The BBSome, a regulator of ciliary membrane protein composition, is required only for the export phase of a process that continuously cycles phospholipase D through cilia.
Abstract
The BBSome is a complex of seven proteins, including BBS4, that is cycled through cilia by intraflagellar transport (IFT). Previous work has shown that the membrane-associated signaling protein phospholipase D (PLD) accumulates abnormally in cilia of Chlamydomonas reinhardtii bbs mutants. Here we show that PLD is a component of wild-type cilia but is enriched ∼150-fold in bbs4 cilia; this accumulation occurs progressively over time and results in altered ciliary lipid composition. When wild-type BBSomes were introduced into bbs cells, PLD was rapidly removed from the mutant cilia, indicating the presence of an efficient BBSome-dependent mechanism for exporting ciliary PLD. This export requires retrograde IFT. Importantly, entry of PLD into cilia is BBSome and IFT independent. Therefore, the BBSome is required only for the export phase of a process that continuously cycles PLD through cilia. Another protein, carbonic anhydrase 6, is initially imported normally into bbs4 cilia but lost with time, suggesting that its loss is a secondary effect of BBSome deficiency.
Introduction
Bardet-Biedl syndrome (BBS; OMIM accession no. 209900) is a rare inherited disorder characterized by retinal degeneration, anosmia, kidney anomalies, polydactyly, hypogonadism, and obesity (Beales, 2005; Blacque and Leroux, 2006; Zaghloul and Katsanis, 2009; Sheffield, 2010). The phenotype of BBS is indicative of defects in the function of cilia, and specifically in cilia-mediated signaling. Mutations in at least 12 genes (BBS1–BBS12) cause the syndrome (Jin and Nachury, 2009). Seven of the BBS gene products (BBS1, 2, 4, 5, 7, 8, and 9) form a biochemically stable complex, the BBSome (Nachury et al., 2007). The BBSome subunits are well conserved in organisms with cilia, indicating that the BBSome fulfills an important ciliary function. Recently, we identified Chlamydomonas reinhardtii mutants for BBS1, BBS4, and BBS7. Because large quantities of cilia can be isolated from C. reinhardtii, it was possible to carry out a direct biochemical comparison of wild-type and bbs mutant cilia. (Because cilia and flagella are essentially identical organelles, here, we refer to the two flagella of C. reinhardtii as cilia.) Remarkably, loss of the BBSome has little effect on the overall composition of cilia or the ciliary axoneme; rather, a small subset of membrane-associated proteins, several of which are predicted to have signaling function, are present at abnormal levels in bbs cilia (Lechtreck et al., 2009). A redistribution of ciliary membrane proteins is also characteristic for Bbs knockout mice (Berbari et al., 2008b; Domire et al., 2011; Seo et al., 2011; Zhang et al., 2011). However, the mechanism by which BBSome deficiency causes changes in ciliary protein composition remains unclear.
Data from Caenorhabditis elegans, C. reinhardtii, and mammalian cells have revealed that only intact BBSomes enter cilia (Blacque et al., 2006; Lechtreck et al., 2009; Seo et al., 2010, 2011). Within cilia, BBSomes are moved by intraflagellar transport (IFT), a process by which large membrane-associated protein particles are transported bidirectionally along the axonemal microtubules (Kozminski et al., 1993; Rosenbaum and Witman, 2002; Blacque et al., 2004; Lechtreck et al., 2009). These particles consist of over 20 different proteins organized into two sub-complexes, termed IFT complexes A and B (Cole and Snell, 2009). Kinesin motors move these particles toward the ciliary tip (anterograde IFT); cytoplasmic dynein 1b (cytoplasmic dynein 2 in mammals) returns the particles to the cell body (retrograde IFT). Ciliary assembly is largely unaffected in C. reinhardtii bbs mutants and in Bbs knockout mice, and organization of IFT complexes A and B appears unaffected in the C. reinhardtii mutants (Mykytyn et al., 2004; Lechtreck et al., 2009). BBS proteins are significantly less abundant than IFT proteins in cilia of C. reinhardtii, and only a subset of IFT particles carries BBSomes (Lechtreck et al., 2009). This suggests that the BBSome is an IFT cargo or cargo adapter dispensable for ciliary assembly and basic IFT in most cell types.
In mammalian cells, BBSome deficiencies affect the presence of ciliary transmembrane proteins, specifically of G protein–coupled receptors (GPCRs), in cilia. Melanin-concentrating hormone receptor 1 (MCHR1) and somatostatin receptor type 3 (SSTR3) fail to localize to neuronal cilia in Bbs2−/− and Bbs4−/− mice (Berbari et al., 2008b). The BBSome interacts with the ciliary targeting motif in the IP3 loop of SSTR3, and IP3SSTR3-GFP fusion proteins translocate into cilia in a BBSome–dependent manner (Berbari et al., 2008a; Jin et al., 2010; Domire et al., 2011). The BBSome could facilitate the transport of proteins from the plasma membrane through the barrier of the ciliary transition zone into the ciliary membrane proper (Nachury et al., 2010). However, recent data show that the localization of some ciliary GPCRs is unaffected by a BBSome deficiency, whereas still others, e.g., dopamine receptor 1 in Bbs4−/− mice and MCHR1 and the hedgehog effector Smoothened in Bbs3−/− mice, accumulate in BBSome-deficient cilia (Domire et al., 2011; Seo et al., 2011; Zhang et al., 2011). This suggests that both import and export of ciliary GPCRs are affected in Bbs mutants.
We previously identified several putative signaling proteins (phospholipase D [PLD], an AMP-regulated protein kinase [AMPK], and a single domain globin [THB1]) that accumulate excessively in the ciliary membranes of C. reinhardtii bbs1, bbs4, and bbs7 mutants (Lechtreck et al., 2009). Although lacking receptor functions, these membrane-associated proteins could modulate ciliary signaling, e.g., by protein phosphorylation and the synthesis of signaling lipids, providing a potential explanation for the disruption of phototactic behavior that is a hallmark of bbs mutants in C. reinhardtii.
To investigate the mechanism by which these signaling proteins are accumulated in the bbs cilia, we performed a detailed analysis of one of the proteins, PLD, in wild type versus the bbs4-1 mutant that is null for BBS4 (Lechtreck et al., 2009). We chose to focus on PLD, both because it has a mammalian orthologue, PLD6, and because of its likely involvement in phospholipid signaling (Munnik et al., 2000). We have found that BBSome deficiency causes a massive redistribution of PLD from the cell body to the ciliary membrane, that the biochemical defects in bbs cilia increase with time but can be rapidly corrected when wild-type BBSomes are introduced into the cytoplasm and cilia, that retrograde IFT acts upstream of the BBSome in the PLD export pathway, that PLD can enter the cilium independently of the BBSome and IFT, and, finally, that BBSome disruption causes secondary defects such as changes in the lipid composition of the ciliary membrane. We conclude that the C. reinhardtii BBSome functions downstream of IFT in the export phase of a process that cycles membrane signaling proteins through the cilium. The results further suggest that the absence of a protein from BBSome-deficient cilia is not necessarily due directly to failure of the BBSome to import the protein into the cilium.
