Summary
ATP-dependent chromatin remodeling enzymes are highly abundant and play pivotal roles regulating DNA-dependent processes. The mechanisms by which they are targeted to specific loci have not been well understood on a genome-wide scale. Here we present evidence that a major targeting mechanism for the Isw2 chromatin remodeling enzyme to specific genomic loci is through sequence-specific transcription factor (TF)-dependent recruitment. Unexpectedly, Isw2 is recruited in a TF-dependent fashion to a large number of loci without TF binding sites. Using the 3C assay, we show that Isw2 can be targeted by Ume6- and TFIIB-dependent DNA looping. These results identify DNA looping as a previously unknown mechanism for the recruitment of a chromatin remodeling enzyme and defines a novel function for DNA looping. We also present evidence suggesting that Ume6-dependent DNA looping is involved in chromatin remodeling and transcriptional repression, revealing a mechanism by which the three-dimensional folding of chromatin affects DNA-dependent processes.
INTRODUCTION
Over the past two decades, an unprecedented amount of information has accumulated on both the structure and function of eukaryotic genomes. DNA sequences and their evolutionary conservation, transcription factor binding sites, nucleosome positions, DNA and histone modification patterns, and transcription initiation and termination sites have been determined at high resolution across many eukaryotic genomes. These studies established linear maps of genomic information that shed light on the regulation of DNA-dependent processes. However, eukaryotic genomes are packaged and function within the three-dimensional space of the nucleus. How this structural arrangement of DNA affects DNA-dependent processes is not well understood.
Efficient three-dimensional packaging of genomes into the relatively small nuclei of eukaryotic cells is achieved at two distinct levels: the compaction of DNA into nucleosomes and the folding of chromatin within the nucleus. Both of these packaging mechanisms are required for normal cellular and developmental processes (Cremer and Cremer, 2001; Rando and Chang, 2009) while defects are associated with complex diseases (Matarazzo et al., 2007; Misteli, 2010; Timme et al., 2011; Wiech et al., 2009; Zardo et al., 2008). Using microscopic approaches, chromosomes within the nuclei of animals, plants, and yeast (Cremer and Cremer, 2010; Duan et al., 2010) have been shown to adopt highly organized nonrandom “territories.” These discrete chromosome conformations have been postulated to regulate DNA-dependent processes. Elucidating mechanisms by which chromatin folding affects DNA-dependent processes will likely reveal important and previously unknown layers of regulation.
The chromosome conformation capture (3C) assay (Dekker et al., 2002) detects DNA loops by measuring the frequency of interactions between any two chromosomal loci, effectively identifying regions that are proximal in three-dimensional space. Using 3C, two general classes of DNA loops have been identified: (i) “chromatin loops” between distal genetic regulatory elements, for example, between a mammalian enhancer or silencer and its target promoter; and (ii) “gene loops,” that specifically place promoter and terminator regions of the same gene in close proximity. To date, chromatin loops and gene loops have been described in human, fly, worm and yeast cells (Ansari and Hampsey, 2005; Duan et al., 2010; Hampsey et al., 2011; Laine et al., 2009; Nemeth et al., 2008; O’Reilly and Greaves, 2007; O’Sullivan et al., 2004; Perkins et al., 2008; Singh and Hampsey, 2007; Tan-Wong et al., 2008; Tan-Wong et al., 2009).
The 3C assay helped identify numerous sequence-specific transcription factors (TFs) (Drissen et al., 2004; Phillips and Corces, 2009; Splinter et al., 2006; Vakoc et al., 2005), general transcription factors (Singh and Hampsey, 2007), RNA 3′-end processing factors (Singh and Hampsey, 2007; Ansari and Hampsey, 2005), and other chromatin bound proteins (Comet et al., 2011; Hadjur et al., 2009; Parelho et al., 2008; Wendt et al., 2008) that are required for the formation and/or maintenance of DNA loops. Functionally, chromatin loops have been linked to transcriptional regulation (Comet et al., 2011; Nemeth et al., 2008; Perkins et al., 2008; Schoenfelder et al., 2010a; Schoenfelder et al., 2010b; Wang et al., 2011) while gene loops have been implicated in transcriptional memory (Laine et al., 2009; Tan-Wong et al., 2009) and in directional transcription from bidirectional promoters (Tan-Wong et al., 2012). However, the molecular mechanisms by which DNA loops affect transcription regulation, memory or promoter directionality remain unknown.
Compaction of DNA into nucleosomes, the most basic repeating unit of chromatin, is achieved by wrapping 147 base pairs (bp) of DNA around an octamer of histone proteins (Luger et al., 1997). The presence of nucleosomes effectively inhibits access of regulatory proteins to the genome by occluding the underlying DNA sequence or preventing translocation of proteins along DNA (Ehrenhofer-Murray, 2004). As a result, all DNA-dependent processes, including transcription, replication, repair, and recombination are significantly affected by the positions of nucleosomes across the genome. One mechanism by which eukaryotic cells modulate chromatin structure is through highly conserved ATP-dependent chromatin remodeling enzymes that utilize the energy of ATP hydrolysis to slide, evict, or replace histones within nucleosomes (Clapier and Cairns, 2009). Because of their ability to alter chromatin structure, chromatin remodeling enzymes are key modulators of DNA-dependent processes (Ehrenhofer-Murray, 2004). As a result, much effort has been put forth to identify loci where ATP-dependent chromatin remodeling enzymes function on a genome-wide scale (Hartley and Madhani, 2009; Rando and Chang, 2009; Tirosh et al., 2010; Whitehouse et al., 2007). In yeast, the Isw2 complex functions around nucleosome free regions (NFRs) to repress transcription by reducing NFR size (Whitehouse et al., 2007; Yadon et al., 2010b), while the RSC complex increases NFR size to activate transcription (Hartley and Madhani, 2009). Similarly, the Swr1 complex targets the 5′-end of genes where it promotes efficient deposition of the histone variant Htz1 (H2A.Z), which becomes acetylated and associated with transcriptional activation (Raisner et al., 2005). In contrast, Chd1 and Isw1 predominately function within the body of genes and are involved in positioning nucleosomes toward the 3′-end (Tirosh et al., 2010) and/or suppressing histone exchange (Smolle et al., 2012).
