an Odyssey infrared-imaging system (LI-COR Biosciences). Primary antibodies against the following proteins/epitopes were used: T7 (Novagen), SRSF1 (AK-96, CSHL), RPL5, RPL11, and RPL23 (GeneTex), MDM2 (2A10, Abcam), p53 (DO-1, EMD Bioscience), β-actin (BD Biosciences), p21 (Abcam), γ-H2AX (Upstate Cell Signaling Solutions), phospho-CHK1 (Cell Signaling), p53 polyclonal (Abcam). Secondary antibodies were HRP-conjugated goat anti-mouse or anti-rabbit (Biorad) for chemiluminescent detection, and IR dye 800 or 680 anti-rabbit or anti-mouse (LI-COR Biosciences) for infrared detection.
Immunoprecipitations
Antibody capture and crosslinking to Dynabeads Protein G (Invitrogen) were performed according to the manufacturer’s specifications. For I-DIRT IPs, heavy and light lysates were combined at equal total-protein concentrations, determined by the Bradford assay, before incubation with beads. All IPs were incubated while rotating for 45 min at 4 ºC and then washed five times with Lysis Buffer.
Mass Spectrometry and Sample Preparation
Immunoprecipitations were subjected to On-Bead Digest (Bish et al., 2008) with 75 ng of sequencing-grade trypsin (Promega) for 10 h at 37 ºC. Tryptic peptides were desalted using ZipTips C18 (Millipore). After elution, peptides were dried down and resuspended in 10 μl of Buffer A (2 % (v/v) acetonitrile, 0.1 % (v/v) formic acid). Heavy amino acid incorporation for I-DIRT was analyzed on a quadrupole time-of-flight mass spectrometer (Q-TOF, Agilent 6520); interacting proteins were identified by 13-step MudPIT ESI LC MSMS on an LTQ Orbitrap XL ion-trap mass spectrometer (Thermo Finnigan). Additional details are given in the Supplemental Experimental Procedures.
Cycloheximide Chase Assay
2×105 cells plated in 6-well plates were treated with 10 μg/mL cycloheximide for the indicated times, followed by lysis and immunoblotting.
RNA Interference
The RPL5 siRNA pool was purchased from Dharmacon. BJ and U2OS cells were reverse-transfected with RNAiMax (Invitrogen) at a final concentration of 20 nM. Thirty-six h after transfection of the siRNA, doxycycline was added for an additional 36 h, and where indicated, 5 nM actinomycin D was added for 8 h. Transduction with SRSF1 shRNA was performed as described (Karni et al., 2007).
Additional siRNA target sequences:
Luciferase: 5’-′CGUACGCGGAAUACUUCGA-3’
RT-PCR Analysis
RT-PCR was performed as described (Karni et al., 2007). For a complete list of primers, see Supplemental Experimental Procedures.
In Vivo Ubiquitination Assays
The ubiquitination assay was adapted from William P Tansey, Cold Spring Harb Protoc; 2006; doi:10.1101/pdb.prot4616.
Senescence-associated β-galactosidase Assay
The SA-β-gal assay was adapted from (Chicas et al., 2010). SRSF1 expression was induced by Dox addition on day 0 and analyzed on day 7.
EdU Cell Proliferation Assay
EdU incorporation was measured using a Click-It EdU Cell Proliferation Assay Kit (Invitrogen). SRSF1 expression was induced by Dox addition on day 0 and analyzed on days 2, 4, and 7. Cells were imaged using a Zeiss Axiovert 200M fluorescent microscope.
Statistical Analysis
Where appropriate, the data are presented as the means ± s.d. Data points were compared using unpaired two-tailed Student’s t-tests, and P-values are indicated in the figure legends.
Supplementary Material
HIGHLIGHTS.
SRSF1 is a component of a ribosomal protein RPL5/MDM2 complex that stabilizes p53.
Through this complex SRSF1 is required for ribosomal-stress-mediated p53 activation.
Overexpression of SRSF1 in primary cells leads to oncogene-induced senescence.
SRSF1-induced senescence is mediated by the ribosomal-stress response and p53.
Acknowledgments
We thank S. Lowe and R. Sordella for helpful comments on the manuscript, and T. Koller and C. Ruse for help with MS analysis and helpful discussions. We thank O. Anczuków-Camarda for help with data analysis. O.F. was supported by the Hearst Foundation and the Seraph Foundation. This work was funded by grant CA13106 from the National Cancer Institute and by the St. Giles Foundation. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, Cheng K, Varmeh S, Kozma SC, Thomas G, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–93. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E, Guil S, Myers M, Moscat J, Cáceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci USA. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy S, Krainer AR. The splicing-factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle MV, Hannan KM, Ng PY, Lee RS, George AJ, Hsu AK, Haupt Y, Hannan RD, Pearson RB. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: implications for targeting mTOR during malignancy. Oncogene. 2012;31:1949–1962. doi: 10.1038/onc.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bish RA, Fregoso OI, Piccini A, Myers MP. Conjugation of complex ubiquitin chains to WRNIP1. J Proteome Res. 2008;7:3481–3489. doi: 10.1021/pr800217q. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2.RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Screaton JR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;1:55–56. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Cáceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Jones SL, Cickowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAPII transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Das S, Anczuków O, Akerman M, Krainer AR. SRSF1 is a critical transcriptional target of MYC. Cell Reports. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic micro RNA precursors. Nature Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre’ M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, Hippo Y, Lowe SW, Krainer AR. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc Natl Acad Sci USA. 2008;105:15323–15327. doi: 10.1073/pnas.0801376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Cáceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y. An ARF independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May E, Jenkins JR, May P. Endogenous HeLa p53 proteins are easily detected in HeLa cells transfected with mouse deletion mutant p53 gene. Oncogene. 1991;6:1363–1365. [PubMed] [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nuñez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group A proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlott T, Reimer S, Jahns A, Ohlenbusch A, Ruschenburg I, Nagel H, Droese M. Point mutations and nucleotide insertions in the MDM2 zinc finger structure of human tumours. J Pathol. 1997;182:54–61. doi: 10.1002/(SICI)1096-9896(199705)182:1<54::AID-PATH815>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR. Arginine methylation controls the subcellular localization and functions of the oncoprotein splicing factor SF2/ASF. Mol Cell Biol. 2010;30:2762–2774. doi: 10.1128/MCB.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AK. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Dai M, Hua L. 5-Fluorouracil activation of p53 involves an MDM2- ribosomal protein interaction. J Biol Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.