Results
PLD redistributes from the cell body into cilia in bbs mutants
We previously reported, based on silver-stained gels, that PLD, AMPK, and THB1 are enriched in C. reinhardtii bbs1, bbs4, and bbs7 cilia (Lechtreck et al., 2009). We have now generated an antibody that is specific for C. reinhardtii PLD (Fig. 1 A) and, using that antibody, are able to confirm that PLD is enriched in cilia of bbs1, bbs4, and bbs7 cells (Fig. 1 B). In addition, we have extended the findings to a newly identified mutant for BBS8 (Fig. 1 B). This mutant has an insertion in the first exon of BBS8 that results in a STOP codon predicted to terminate the normally 530-aa protein at aa 38 (Fig. S1 A). Therefore, this mutant is likely to be null for BBS8. Its isolated cilia lack BBS4, indicating that the BBSome without BBS8 is either unstable as in bbs1-1 or is unable to enter the cilium as in bbs7-1 (Lechtreck et al., 2009). To distinguish between these two possibilities, we assessed the level of BBS4 in the cytoplasm of the bbs8-1 mutant; BBS4 was strongly reduced, indicating that the BBSome is largely unstable in the absence of BBS8 (Fig. S2 B). All four bbs mutants have normal length cilia (Fig. S2 A). Anterograde and retrograde IFT velocities as determined by differential interference contrast (DIC) microscopy are similar to that of wild type (Fig. S2 C); anterograde IFT frequency is also similar to wild type but retrograde IFT frequency appears to be slightly reduced (Fig. S2 D), although this was more difficult to measure because the retrograde particles are near the limit of detection for DIC. The levels of IFT particle proteins in the bbs cilia are similar to wild type (Fig. 1 C; and see Figs. 3 B and 6 A) or slightly elevated (Fig. S2 B and see Fig. 5 C); this slight increase, if real, may be an attempt by the mutant cells to compensate for loss of the BBSome. These results are consistent with our previous observation using immunofluorescence microscopy that the ciliary distribution of IFT complex A protein IFT139 and complex B protein IFT46 is unaffected in bbs4-1 (Lechtreck et al., 2009).
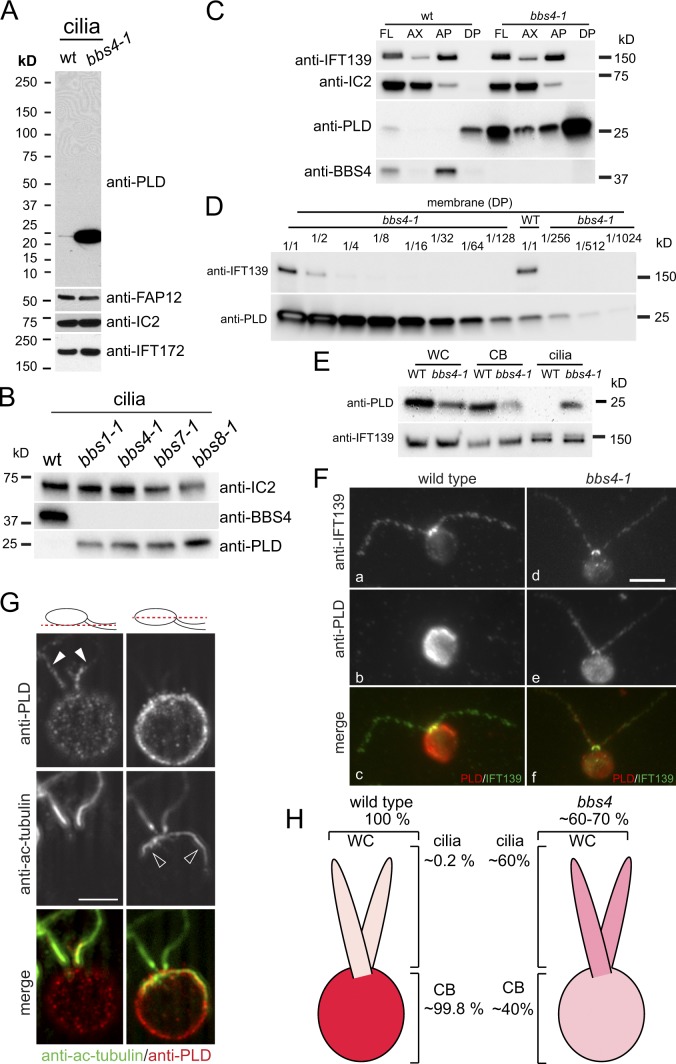
Figure 1.
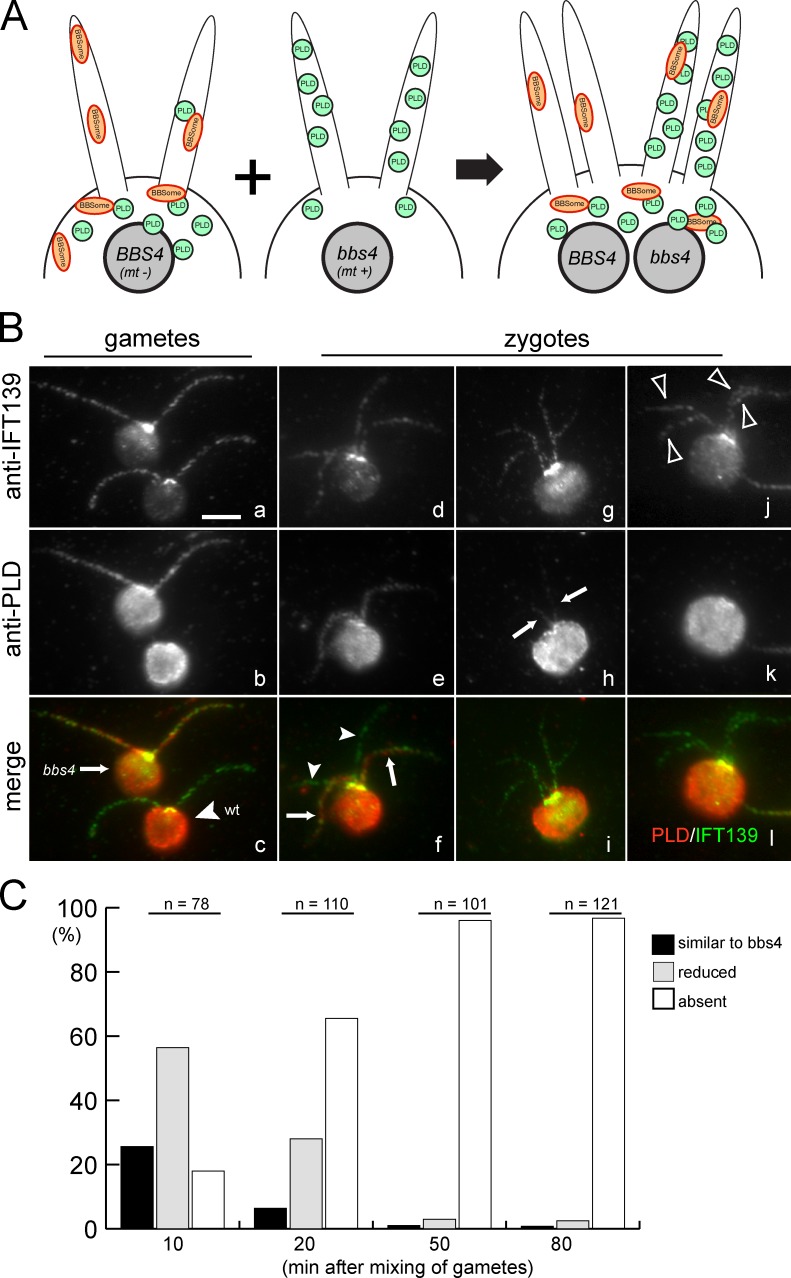
BBSome deficiency causes PLD redistribution from the cell body into cilia. (A) Western blot of isolated wild-type and bbs4-1 cilia probed with antibodies to PLD and, as loading controls, antibodies to the membrane-associated protein FAP12, the axonemal dynein subunit IC2, and the IFT complex B protein IFT172. (B) Western blot of isolated cilia from wild type (wt) and bbs1-1, bbs4-1, bbs7-1, and bbs8-1 probed with antibodies to BBS4 and PLD. IC2 was used as loading control. (C) Western blots of wild-type and bbs4-1 cilia (FL) and ciliary fractions (Ax, axonemes; AP and DP, aqueous and detergent phase from Triton X-114 phase partitioning, respectively) were probed with antibodies to PLD and BBS4, and with antibodies to the axonemal