Chromatin remodeling enzymes are highly abundant in all eukaryotes, estimated at one remodeler per ~14 nucleosomes (Erdel et al., 2010) in human cells. Despite their abundance, remodeling factors affect chromatin structure only at select loci. Therefore, understanding how such abundant factors function only at specific loci is a fundamental issue. To date, two distinct mechanisms have been implicated in the targeting of chromatin remodeling enzymes to specific loci: (i) direct recruitment by covalently modified N-terminal histone tails; and (ii) physical interactions with TFs. The Swi/Snf and Chd1 classes of remodeling factors contain bromo- and chromodomains, respectively, which are capable of recognizing acetylated and methylated histone lysine residues (Clapier and Cairns, 2009). However, the extent to which these interactions contribute to targeting of these factors across the genome remains to be determined. In contrast, Isw2 is targeted directly by the TF Ume6 to its binding sites at a small number of loci (Gelbart et al., 2005; Goldmark et al., 2000). Whether Ume6 targets Isw2 genome-wide, however, has not been determined. Similarly, the TFs Abf1 and Reb1 have been implicated in targeting RSC to a subset of loci (Hartley and Madhani, 2009), although direct evidence for either of these TFs in RSC targeting has not been demonstrated.
In this study, we sought to determine the mechanisms by which the Isw2 chromatin remodeling enzyme is targeted to specific loci across the S. cerevisiae genome. Here we report the first comprehensive genome-wide view of TF-mediated Isw2 targeting. Unexpectedly, Isw2 was targeted in a TF-dependent fashion to a large number of loci without annotated TF binding sites. This led to the discovery that Isw2 is targeted to specific loci via DNA looping in a manner dependent upon the general transcription factor TFIIB and the sequence-specific TF Ume6. This work defines a previously unknown mechanism to target a chromatin remodeling factor and identifies a novel physiological role for DNA looping. Furthermore, we provide evidence suggesting that Ume6-dependent DNA looping mediates chromatin remodeling and transcriptional repression. Therefore, we have uncovered a mechanism by which the three-dimensional spatial organization of genomes affects chromatin structure and DNA-dependent processes on a genome-wide scale.
RESULTS
The Binding Sites of Multiple TFs are Enriched at Isw2 Targets
In budding yeast, S. cerevisiae, the highly conserved ATP-dependent chromatin remodeling enzyme Isw2 is enriched at ~2,100 loci genome-wide (Whitehouse et al., 2007). These targets can be categorized into three major classes: the 5′-end of genes, the 3′-end of genes, and upstream of tRNA genes (Gelbart et al., 2005; Goldmark et al., 2000; Whitehouse et al., 2007). At the 5′- and 3′-ends of genes, Isw2 utilizes the energy of ATP hydrolysis to slide nucleosomes along DNA and reduce the size of NFRs to repress both coding and non-coding RNA transcription (Whitehouse et al., 2007; Yadon et al., 2010b). Upstream of tRNA genes, Isw2 is required for the periodic integration pattern of the Ty1 retrotransposon (Bachman et al., 2005; Gelbart et al., 2005). TF-dependent targeting can explain Isw2 enrichment at only a few loci: Ume6 for the promoters of early meiotic genes during mitotic growth (~190 genes) (Goldmark et al., 2000), α2-MCM1 for the promoters of MATa-specific genes in MATα cells (~5 genes) (Bachman et al., 2005; Gelbart et al., 2005), and Bdp1 at tRNA genes (~140 genes) (Bachman et al., 2005; Gelbart et al., 2005). It is therefore apparent that these three TFs account for targeting of Isw2 to only a small fraction of loci genome-wide. Considering that other TFs, including Abf1, Reb1, Pho4, and Gal4, have also been implicated in the recruitment of other classes of chromatin remodeling enzymes (Adkins et al., 2007; Badis et al., 2008; Bryant et al., 2008; Hartley and Madhani, 2009), we postulated that additional TFs may be involved in Isw2 targeting.
To address this possibility, we performed a statistical analysis to curate a list of annotated sequence-specific TF binding sites that were preferentially enriched around Isw2 targets. A total of 15 TFs with a p-value ≤0.01 were identified (Figure S1A). Importantly, Ume6 was found to be the most highly enriched TF, validating our strategy to identify TFs involved in Isw2 targeting.
TFs Target Isw2 to Specific Loci
Next, we assessed the role of these TFs in the targeting of Isw2 genome-wide. To this end, we performed chromatin immunoprecipitation of Isw2 on genome-wide tilling microarrays (Isw2 ChIP-chip) in strains containing a null TF deletion, Δume6, Δnrg1, Δcin5, or Δsok2. Each TF was chosen because its binding sites are highly enriched at Isw2 targets (Figure S1A) and because they have a large number of binding sites genome-wide. The Δume6 mutant was included as a positive control.
Previous studies from our lab demonstrated that a catalytically inactive Isw2 mutant, Isw2-K215R, is preferentially enriched at Isw2 target sites while wild type (WT) Isw2 is nonspecifically bound across the genome (Gelbart et al., 2005). Isw2 targets have been defined as sites where Isw2-K215R is enriched relative to WT Isw2 (Fazzio et al., 2005; Gelbart et al., 2005; Whitehouse et al., 2007). We therefore identified Isw2 targets by competitively hybridizing DNA fragments co-immunoprecipitating with Isw2-K215R against those with WT Isw2. Visualization of the average log2 signal in each strain revealed well-defined peaks of Isw2 enrichment at many loci in all strains (Figure S1B, C). The enrichment of Isw2 ChIP signals observed around the Ume6 binding sites of the previously defined Isw2 target genes (Goldmark et al., 2000) INO1 (Figure S1B) and SIP4 (Figure S1C) observed in UME6 strains is selectively lost in Δume6 strains, validating our strategy.