protein IC2 and the matrix protein IFT139 as loading controls. (D) Western blot of isolated ciliary membrane from wild type and bbs4-1 probed with anti-PLD. The bbs4-1 sample was diluted as indicated. IFT139, small amounts of which can be detected in the ciliary membrane, was used as a loading control. The signal strength obtained from the undiluted wild-type sample is between that of the 1:128 and 1:256 dilution of the bbs4-1 sample. (E) Western blots of whole cells (WC), deciliated cell bodies (CB), and isolated cilia (cilia) from wild type and bbs4-1 were probed with anti-PLD. Equivalent amounts of cells and cilia were loaded (i.e., one whole cell, one cell body, and two cilia). The IFT particle protein IFT139 was used as a loading control. (F) Wild-type and bbs4-1 gametes were stained with anti-IFT139 and anti-PLD. PLD was readily detected in cilia from bbs4-1 but not detected in wild-type cilia. Bar, 5 µm. (G) Two optical sections of a bbs4-1 vegetative cell stained with anti-PLD and anti-acetylated tubulin. Merged images are shown in color. The positions of the optical sections are indicated in the diagrams. Solid arrowheads, cilia; open arrowheads, microtubular roots. Bar, 5 µm. (H) Schematic presentation of PLD distribution in wild-type and bbs4-1 mutant cells.
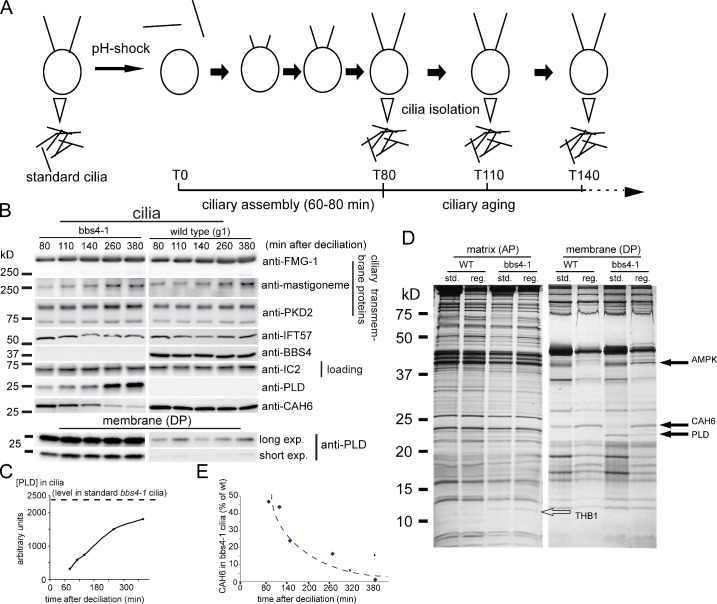
Figure 3.
The biochemical defects in bbs4-1 cilia increase with time. (A) Schematic presentation of the experimental design for obtaining age-sorted cilia from C. reinhardtii. (B) Western blots comparing regenerated wild-type and bbs4-1 cilia isolated at various time points after deciliation. (top) Western blots were probed with anti-FMG-1, anti-mastigoneme protein, anti-PKD2, anti-IFT57, anti-BBS4, anti-IC2 as a loading control, anti-PLD, and anti-CAH6. Ciliary length measurements for a similar experiment are shown in Fig. S3 E. (bottom) Long and short exposures of a Western blot of isolated ciliary membrane fractions from the same experiment probed with anti-PLD. (C) Graph showing the accumulation of PLD with time in bbs4-1 cilia; the dashed line corresponds to PLD levels in standard bbs4-1 cilia, i.e., cilia harvested without prior deciliation at least 6 h after the beginning of the light phase. (D) Silver-stained gel comparing matrix and membrane isolated from standard and 2-h-old regenerated wild-type and bbs4-1 cilia. Solid arrows, proteins altered in bbs4-1 ciliary membrane; open arrow, THB1. Note the differences in composition in the membrane of young and old cilia; no major differences were observed between young and old axonemal (not depicted) and matrix fractions, indicating that age-induced changes in ciliary protein composition are largely restricted to the ciliary membrane. (E) Graph showing the loss of CAH6 in bbs4-1 cilia as a percentage of that in wild-type cilia over time. The data shown in C and E are from a single representative experiment out of two repeats.
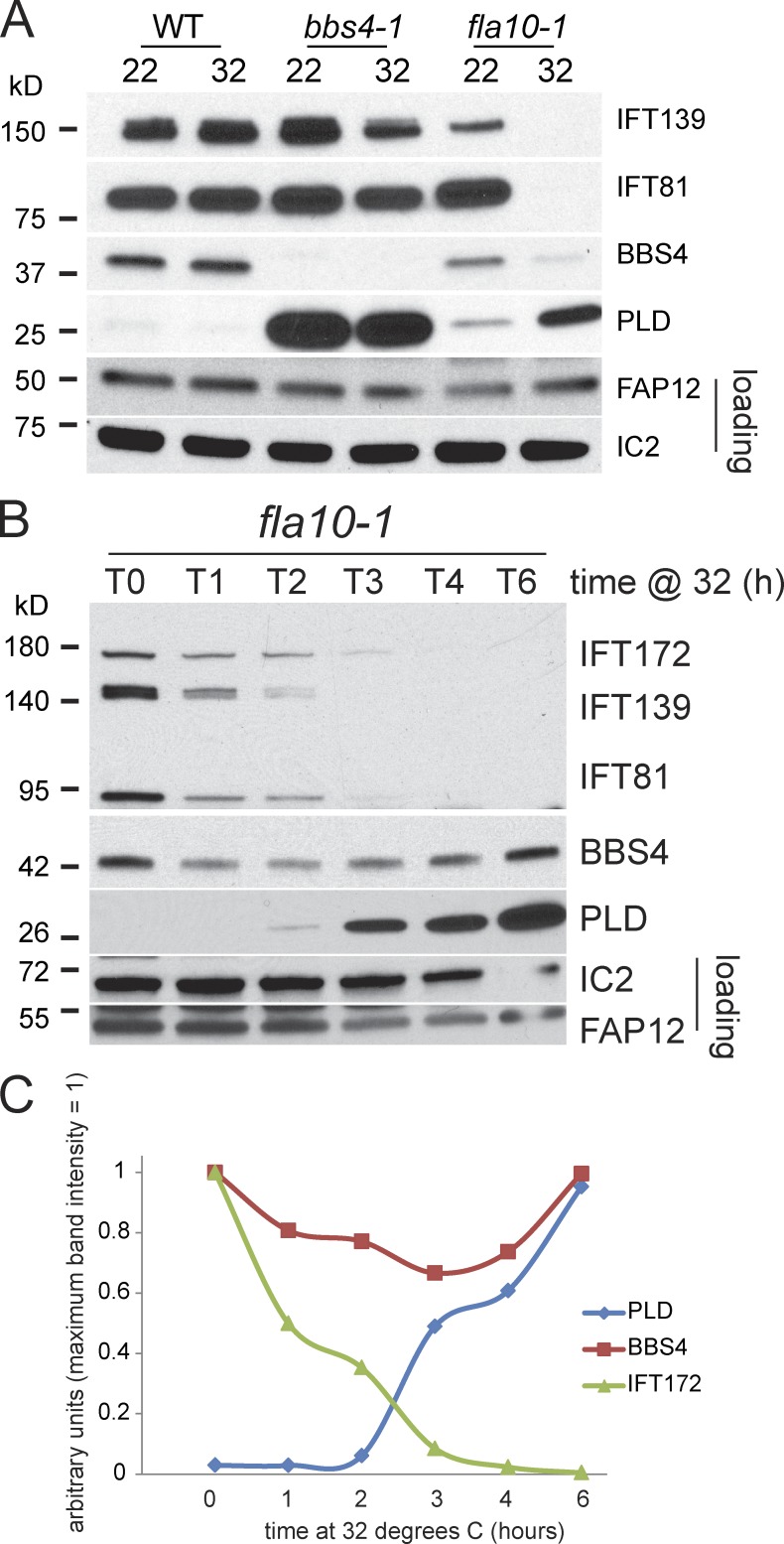
Figure 6.
PLD accumulates in cilia after depletion of IFT proteins. (A) Western blot of cilia isolated from wild type, bbs4-1, and fla10-1 at 22°C and after 2.5 h at 32°C. The blot was probed with antibodies to the proteins indicated. Depletion of IFT proteins and accumulation of PLD in response to the temperature shift was observed only in cilia from fla10-1. (B) Western blot comparing fla10-1 cilia isolated at various time points from 0 to 6 h (T0–T6) after shifting the cells to 32°C. The blot was probed with antibodies to the proteins indicated. Note progressive depletion of IFT proteins and concomitant accumulation of PLD. (C) Relative amounts of PLD, BBS4, and IFT172 in the Western blot analysis shown in B. Maximum band intensity for each protein was set to 1. At later time points the mean length of fla10-1 cilia was reduced (Fig. S3 D). Because similar amounts of protein were loaded for each sample, the slight increase observed for BBS4 during the second half of the incubation time is likely to reflect the higher number of cilia loaded.
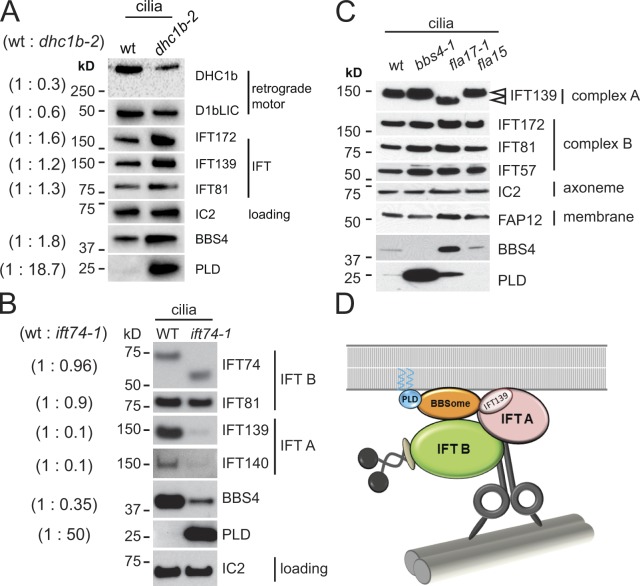
Figure 5.
Ciliary PLD is elevated in retrograde IFT mutants. (A) Western blot of isolated wild-type and dhc1b-2 cilia probed with antibodies to the proteins indicated. The amounts of the dynein 1b heavy chain, DHC1b, and light intermediate chain, D1bLIC, are reduced in the mutant cilia because of the instability of the dynein motor complex. BBS4 and in particular PLD are enriched in mutant cilia. IC2 was used as a loading control. For quantification of relative protein levels, values for band intensities were first normalized to the IC2 band intensity and the ratios (left, parentheses) were then calculated. (B) Western blot of cilia isolated from wild type and ift74-1 probed with antibodies to the proteins indicated. In ift74-1 the IFT complex B protein IFT74 is truncated. Note reduced amounts of IFT complex A proteins and BBS4 and increased levels of PLD in the mutant cilia. Relative protein levels (in parentheses) were determined as in A. (C) Western blot probing isolated cilia from wild type, bbs4-1, fla17-1 (expressing a truncated IFT139; arrowheads), and fla15-1. Note increase of BBS4 and PLD in fla17-1. (D) Summary diagram showing the IFT motors, IFT complexes A and B, the BBSome, and PLD. The BBSome functions as an IFT cargo adapter linking PLD and other cargoes to retrograde IFT particles. PLD is predicted to be myristoylated and palmitoylated (Fig. S4 C), offering a possible explanation for its membrane association. Dynein is shown linking the IFT particle to a doublet microtubule, which provides a track for IFT.
Importantly, PLD can also be detected in wild-type cilia, particularly in the ciliary membrane, by means of Western blots probed with the anti-PLD antibody, indicating that PLD is a bona fide ciliary protein (Fig. 1, A and C). In wild type and bbs4-1, ciliary PLD is largely associated with the ciliary membrane, defined as the detergent phase of Triton X-114 phase partitioning (Fig. 1 C). To estimate just how great the increase is in the mutant cilia, we used Western blotting to quantitate the relative amounts of PLD in isolated membranes of wild-type and bbs4 mutant cilia. We determined that PLD is ∼150–200× more abundant in the mutant versus wild-type ciliary membrane (Fig. 1 D). A similar enrichment was observed in cilia of mutant versus wild-type gametes (unpublished data).