We next systematically identified regions in which Isw2 targeting is affected in each of the TF mutants (see Materials and Methods for details). A total of 563, 194, 341, and 226 regions with decreased Isw2 signals were identified in Δume6, Δnrg1, Δcin5, and Δsok2 strains, respectively (hereafter referred to as Ume6-, Nrg1-, Cin5-, and Sok2-dependent Isw2 targets). Visual inspection of the results (e.g. Figure S1B, C) confirms that our annotation accurately marks regions in which Isw2 targeting is reduced in the mutants relative to WT. It should be noted that the number of regions annotated in each mutant is an underestimate, as our algorithm utilized conservative parameters, and chromosomes II, V, and XII were removed from the analysis. Nonetheless, these data revealed, for the first time, a role for TFs in targeting a chromatin remodeling enzyme on a genome-wide scale.
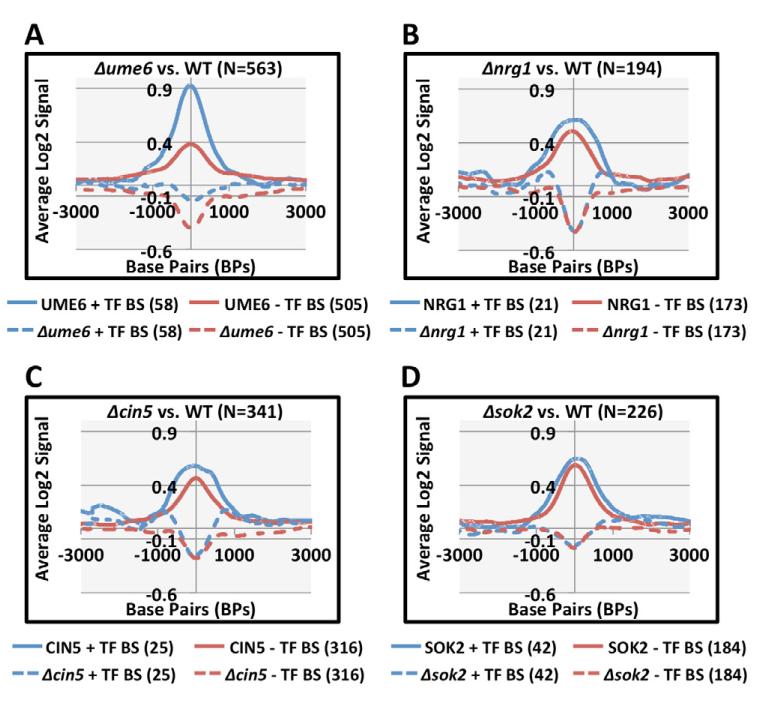
Among annotated Ume6 binding sites, 49% (70 of 142) exhibit Ume6-dependent Isw2 ChIP signals. In contrast, only 21% (30 of 167), 10% (36 of 355), and 29% (160 of 544) of Nrg1, Cin5, and Sok2 binding sites show Isw2 signals that are dependent on the corresponding TFs. This could be partly explained by the stringent criteria used to identify regions with decreased Isw2 ChIP signal. On the other hand, only a small fraction of the total TF-dependent Isw2 targets contained annotated binding sites for the corresponding TF (Figure 1): 10% (58 of 563) of the Ume6-dependent targets, 11% (21 of 194) of the Nrg1-dependent targets, 7% (25 of 341) of the Cin5-dependent targets, and 19% (42 of 226) of the Sok2-dependent targets. Interestingly, the average Isw2 ChIP signal in UME6 strains is much higher around Ume6-dependent Isw2 targets containing an annotated Ume6 binding site than those without it (Figure 1A, solid blue and red lines, respectively). Nevertheless, the average Isw2 ChIP signals were strongly reduced in Δume6 strains regardless of the presence or absence of an Ume6 binding site (Figure 1A, dashed lines). Similar trends were seen around Nrg1- (Figure 1B), Cin5- (Figure 1C), and Sok2-dependent regions (Figure 1D), although the average Isw2 ChIP signals at regions with or without TF binding sites were similar. These results are consistent with a model in which TFs are required for the targeting of chromatin remodeling factors to specific loci. Importantly, they also reveal that there are many Isw2 targets across the S. cerevisiae genome that cannot be explained by a simple model of TF-dependent targeting to its binding site. We therefore sought to determine the mechanisms for TF-dependent Isw2 targeting to regions without a corresponding TF binding site. In the following analysis we used Ume6-dependent targeting of Isw2 as a model, as its role in Isw2 targeting has been well characterized (Goldmark et al., 2000).
Figure 1. Ume6, Nrg1, Cin5, and Sok2 May Target Isw2 By Unknown Mechanisms.
Isw2 ChIP signals in TF mutant strains. TF-dependent Isw2 targets were aligned by their midpoint and the average log2 Isw2 ChIP signal in WT (solid lines) or TF null mutants (dashed lines) is displayed across 6,000 bp. Regions containing an annotated binding site are displayed in blue and regions without an annotated binding site displayed in red. (A) Δume6 vs. WT. (B) Δnrg1 vs. WT. (C) Δcin5 vs. WT. (D) Δsok2 vs. WT. The numbers in parenthesis denote the number of loci analyzed.