To distinguish whether the observed enrichment of PLD in the cilia of bbs mutants is a result of a general increase in PLD levels in the mutant cells or to defects in ciliary trafficking of PLD, we determined the distribution of PLD in wild-type and bbs mutant cells and cilia. Western blotting revealed that the total amount of PLD in bbs4-1 cells actually was reduced by ∼40% in comparison to wild-type cells (Fig. 1, E and H). Moreover, in wild type, ∼0.2% of the total PLD is in the cilia, whereas in the mutant, ∼60% of the total PLD is in the cilia.
Immunofluorescence microscopy confirmed our biochemical data: PLD was readily detected in a spotted distribution along the length of bbs4-1 mutant cilia but was barely detectable in wild-type cilia (Fig. 1 F and see Fig. 4 B). Within mutant cilia the signal strength was more pronounced in the proximal region of the cilium. Labeling by the anti-PLD antibody was similar in wild-type and bbs4-1 cell bodies, albeit signal strength was reduced in the mutant. Label was distributed in a spotted pattern at the cell periphery (Fig. 1 G for bbs4-1); this could represent vesicles underlying the plasma membrane. In summary, the bbs4-1 mutation causes a dramatic redistribution of PLD from the cell body into cilia (Fig. 1 H).
Figure 4.
PLD is removed from cilia of bbs4-1 cells when BBSomes are introduced by fusion with wild-type cells. (A) Schematic presentation of the PLD ciliary export assay. In wild-type gametes BBSomes are present in both cytoplasm and cilia, and nearly all of the cell’s PLD is in the cell body; bbs4-1 gametes lack BBSomes and have undergone a massive redistribution of PLD from the cell body to the cilia. Wild-type (BBS4, mt−) and bbs4-1 (bbs4, mt+) gametes were mixed and allowed to fuse. The resulting zygotes have four cilia (two each from the wild-type and mutant gametes); BBSomes are present in the shared cytoplasm. (B) Gametes (a–c) and quadriciliated zygotes (d–l) were stained with antibodies to PLD and to IFT139 to visualize the cilia. Arrow in c, bbs4 gamete; arrowhead in c, wild-type gamete; arrows in f and h, cilia containing PLD derived from mutant gametes; arrowheads in f, cilia lacking PLD derived from a wild-type gamete; arrowheads in j, four cilia of a zygote without detectable PLD. Bar, 5 µm. (C) Quadriciliated zygotes were scored by eye for the amount of PLD in their cilia. Black bars, quadriciliated cells with two wild-type cilia and two cilia with levels of PLD characteristic of the bbs4-1 gamete; gray bars, quadriciliated cells with two wild-type cilia and two cilia with detectable but reduced amounts of PLD; white bars, quadriciliated cells without detectable PLD in their cilia.
The lipid composition of bbs4 cilia is altered
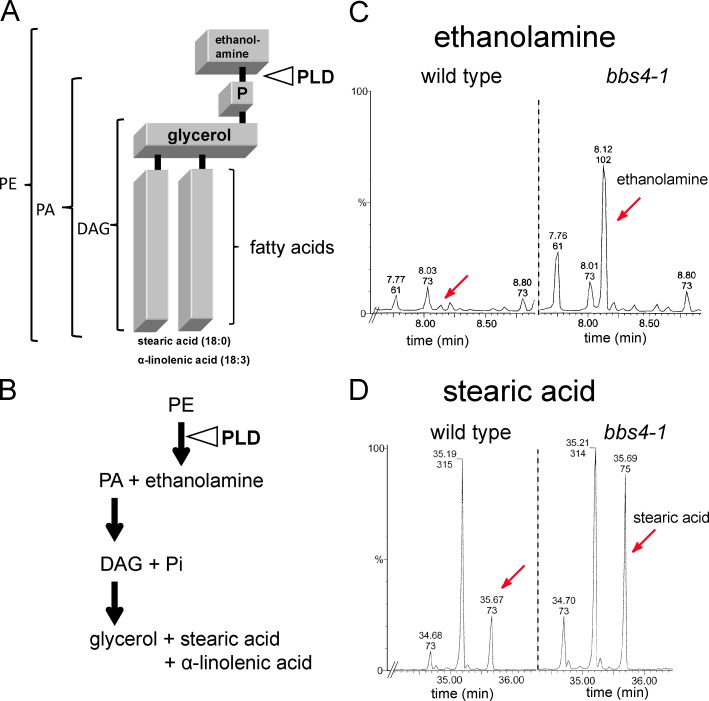
To assess whether the accumulation of PLD in bbs cilia affects the lipid composition of the ciliary membrane, we performed a metabolomic and lipidomic analysis of purified wild-type and mutant cilia (Table 1 and Fig. 2). Phosphatidylethanolamine (PE; Fig. 2, A and B), the substrate of PLD in C. reinhardtii (Munnik et al., 2000), was reduced by ∼30–50% in bbs4-1 cilia. Phosphatidic acid (PA) and ethanolamine, the products of PLD activity, were increased by 50% and 4,000%, respectively (Table 1 and Fig. 2 C). DAG and stearic acid, putative downstream products of PA enzymatic degradation, were each increased by ∼500%; a two- to threefold increase was also observed for glycerol and glycerol phosphate (Fig. 2 D and not depicted). The changes in PE, PA, and DAG occur predominately in species carrying 18:3/18:0 fatty acids, indicating that they are linked, i.e., that DAG 18:3/18:0 is formed by dephosphorylation of PA 18:3/18:0 (Table 1). The amounts of other membrane lipids, including that of the major ciliary membrane lipids ergosterol and stigmasterol, which account for ∼32% and ∼20% of the total ciliary lipid, respectively, were similar in both strains (unpublished data). In conclusion, the lipid composition of the ciliary membrane of the bbs4-1 mutant is altered and the observed changes are as predicted for increased PLD activity. The data also reveal that only a limited number of phospholipid species are present in the ciliary membrane of C. reinhardtii (Table 1).
Table 1.
Quantitative analysis of phospholipids and DAG in wild-type and bbs4-1 cilia
| Phospholipid | Total phospholipid in wild-type cilia | Total phospholipid in bbs4-1 cilia | Change in bbs4-1 cilia |
| % | % | % | |
| DAG 18:1/18:1 | 1.35 | 1.95 | +44 |
| DAG 18:3/18:0 | 1.55 | 9.29 | +500 |
| PA 18:3/18:0 | 11.36 | 16.84 | +50 |
| PE 18:0/18:3 | 49.64 | 35.12 | −29 |
| PE 18:1/18:3 | 1.77 | 1 | −43 |
| PG 16:0/16:0 | 20 | 22.6 | +13 |
| PG 16:1/16:1 | 3 | 3.74 | +25 |
| PG 18:0/16:0 | 0.46 | 0.46 | +1 |
| PG 18:1/16:0 | 8.68 | 7.3 | −16 |
| PI 18:1/16:0 | 2.89 | 1.7 | −41 |
The distribution of phospholipids in cilia of wild type and bbs4-1 as a percentage of total phospholipid. PG, phosphatidylglycerol; PI, phosphatidylinositol.
Figure 2.
The lipid composition of bbs4-1 cilia is altered. (A) Schematic presentation of the PLD substrate PE 18:0/18:3 and the site of PLD cleavage. (B) Schematic presentation of the enzymatic breakdown of PE into PA, ethanolamine, DAG, and downstream products. (C and D) Gas chromatography–mass spectrometry profiles showing the elution times for small metabolites from wild-type and bbs4-1 cilia. Ethanolamine (peaks marked by red arrows in C) is enriched ∼40× and free stearic acid (peaks marked by red arrows in D) is enriched ∼6× in bbs4-1 cilia. Accuracy of quantitation is ±10%.
The defects in protein content of bbs4 cilia increase with time
To distinguish whether the abnormal amounts of PLD in bbs cilia are deposited there during initial ciliary assembly or accumulate there with time after ciliary formation, we took advantage of the ability of C. reinhardtii to fully assemble cilia de novo within ∼60 min after amputation of the cilia by pH shock (Fig. 3 and Fig. S3 E). We simultaneously deciliated large populations of wild-type and bbs4-1 cells and allowed them to synchronously regenerate their cilia; we then isolated the recently formed cilia at time points ranging from 80 to 380 min after deciliation and compared them by SDS-PAGE and Western blotting (Fig. 3 A). At the loadings used, PLD was not detected in wild-type whole cilia (Fig. 3 B, top), but small amounts of PLD were consistently detected in isolated wild-type ciliary membranes, where the PLD was maintained at a relatively constant low level during the time period analyzed (Fig. 3 B, bottom). In contrast, PLD was detectable in the newly assembled mutant whole cilia at least as early as 80 min after deciliation; it then accumulated steadily over the course of several hours as the cilia aged (Fig. 3, B [top] and C). The data indicate that PLD accumulation in mutant cilia is a dynamic process that continues in full-length steady-state cilia. The amounts and kinetics of accumulation of several ciliary transmembrane proteins, specifically PKD2, FMG1, and the mastigoneme protein, were similar in wild-type and mutant cilia, suggesting that the transport of these proteins is unaffected by BBS4 deficiency.
In the course of the cell cycle, cilia are normally formed after division, at least 6 h before our synchronously grown cells were used for biochemical analyses. We here term such 6-hour-old cilia “standard cilia.” Comparison of regenerated and standard cilia from wild type and bbs4-1 using silver-stained gels revealed a protein of ∼24 kD that was present in wild-type cilia and in newly regenerated bbs4 cilia but was significantly reduced in standard bbs4 cilia (Fig. 3 D). The protein was tentatively identified as carbonic anhydrase 6 (CAH6) based on recovery of a single peptide by mass spectrometry (Fig. S1 B); the identification was confirmed using an antibody to C. reinhardtii CAH6 (Fig. 3 B). Quantitative analysis of age-sorted cilia showed that at 80 min after deciliation the mutant cilia contained ∼50% of the CAH6 present in comparable wild-type samples (Fig. 3, B and E). Over the next five hours CAH6 was progressively lost from mutant cilia, whereas little reduction of CAH6 was observed in wild-type cilia (Fig. 3, B, D, and E). Because a substantial amount of CAH6 is present in newly formed mutant cilia, it is unlikely that BBSome defects inhibit CAH6 ciliary import. The data rather suggest that bbs cilia increasingly lose the ability to retain CAH6. Thus, CAH6 loss from bbs cilia appears to be caused indirectly by the BBSome defect.
Excess PLD is rapidly removed from bbs cilia when BBSomes are reintroduced
The progressive redistribution of PLD from the cell body into cilia of bbs mutants could be caused by a failure of PLD ciliary export. Therefore, to test directly whether BBSomes are required to remove PLD from cilia, bbs4 and wild-type gametes were mixed and the distribution of PLD in cilia of the resulting zygotes was analyzed at various time points after mating. The zygotes possess four cilia, two derived from the mutant gametes and two from the wild-type gametes (Fig. 4 A). After cell fusion, BBSomes contributed by the wild-type cytoplasm become immediately available to bbs4 mutant cilia. With immunofluorescence microscopy, PLD is readily detectable in cilia of mutant but not wild-type gametes (Fig. 4 B, a–c). Ten minutes after mixing of the gametes, some zygotes were observed that possessed two cilia with PLD derived from the bbs4 parent and two cilia without detectable PLD derived from the wild-type parent (Fig. 4, B [d–f] and C). However, even at this early time point, PLD was reduced or not detectable in cilia of the majority of zygotes (Fig. 4, B [g–l] and C). The share of zygotes without detectable ciliary PLD increased progressively from 18% at 10 min to 96% at 50 min after mixing of the gametes (Fig. 4 C). The time span required for gametes to complete fusion after mixing is variable—some zygotes may form as early as 15 s after mixing, with typically 50% of the gametes having completed fusion after 5 min in good mating reactions (Hunnicutt and Snell, 1991). The presence of a few zygotes with cilia still containing PLD at later time points can be explained by delayed fusion of some gametes. In conclusion, PLD is removed in <10 min from the formerly mutant cilia once BBSomes become available. Therefore, an efficient pathway for PLD export from cilia is present in C. reinhardtii and this pathway requires the BBSome.
PLD export from cilia requires retrograde IFT
We previously showed that the BBSome cycles in association with IFT through cilia (Lechtreck et al., 2009). To determine if this association is important for BBSome function and normal export of PLD from the cilium, we analyzed the mutant strain dhc1b-2 (previously referred to as dhc1bts). This strain contains a hypomorphic allele of the gene encoding the heavy chain of dynein 1b, the motor for retrograde IFT (Lechtreck et al., 2009; Witman, 2012). At the permissive temperature (21°C) used for all experiments involving dhc1b-2 here, ∼60% of the mutant cells assembled two cilia that were slightly shorter than those of wild type (Fig. S3, A and B). The velocity and frequency of retrograde IFT and the amounts of the retrograde dynein subunits DHC1b and D1bLIC were reduced in the dhc1b-2 cilia (Fig. 5 A and Fig. S2, C and D). The amount of BBS4 was elevated in dhc1b-2 cilia, as expected for a protein cycled through the cilium by IFT. However, despite the presence of BBS4/BBSomes, PLD was highly enriched in the mutant cilia (Fig. 5 A), indicating that the BBSome-dependent export of PLD from cilia requires the retrograde motor for IFT.
As an independent test of the involvement of IFT in export of PLD, we analyzed the mutant ift74-1, which expresses a truncated version of the IFT complex B protein IFT74 (Fig. 5 B; unpublished data). The truncated IFT74 is assembled into complex B and transported into the cilium as part of the IFT particle; the ciliary levels of the other complex B proteins are near normal, but the amount of IFT complex A is greatly reduced. Compared with wild-type cilia, BBS4 was reduced by ∼65% and PLD was strongly accumulated in ift74-1 cilia (Fig. 5 B). The data suggest that entry of BBSomes into cilia requires IFT74 and/or IFT complex A and that the residual IFT and BBSomes in ift74-1 cilia are insufficient to remove PLD from cilia. The result is consistent with a recent observation in C. elegans that a mutation in complex A protein IFT144 abrogates entry of BBSomes into cilia (Wei et al., 2012).