Ume6 Targets Isw2 Through a Novel Mechanism(s)
Our analyses above revealed that only 58 out of 563 Ume6-dependent Isw2 targets contained an annotated Ume6 binding site. One possible explanation is miss-annotation of Ume6 binding sites. To address this possibility, genome-wide Ume6 ChIP-chip was performed. As expected, the majority of annotated Ume6 binding sites have high Ume6 ChIP signals surrounding the binding sites (Figure S2A and S2B, red arrows). However, at a small number of loci either Ume6 binding sites were missed (Figure S2C) or no enrichment of Ume6 is observed at annotated binding sites (Figure S2B, green arrow). Globally, Isw2 targets with an annotated binding site exhibit on average much higher levels of Ume6 ChIP signal compared to those without an annotated binding site (Figure S2D). Together, these data establish that there are a large number of Ume6-dependent Isw2 targets with no evidence of Ume6 binding. Importantly, these results strongly suggest that Isw2 is targeted to a large number of loci through a previously unknown, Ume6-dependent mechanism(s).
TFIIB is Required for Isw2 Targeting
While analyzing the genome-wide targeting of Isw2, we noted that Isw2 targets both the 5′- and 3′-end of the same gene at a highly statistically significant frequency (Figure 2A). Recent studies examining the spatial organization of the S. cerevisiae genome using the chromosome confirmation capture (3C) assay have shown that many genes juxtapose their promoter and terminator regions via gene looping (Ansari and Hampsey, 2005; El Kaderi et al., 2009; Hampsey et al., 2011; Laine et al., 2009; O’Sullivan et al., 2004; Singh and Hampsey, 2007; Tan-Wong et al., 2009). Based on these data, we hypothesized that gene looping between the 5′- and 3′-end of the same gene, of which an Ume6 binding site is only present at the 5′-end, mediates Ume6-dependent targeting of Isw2 to both ends of the gene.
Figure 2. Evidence for DNA Looping-Dependent Targeting of Isw2.
(A) Venn diagram displaying the statistically significant overrepresentation of Isw2 targets that are located at both the 5′- and 3′-ends of the same genes. P-value was calculated with a hypergeometric distribution using non-dubious genes only. (B-D) Isw2 and Ume6 binding at Isw2 targets. The average log2 Isw2 ChIP signals for WT (black), Δume6 (red), and sua7-1 (orange) strains (Average Log2 Ratio – Isw2 ChIP), and the average log2 Ume6 ChIP signals for WT (black) strains (Average Log2 Ratio – Ume6 ChIP) are displayed. Red and orange boxes in Significant Regions indicate regions having significantly different Isw2 ChIP signal between WT and Δume6 or sua7-1 strains, respectively. Non-dubious genes are displayed as black (Watson strand) and grey (Crick strand) boxes in Genomic Features. Loss of Isw2 ChIP enrichment is highlighted by arrows (red, Ume6 binding sites, and black, no Ume6 binding site) (B) ChrXIII:300500-314500 (C) ChrI:30000-50000 (D) ChrVII:602000-632000
To test this hypothesis, we performed Isw2 ChIP-chip using the sua7-1 mutant. This mutant contains a single point mutation, encoding an E62K replacement, in the general transcription factor TFIIB that abrogates gene looping in yeast (Singh and Hampsey, 2007). Supporting our model, Isw2 ChIP signals in the sua7-1 mutant were strongly decreased at the 3′-end of a few genes containing an Ume6 binding site at the 5′-end (Figure 2B). Unexpectedly, however, it was far more common to observe a decrease in both TFIIB- and Ume6-dependent Isw2 signals far from Ume6 binding sites and across many genes (Figure 2C and D). These observations suggested that TFIIB-dependent DNA looping facilitates TF-dependent targeting of Isw2.
The Isw2 ChIP signals in WT and sua7-1 strains were then compared genome-wide. A total of 454 regions were identified as TFIIB-dependent Isw2 targets, which, for the first time, revealed that the general transcription factor TFIIB has a global effect on the targeting of Isw2. If DNA looping indeed mediates Ume6-dependent targeting of Isw2 between two distant loci, one with and another without an Ume6 binding site, we would expect to see a decrease in Isw2 ChIP signals in both Δume6 and sua7-1 at targets without an Ume6 binding site. On the other hand, Isw2 ChIP signals would decrease only in the Δume6 mutant at Ume6 binding sites. We found 255 Isw2 targets that satisfy these criteria (Figure 3A). We refer to Ume6-dependent Isw2 targets containing an annotated Ume6 binding site as “canonical” targets and Ume6- and TFIIB-dependent Isw2 targets without an annotated Ume6 binding site as “ectopic” targets. The highly statistically significant overlap between Ume6- and TFIIB-dependent Isw2 targets (p-value<10−50) demonstrates that ectopic targets represent a major class amongst both Ume6- and TFIIB-dependent Isw2 targets. Analysis of all Ume6-dependent Isw2 targets (Figure 3B) further supports our model that DNA looping facilitates TF-dependent targeting of Isw2 and demonstrates that there are a large number of uncharacterized Ume6-dependent Isw2 targets that do not have Ume6 binding nor are TFIIB-dependent.
Figure 3. Ume6- and TFIIB-Dependent Isw2 Targets Extensively Overlap.
(A) Venn diagram displaying the number of UME6- and SUA7-dependent Isw2 target regions that directly overlap (“Ectopic”). P-value was calculated using a hypergeometric distribution. (B) Isw2 and Ume6 ChIP signals around Ume6-dependent Isw2 targets. The average log2 ChIP signals of Isw2 in WT, Δume6, and sua7-1 strains, as well as the average log2 ChIP signals of Ume6 in WT strains are displayed for a 3000 bp window centered around the midpoint of the targets. Canonical and ectopic Isw2 targets are marked on the right. Targets that directly overlap with Isw2-dependent chromatin remodeling (Whitehouse et al., 2007) are denoted on the far right.
To rule out the possibility that the sua7-1 mutation directly or indirectly alters Ume6 binding, Ume6 ChIP-chip was performed in a sua7-1 mutant. The average log2 signal in the sua7-1 mutant was indistinguishable from SUA7 genome-wide (data not shown). This result excludes the possibility that TFIIB indirectly affects Isw2 targeting through Ume6 binding.