IFT complex A has an important role in retrograde IFT (Pedersen and Rosenbaum, 2008; Pedersen and Christensen, 2012). To more directly investigate if complex A has a role in BBSome transport and PLD export, we analyzed strains fla15-1 and fla17-1, which carry mutations in the complex A proteins IFT144 and IFT139, respectively (Piperno et al., 1998; Iomini et al., 2009). The fla15-1 strain has a point mutation that converts a highly conserved cysteine in IFT144 to an arginine. The fla17-1 strain has an in-frame deletion of three exons of the IFT139 gene and produces a smaller-than-normal version of IFT139 (Fig. 5 C; Iomini et al., 2009). These fla mutants fail to assemble cilia at elevated temperatures. At the permissive temperature used here IFT is sufficient for ciliary assembly but the velocity and frequency of retrograde IFT is reduced and the cilia develop small bulges containing complex B components, indicative of a defect in returning cargo to the cell body (Iomini et al., 2009). Compared with wild type, BBS4 was highly enriched in cilia of fla17-1 (Fig. 5 C), suggesting that the BBSome also is not returned normally to the mutant cell body. Despite the abundance of BBSomes, PLD also was highly enriched in fla17-1 cilia (Fig. 5 C), indicating that PLD export from cilia is dependent on IFT complex A. In contrast, the amounts of BBS4 and PLD were near normal in cilia of fla15-1 (Fig. 5 C). The data suggest that IFT139 is involved in retrograde transport of BBSomes and removal of PLD from cilia.
PLD can enter cilia independently of IFT
Our data suggest that retrograde IFT particles transport PLD in a BBSome-dependent manner. Because PLD enters cilia through a BBSome-independent process, we investigated if IFT is required for entry of PLD into cilia. We took advantage of the fla10-1 mutant, which carries a temperature-sensitive mutation in one of the heavy chains of the anterograde IFT motor kinesin-2, so that IFT can be rapidly turned off by a temperature shift (Kozminski et al., 1995). At the permissive temperature (22°C), fla10-1 cilia contained slightly elevated amounts of PLD, which can be explained by the somewhat reduced amounts of IFT complex A and BBSomes (as indicated by reduced amounts of IFT139 and BBS4, respectively) in the mutant cilia (Fig. 6 A). Cilia isolated from fla10-1 that had been incubated for 2.5 h at the nonpermissive temperature (32°C) were largely depleted of IFT81 (complex B) and IFT139 (complex A). This loss of IFT proteins at the restrictive temperature was accompanied by an increase in ciliary PLD; in contrast, the amounts of PLD in wild-type and bbs4-1 cilia were essentially unaffected by the temperature shift. To determine the temporal sequence of IFT depletion and PLD accumulation in fla10-1 cilia, we analyzed cilia isolated at various time points (1–6 h) after shifting fla10-1 cells to 32°C (Fig. 6 B). During the incubation time the number of mutant cells carrying one or two cilia decreased from >90% to ∼70% and the mean length of the cilia that they did have decreased from ∼11 to ∼6 µm; the number and length of cilia of wild type was unaffected at 32°C (Fig. S3, C and D). Starting at ∼2 h at 32°C, by which time the level of IFT proteins was considerably reduced, PLD began to accumulate in fla10-1 cilia. Importantly, PLD continued to accumulate in fla10-1 cilia after 4 h at 32°C when IFT proteins were nearly undetectable; after ∼6 h at 32°C it reached a level approaching that of bbs mutants (>30× above the level before temperature shift; Fig. 6 C). The data indicate that PLD entry into cilia does not require IFT. Note that despite the depletion of IFT proteins, BBS4 remained present in fla10-1 cilia. The accumulation of PLD in the presence of BBS4 provides additional evidence that BBSomes alone are insufficient to control PLD levels in cilia in the absence of IFT.
Discussion
BBSome deficiency has been shown to alter the membrane protein composition of cilia in C. reinhardtii and mammals (Berbari et al., 2008b; Lechtreck et al., 2009; Domire et al., 2011; Seo et al., 2011; Zhang et al., 2011). We showed previously that BBSomes travel through cilia in association with a subset of IFT particles (Lechtreck et al., 2009). These observations raise the possibility that BBSomes contribute to IFT-mediated transport of ciliary membrane proteins. Here we analyzed the role of the BBSome and IFT in the ciliary transport of PLD, a membrane-associated protein that accumulates in cilia of C. reinhardtii bbs mutants. Our data reveal that PLD continuously enters the cilium in a BBSome- and IFT-independent manner but that export of PLD from the cilium is dependent on the BBSome, probably functioning as a cargo adaptor for IFT.
PLD cycles through the cilium and redistributes from the cell body to the cilium in the absence of the BBSome or IFT
PLD is abundant in wild-type cell bodies and ∼0.2% of the PLD is present in wild-type cilia. However, in bbs4-1 mutants there is a massive redistribution of PLD from the cell body into the cilia, such that about half of the PLD is now contained in the cilia. Taking into account the relative volumes of the cell body (>500 µm3) and the cilium (<0.5 µm3; and ignoring possible differences in the fraction of each that is accessible to PLD), PLD in bbs4-1 cells is enriched ∼1,000× in the two cilia relative to the cell body. Because this concentration occurs in bbs mutants, PLD must be able to move into the cilia in the absence of BBSomes. A similar great enrichment was observed in cilia of the conditional mutant fla10-1 after IFT proteins (but not BBS4) were depleted by incubation at the nonpermissive temperature. This strongly suggests that entry of PLD into cilia can occur even in the absence of IFT. Recently, Kee et al. (2012) reported that small molecules and proteins ≤40 kD can freely pass from the cytoplasm into cilia. Thus, PLD, which is only 25 kD, may enter the cilia slowly by diffusion and then become trapped there in the absence of an active export mechanism.
Because PLD accumulates progressively over time in BBSome- and IFT-deficient cilia, it is likely that the protein is continuously cycled through the cilium in wild-type cells, with IFT-dependent export mediated by the BBSome. The high efficiency of this PLD export pathway, as demonstrated by the rapid clearing of accumulated PLD when BBSomes are reintroduced into bbs mutant cilia, suggests that PLD entering wild-type cilia will be rapidly picked up and removed from the organelle. A system in which the movement of a protein into the cilium is BBSome independent but its export is BBSome dependent would allow fine tuning of the amount of the protein in the cilium by regulating the number of BBSomes that enter the cilium or by regulating the affinity of the BBSome for the protein.