DNA Looping Targets Isw2 to Specific Loci
To directly test our model that DNA looping mediates Isw2 targeting, we next performed the 3C assay at three loci to examine whether DNA looping takes place between canonical and ectopic Isw2 targets. At all loci tested, strong 3C signals were detected between the canonical and ectopic loci in WT cells (Figure 4). Importantly, quantification of the 3C signals using primer pairs that spanned across each locus revealed strong peaks of 3C signals specifically between the canonical and ectopic Isw2 targets (Fig. 4A-C, primer pairs F9-F4, F5-F3, and F9-F2, respectively). The 3C signals between the canonical and ectopic Isw2 targets were dependent upon cross-linking, ligation, and restriction digestion (Figure S3), showing that the signals do indeed reflect DNA looping. These results are consistent with our model that DNA looping mediates TF-dependent Isw2 targeting to ectopic loci.
Figure 4. DNA Loops are Formed Between Canonical and Ectopic Isw2 Targets in WT Strains.
The 3C assay between canonical and ectopic Isw2 targets. Schematic diagrams of each locus analyzed by 3C are displayed at the top of each panel. Watson and Crick genes are displayed as black and grey boxes, respectively. Ectopic Isw2 targets and Ume6 binding sites are marked in red. Red arrowheads depict primer locations and directionality. Dashed vertical lines depict MspI restriction sites used in the 3C assay. Double headed, dashed arrows indicate looping between canonical and ectopic Isw2 targets. Ethidium bromide staining of 3C and control PCR products are displayed. 3C signals were normalized to the corresponding primer pair control PCR signals. The asterisk indicates a non-specific band and was not included in the quantitation. (A) SPO20-YMR018W (B) BDH1-ACS1 (C) ERG25-YGR067C
Our model predicts that the 3C signals between the canonical and ectopic Isw2 targets would be significantly reduced in the sua7-1 strain. Indeed, a statistically significant loss of 3C signals was observed for the canonical and ectopic primer pairs of each locus tested (Figure 5A-C). These results demonstrate for the first time that TFIIB is required for DNA looping not only between the 5′ and 3′ ends of the same gene (Figure 5A), but also between two loci that are separated by several genes (Figure 5B-C). Together, these results strongly support our model that DNA looping facilitates Isw2 targeting to ectopic loci across the yeast genome.
Figure 5. DNA Looping Between Canonical and Ectopic Isw2 Targets is dependent on TFIIB and Ume6.
3C assay between Isw2 targets in TF and Isw2-K215R mutants. 3C PCR products (top) and quantitation (bottom) at the canonical and ectopic sites of the indicated loci are shown. (A-C) WT and sua7-1 strains. (D-F) WT, Δume6, and Isw2-K215R strains. (A) and (D) SPO20-YMR018W locus. (B) and (E) BDH1-ACS1 locus. (C) and (F) ERG25-YGR067C locus. Error bars represent the SEM of three independent experiments.
Ume6, But Not Isw2, is Required for DNA Looping
Because a very significant fraction of Isw2 ChIP signals at Ume6-dependent targets are also dependent on TFIIB (Figure 3A), we considered the possibility that Ume6 may also be involved in looping. To test this model, the 3C assay was performed between canonical and ectopic Isw2 targets in Δume6 strains. Strikingly, at each of the three loci tested, the 3C signal was nearly completely abrogated (Fig. 5D-F). These results show, for the first time, that a transcriptional repressor can play an essential role in mediating DNA looping in S. cerevisiae.
We next tested whether Isw2 affects DNA looping. We reasoned that Isw2-dependent reduction in NFR sizes at canonical and/or ectopic targets might affect the efficiency of DNA looping. However, in contrast to sua7-1 and Δume6 mutations, neither the catalytically inactive Isw2-K215R mutation (Figure 5D-F), which abrogates the ATPase activity of Isw2 (Eden et al., 2007) nor a Δisw2 mutation (Figure S4), had any significant effect on the 3C signals. Together, these results suggest that DNA looping occurs at a large number of loci across the S. cerevisiae genome in a manner that is dependent on both TFIIB and Ume6, and this class of DNA looping is required for the targeting of Isw2 to specific loci (Figure 7).
Figure 7. Schematic Representation of Major Targeting Mechanism(s) of Isw2.
Isw2 is targeted to canonical sites via physical interactions with Ume6. In contrast, Isw2 is targeted to ectopic sites via Ume6- and TFIIB-dependent DNA-looping. A light blue oval represents the transcription preinitiation complex.
Ume6-Dependent DNA Looping Mediates Chromatin Remodeling and Transcriptional Repression
We next sought to address the biological consequences for DNA looping-dependent targeting of Isw2. We first examined whether canonical and ectopic Isw2 target loci exhibit Isw2-dependent chromatin remodeling. Our analysis revealed that Isw2-dependent chromatin remodeling takes place at both classes of Isw2 targets (Figure 3B, far right panel). Interestingly, the fraction of Ume6-dependent Isw2 targets that exhibit Isw2-dependent chromatin remodeling is much higher at canonical Isw2 targets than ectopic targets. We next looked for statistically enriched GO process terms (Eden et al., 2007) connected with genes associated with canonical (57 genes) and ectopic (275 genes) Isw2 targets. Consistent with previous reports (Fazzio et al., 2001; Goldmark et al., 2000), this analysis revealed that canonical Isw2 target genes are primarily enriched for meiosis (p-value=3.95E-05), carbohydrate derivative catabolic processes (p-value=5.68E-05), DNA recombination (p-value=5.72E-05), reciprocal meiotic recombination (p-value=9.84E-05), and chromosome organization involved in meiosis (p-value=9.84E-05). On the other hand, ectopic Isw2 targets are enriched for cytoplasmic translation (p-value=2.62E-10), translation (p-value=8.47E-07), biosynthetic processes (p-value=2.82E-06), and glucose metabolic process (p-value=6.82E-06). This result suggests that canonical Ume6-dependent targeting and ectopic DNA looping-dependent targeting have distinct gene specificities.
To examine the effects of Ume6-dependent DNA looping on transcription, we analyzed previously published transcript array data of Δume6 strains (Fazzio et al., 2001). This analysis revealed an up-regulation of 41% (24 of 58) of canonical Isw2 target genes, and 11% (27 of 245) of ectopic Isw2 target genes, using a previously employed 1.7-fold cut off (Fazzio et al., 2001) (Figure 6). The fact that a smaller fraction of genes are de-repressed and exhibit chromatin remodeling at ectopic targets is consistent with our data that Isw2 ChIP signals are generally weaker at these sites compared to canonical targets (Figure 1A). Together, these results support a model in which Ume6-dependent DNA looping is required for chromatin remodeling and transcriptional repression at a subset of genes that do not have Ume6 binding sites at their promoters, indicating, for the first time, a functional role for repressor-mediated DNA looping.
Figure 6. Ume6-Dependent DNA Looping is Associated with Transcriptional Repression.
(A) Heat maps showing the fold transcriptional de-repression (Fazzio et al., 2001) of Δume6 mutation at Isw2 target genes. Canonical (n=46) and ectopic (n=232) Isw2 target genes are aligned according to the fold de-repression. Gene de-repressed ≥ 1.7-fold (Fazzio et al., 2001) are denoted by black bars. (B) A table showing top five most highly de-repressed canonical or ectopic Isw2 target genes.
DISCUSSION
TF-Dependent Recruitment is the Major Targeting Mechanism of an ATP-Dependent Chromatin Remodeling Enzyme
ATP-dependent chromatin remodeling enzymes are highly conserved protein complexes that play essential roles in numerous cellular and developmental processes. Although they are generally highly abundant, these enzymes affect chromatin structure at specific genomic loci. To understand how they function in vivo, elucidating the mechanisms for targeting these enzymes to specific loci is essential. However, only a few TFs have been shown to target chromatin remodeling enzymes to a small number of loci, and the extent to which the conventional TF-dependent recruitment model could explain chromatin remodeling enzyme targeting in vivo has not been determined. Here we present the first genome-wide analysis of TF-dependent targeting of the ATP-dependent chromatin remodeling enzyme Isw2. We identified Ume6, Nrg1, Cin5, and Sok2 as global mediators of Isw2 targeting and established that TF-dependent targeting is a primary mechanism for the recruitment of a chromatin remodeling enzyme genome-wide.
We have shown that the physical interaction of Ume6 and Isw2 mediates the targeting of Isw2 to Ume6 binding sites (Goldmark et al., 2000). The mechanisms for Nrg1, Cin5, and Sok2-dependent targeting of Isw2 are not known. A recent study identified ~20 sequence-specific TFs that are likely involved in targeting of the transcriptional co-repressor complex Tup1-Ssn6 (Hanlon et al., 2011). Strikingly, 11 of the 15 sequence-specific TFs (Nrg1, Sut1, Skn7, Phd1, Sok2, Cin5, Sko1, Swi6, Rox1, Swi4, and Rfx1), whose binding sites are enriched at the 5′-end of Isw2 target genes (Table S1), were implicated in Tup1-Ssn6 targeting. We have indeed shown that the MATα-specific transcriptional repressor α2 is required for Isw2 targeting to MATa-specific gene promoters (Gelbart et al., 2005). α2 physically interacts with Tup1 and is required for the targeting of Tup1 to MATa-specific gene promoters (Komachi and Johnson, 1997). It is therefore likely that Nrg1, Cin5, and Sok2, and possibly also Sut1, Skn7, Phd1, Sko1, Swi6, Rox1, Swi4, and Rfx1, are involved in Isw2 targeting via their interactions with Tup1. It has been further suggested (Hanlon et al., 2011) that Tup1 could be simultaneously recruited to a single locus by multiple cofactors. This is also consistent with our observation that many of the Isw2 target regions with decreased Isw2 ChIP signal in Δnrg1, Δcin5, and Δsok2 strains overlap (data not shown).
Isw2 Targeting: A novel Functional Role for DNA Looping
Our work unexpectedly revealed that a conventional model of TF-dependent targeting, in which TFs recruit remodeling factors to the immediate vicinity of their binding sites, can account for only a small fraction of Isw2 targets. Instead, we found that Ume6- and TFIIB-dependent DNA looping is required to target Isw2 to many loci genome-wide, revealing both a novel functional role for DNA looping and a mechanism for targeting an ATP-dependent chromatin remodeling enzyme.
DNA looping has been observed in many different eukaryotic organisms and has been associated with diverse functional roles. In mammalian cells, DNA looping is correlated with transcriptional regulation (Comet et al., 2011; Nemeth et al., 2008; Perkins et al., 2008; Schoenfelder et al., 2010a; Schoenfelder et al., 2010b; Wang et al., 2011). On the other hand, in yeast, gene looping has been implicated in transcriptional memory (Laine et al., 2009; Tan-Wong et al., 2009), and in promoter directionality (Tan-Wong et al., 2012). However, how DNA looping affects transcription remains to be determined. Our work revealed a previously unknown mechanism by which DNA looping can regulate DNA-dependent processes.
The fact that 255 ectopic Isw2 targets were identified suggests that DNA looping plays a major role in facilitating TF-dependent targeting of Isw2. Unfortunately, the low throughput and laborious nature of the 3C assay has precluded a genome-wide characterization of DNA looping-dependent Isw2 targeting at this time. Recently, a chromosome interaction map in yeast using a modified 4C (chromosome conformation capture-on-chip) assay combined with massively parallel sequencing was published (Duan et al., 2010). However, the low resolution (on the order of ~3-4 kilobases) of this data set precluded its use in the analysis of how ectopic Isw2 targets interact with other loci in a genome-wide manner. Genome-wide, high-resolution chromatin interaction maps would greatly facilitate our future studies to investigate the functions and dynamics of interactions between Isw2 targets under various conditions.
To date, all DNA looping events described in yeast have been shown to depend on transcriptional activators that function at the 5′ end of genes, and RNA processing factors that function at the 3′ end of the same genes (Ansari and Hampsey, 2005; El Kaderi et al., 2009; Hampsey et al., 2011; Laine et al., 2009; Singh and Hampsey, 2007). In mammalian cells, DNA looping at both the BRCA1 and CD68 genes in breast tumor cell lines and B- and T-lymphocytes, respectively, is associated with transcriptional repression (O’Reilly and Greaves, 2007; Tan-Wong et al., 2008). Here we show, similar to mammalian DNA looping functions, that Ume6-dependent DNA looping is associated with transcriptional repression in S. cerevisiae. Together with the fact that DNA looping takes place across several genes, it is likely that Ume6-dependent DNA looping we have identified represents a class of DNA looping that is distinct from gene looping previously reported in yeast.
One intriguing question is whether there are distinct consequences associated with TF-dependent Isw2 targeting to canonical targets versus DNA looping-dependent ectopic targets. Our GO term analysis revealed that canonical and ectopic Isw2 targets are enriched for different classes of genes, suggesting different specificities for the two mechanisms. Furthermore, we expect DNA looping to be more transient and dynamic than binding of TFs to their recognition sites. If this is the case, one interesting possibility is that, at any given moment, Isw2 would be targeted to canonical sites in most cells within a population, whereas Isw2 would be targeted more transiently and to a smaller fraction of cells at ectopic sites. This possibility is consistent with our observations that Isw2 ChIP signals are typically weaker and the degree of transcriptional de-repression in Δume6 cells is generally smaller at ectopic targets. Therefore, DNA looping-dependent Isw2 targeting may result in more variable transcriptional levels of target genes within a population.
Our analyses also identified a large number of Ume6-dependent Isw2 targets that neither contain an annotated Ume6 binding site nor are dependent on TFIIB-mediated DNA looping (Figure 3). This suggests that there are still unidentified mechanisms for Ume6-dependent targeting of Isw2 to these loci. One possibility is TFIIB-independent DNA looping. In this case, Ume6 alone may be sufficient for DNA looping under certain circumstances. It is also possible that Ume6 functions together with other TFs to facilitate DNA looping. The fact that a basal transcription factor (TFIIB) and a sequence-specific DNA binding repressor (Ume6) play integral roles in DNA looping supports the possibility that other unidentified TFs may be involved in this process.
In summary, our results suggest that TF-dependent recruitment plays the major role in Isw2 targeting through at least two distinct mechanisms (Figure 7). TFs can directly target Isw2 to their binding sites (canonical targets), likely through physical interaction. On the other hand, DNA looping, which is mediated by TFIIB, Ume6, and possibly other TFs, can also target Isw2 to a large number of loci across the genome where the binding sites of the TFs are absent (ectopic targets). This mode of targeting is likely more dynamic than the direct Isw2 targeting. Our model provides one mechanism by which the three dimensional folding of chromatin fiber affects DNA-dependent processes.
EXPERIMENTAL PROCEDURES
Isw2 Targets and Transcription Factor Enrichment
Isw2 target genes are defined and classified as Isw2-K215R enriched non-dubious genes (Whitehouse et al., 2007). TF binding sites are as previously reported (Harbison et al., 2004) using the binding threshold of p<0.005 and no conservation criteria. A total of 5642 intergenic regions upstream of non-dubious genes was used to determine the hypergeometric distribution of 1020 intergenic regions upstream of genes with Isw2 enrichment at the 5′-end and containing at least one annotated binding site for a particular TF.
Isw2 and Ume6 ChIP-chip
Isw2 and Ume6 ChIP-chip was performed using 3x-Flag-tagged Isw2 or 3x-Flag-tagged Ume6 as described (Yadon et al., 2010a). Equal concentrations of WT Isw2 and Isw2-K215R or input and Ume6 were competitively hybridized onto custom Nimblegen microarrays according to the manufactures protocol. Log2 ratios and normalization was performed individually for forward and reverse strands using Ringo (Toedling et al., 2007) and chipchipnorm (nlag=30) (Peng et al., 2007) R packages followed by median adjustment and pseudomedian smoothing (150 bp window) (Royce et al., 2007). Figures display the average log2 ratio of forward and reverse strands for all replicates combined. ChIP signals of chromomsomes II, V, and XII revealed gross abnormal increases and/or decreases of Isw2 enrichment chromosome wide in the Δume6 strain and were discarded. We speculate this is partially the result of chromosomal duplications and/or deletions, as previously reported (Fazzio et al., 2001). Raw and normalized data are available for download at http://labs.fhcrc.org/tsukiyama and are deposited into Gene Expression Omnibus database (http://ncbi.nlm.nih.gov/projects/geo/) under accession number GSE39542.
Changes in Isw2 Targeting
Changes in Isw2 enrichment were measured using LIMMA (Smyth, 2004) by identifying probes with significantly different signals between WT and mutant strains, utilizing an object containing the normalized log2 ratio from each hybridization (forward and reverse strands for each comparison was performed separately). Consecutive probes, totaling at least 250 base pairs (bp) in length, each with a statistically significant (p-value≤0.05) reduction of Isw2 signal in each mutant relative to WT, and averaging at least 1.65 fold-change, were denoted. Reported regions represent the directly overlapping areas of forward and reverse comparisons. Isw2 target genes, 5- or 3′, represent regions directly overlapping with an annotated transcription start or transcription termination sites, respectively. Annotated regions are available for download at http://labs.fhcrc.org/tsukiyama.
Chromosome Confirmation Capture (3C)
DNA loops were analyzed by a modified version of 3C (Dekker, 2006; Dekker et al., 2002), as described elsewhere (Singh et al., 2009), using the restriction enzyme MspI. 3C PCR reactions were performed using the indicated tandem primer pairs (Table S1) for 40 cycles. Primer pair efficiencies were determined as previously described (Dekker, 2006). Control PCR products were generated as previously described (Ahn et al., 2004) after 25 cycle of PCR. PCR products were fractionated in a 1.5% agarose gel, visualized and quantified by ethidium bromide staining using an AlphaImager 2000. PCR primer sequences are listed in Table S1.
Supplementary Material
HIGHLIGHTS.
Transcription factor (TF)-dependent recruitment is the major Isw2 targeting mechanism
The majority of TF-dependent Isw2 targets do not have the TF binding site
DNA looping plays a major role in Ume6-dependent Isw2 targeting
DNA looping is associated with chromatin remodeling and transcriptional repression
ACKNOWLEDGMENTS
We thank Steve Hahn, Stephen Tapscott, Barbara Wakimoto, Sue Biggins, J. Rodriguez, N. Bogenschutz, T. Cunningham, L. Lee, J. McKnight, L. Haselden, E. Alcid and the Biggns lab members for helpful discussions and suggestions.
This research was supported by NIH grant RO1 GM058465 to T.T. and by RO1 GM039484 to M.H. A.N.Y. was supported by Developmental Biology Predoctoral Training Grant T32HD007183 from the National Institutes of Child Health and Human Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adkins MW, Williams SK, Linger J, Tyler JK. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Molecular and cellular biology. 2007;27:6372–6382. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Molecular cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes & development. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes & development. 2005;19:955–964. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Molecular cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS biology. 2008;6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2294–2299. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature reviews. Genetics. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor perspectives in biology. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nature methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes & development. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS computational biology. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. European journal of biochemistry/FEBS. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. The Journal of biological chemistry. 2009;284:25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Schubert T, Marth C, Langst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19873–19878. doi: 10.1073/pnas.1003438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Gelbart ME, Tsukiyama T. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Molecular and cellular biology. 2005;25:9165–9174. doi: 10.1128/MCB.25.21.9165-9174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread Collaboration of Isw2 and Sin3-Rpd3 Chromatin Remodeling Complexes in Transcriptional Repression. Molecular and cellular biology. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes & development. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Advances in enzyme regulation. 2011;51:118–125. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon SE, Rizzo JM, Tatomer DC, Lieb JD, Buck MJ. The stress response factors Yap6, Cin5, Phd1, and Skn7 direct targeting of the conserved co-repressor Tup1-Ssn6 in S. cerevisiae. PloS one. 2011;6:e19060. doi: 10.1371/journal.pone.0019060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komachi K, Johnson AD. Residues in the WD repeats of Tup1 required for interaction with alpha2. Molecular and cellular biology. 1997;17:6023–6028. doi: 10.1128/mcb.17.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes & development. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Matarazzo MR, Boyle S, D’Esposito M, Bickmore WA. Chromosome territory reorganization in a human disease with altered DNA methylation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16546–16551. doi: 10.1073/pnas.0702924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harbor perspectives in biology. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Guibert S, Tiwari VK, Ohlsson R, Langst G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. The EMBO journal. 2008;27:1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly D, Greaves DR. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics. 2007;90:407–415. doi: 10.1016/j.ygeno.2007.04.010. [DOI] [PubMed] [Google Scholar]
- O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nature genetics. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Molecular cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annual review of biochemistry. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce TE, Carriero NJ, Gerstein MB. An efficient pseudomedian filter for tiling microrrays. BMC bioinformatics. 2007;8:186. doi: 10.1186/1471-2105-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Current opinion in genetics & development. 2010a;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010b;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Ansari A, Hampsey M. Detection of gene loops by 3C in yeast. Methods. 2009;48:361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Molecular cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nature structural & molecular biology. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes & development. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes & development. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. Gene Loops Enhance Transcriptional Directionality. Science. 2012 doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme S, Schmitt E, Stein S, Schwarz-Finsterle J, Wagner J, Walch A, Werner M, Hausmann M, Wiech T. Nuclear position and shape deformation of chromosome 8 territories in pancreatic ductal adenocarcinoma. Analytical cellular pathology. 2011;34:21–33. doi: 10.3233/ACP-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome biology. 2010;11:R49. doi: 10.1186/gb-2010-11-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo--an R/Bioconductor package for analyzing ChIP-chip readouts. BMC bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Molecular cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Wiech T, Stein S, Lachenmaier V, Schmitt E, Schwarz-Finsterle J, Wiech E, Hildenbrand G, Werner M, Hausmann M. Spatial allelic imbalance of BCL2 genes and chromosome 18 territories in nonneoplastic and neoplastic cervical squamous epithelium. European biophysics journal: EBJ. 2009;38:793–806. doi: 10.1007/s00249-009-0474-5. [DOI] [PubMed] [Google Scholar]
- Yadon AN, Van De Mark D, Basom R, Delrow J, Whitehouse I, Tsukiyama T. Chromatin Remodeling Around NFRs Leads to Repression of Non-Coding RNA Transcription. Molecular and cellular biology. 2010a doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon AN, Van de Mark D, Basom R, Delrow J, Whitehouse I, Tsukiyama T. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Molecular and cellular biology. 2010b;30:5110–5122. doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardo G, Cimino G, Nervi C. Epigenetic plasticity of chromatin in embryonic and hematopoietic stem/progenitor cells: therapeutic potential of cell reprogramming. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2008;22:1503–1518. doi: 10.1038/leu.2008.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.