A similar role for the BBSome in mammalian primary cilia is suggested by recent observations on dopamine receptor 1, a GPCR that is rapidly transported to and from cilia in response to environmental cues (Domire et al., 2011). Import of dopamine receptor 1 into cilia is BBSome independent whereas translocation of the protein from cilia requires the BBSome (Domire et al., 2011; Zhang et al., 2011). In addition, very recent results indicate that accumulation of the hedgehog signaling proteins Smoothened and Patched 1 occur in cilia of cells from Bbs−/− mice (Zhang et al., 2012). Like PLD in C. reinhardtii, Smoothened is believed to continuously shuttle through the cilium (Ocbina and Anderson, 2008). Thus, BBSome-mediated export may be a feature of many membrane-associated proteins that undergo such cycling.
BBSome function depends on retrograde IFT
To investigate the role of IFT in the BBSome-dependent export of PLD from the cilium, we analyzed the distribution of BBS4 and PLD in cilia isolated from the dynein 1b mutant dhc1b-2, which is characterized by reduced retrograde movement of IFT particles. Using silver-stained 1D and 2D gels, we previously determined that THB1 is enriched in dhc1b-2 cilia, but we could not ascertain the levels of PLD in these cilia (Lechtreck et al., 2009). Now, using our anti-PLD antibody and Western blotting, we found that PLD also is significantly accumulated in dhc1b-2 cilia despite the presence of BBSomes. Likewise, BBSomes are present albeit reduced during PLD accumulation in fla10-1 cilia at restrictive temperature. Therefore, the presence of BBSomes in cilia is insufficient to remove PLD when IFT is defective. Because the primary effect of the dhc1b-2 mutation is on retrograde IFT, these results indicate that retrograde IFT is critical for PLD export from the cilium. Inasmuch as loss of the BBSome has little if any effect on IFT (Figs. 1 C, 3 B, 6 A, and S2; Lechtreck et al., 2009) but defects in retrograde IFT affect export of both the BBSome and PLD, IFT must function upstream of the BBSome in the PLD export pathway; i.e., retrograde IFT moves the BBSome out of the cilium, and the BBSome in turn carries PLD from the cilium (Fig. 5 D).
Export of Smoothened from mammalian primary cilia similarly is dependent on the retrograde IFT motor, as indicated by the accumulation of Smoothened in cilia of mouse cells having a mutation in the dynein 2 heavy chain Dync2h1 (Ocbina and Anderson, 2008) or in which the same protein was knocked down by shRNA (Kim et al., 2009) or inhibited pharmacologically (Firestone et al., 2012). Thus, IFT-dependent BBSome-mediated export of signaling molecules from the cilium may be widespread. However, another hedgehog signaling protein, Gli2, which also is believed to cycle though the cilium (Kim et al., 2009), does not accumulate in Bbs−/− mice (Zhang et al., 2012), suggesting the existence of distinct export (and import) mechanisms for different ciliary proteins.
IFT complex A is critical for BBSome export
The aforementioned observations suggest that coupling of PLD to retrograde IFT particles requires the BBSome. To explore how the BBSome interacts with the IFT machinery, we used mutants defective in IFT particle proteins. IFT complex A proteins and BBS4 are similarly decreased in cilia of ift74-1, which expresses a truncated complex B IFT74 protein. PLD is highly enriched in ift74-1 cilia, indicating that the residual BBSomes are insufficient for its export. This suggests that IFT74 is critical for the entry of IFT complex A into cilia and that complex A could mediate binding of the BBSome to IFT complex B and the associated dynein motors during retrograde IFT. To test for a possible role of IFT complex A as a docking site for the BBSome, we analyzed fla15 and fla17-1, which are defective in the IFT complex A proteins IFT144 and IFT139, respectively, and have reduced retrograde IFT (Piperno et al., 1998; Iomini et al., 2009). The distributions of BBS4 and PLD were nearly normal in fla15, whereas PLD and in particular BBS4 were enriched in cilia of fla17-1. Therefore, IFT139 could be involved in binding the BBSome to IFT complex A during retrograde IFT, rendering it critical for the export of the BBSome and PLD from cilia (Fig. 5 D). It may be relevant that the Tubby family protein TULP3 similarly interacts with complex A in mammals to promote ciliary trafficking of a subset of GPCRs, including MCHR1, and that retrograde transport of TULP3 specifically requires IFT139 (Mukhopadhyay et al., 2010).
In summary, PLD accumulates in cilia of mutants with defective BBSomes (bbs4-1, bbs1-1, bbs7-1, and bbs8-1; Lechtreck et al., 2009), with reduced amounts of BBSomes in cilia (ift74-1), and with reduced retrograde IFT of the BBSome (dhc1b-2 and fla17-1). We propose that the BBSome functions as a cargo adapter linking PLD to retrograde IFT particles to ensure export of PLD from cilia. Further work will be necessary to understand how PLD is delivered to the cilium, how PLD interacts with the BBSome during the export phase of its cycle, and how the BBSome interacts with IFT complexes A and B.
BBSome function in IFT assembly and turnaround
In C. elegans mutants null for bbs-7 and bbs-8, IFT complex A separates from complex B and the two complexes are transported anterogradely by kinesin-2 and OSM-3, respectively (Ou et al., 2005). These observations led to the hypothesis that the BBSome couples complex A to complex B and coordinates the activity of the two anterograde IFT motors. However, more recently it has been reported that a hypomorphic mutation at a conserved site in the complex A protein DYF-2 (the C. elegans homologue of IFT144) causes complete dissociation between the BBSome and moving IFT particles, yet complexes A and B remain associated in anterograde IFT (Wei et al., 2012). These results indicate that in C. elegans the BBSome is not essential for stabilizing the binding of complex A to complex B, in good agreement with our previous observation that the colocalization of complexes A and B is not affected in cilia of C. reinhardtii bbs4-1 mutants (Lechtreck et al., 2009). In the C. elegans IFT144 mutant, the BBSome is still formed but remains at the base of the cilium. Moreover, complex B but not complex A accumulates at the tip of the cilium, suggesting that complex B fails to be coupled to complex A for retrograde IFT. Based on these observations, Wei et al. (2012) proposed that in wild-type C. elegans the BBSome regulates assembly of complexes A and B into intact IFT particles at the base of the cilium, and is then carried as cargo by IFT to the tip of the cilium, where it has a similar role in reorganizing the IFT particle for retrograde transport. In the mutant, the BBSome still functions to assemble the IFT particle at the base of the cilium, but because it is no longer transported to the ciliary tip, IFT particle reorganization is defective and complex B accumulates there.
We previously reported that in C. reinhardtii the BBSome is not an integral part of the IFT machinery but rather appears to be a cargo or cargo adapter, a conclusion now supported by the observations of Wei et al. (2012). However, in C. reinhardtii loss of the BBSome does not seem to affect the assembly of complexes A and B (Lechtreck et al., 2009), does not lead to an accumulation of complex B at the tip of the cilium (Lechtreck et al., 2009), and does not obviously alter the balance of complex A to complex B within the cilium as would be expected if complex B were being left behind during retrograde transport (Figs. 5 C, 6 A, and S2 B). Therefore, in contrast to the situation in C. elegans, it is unlikely that the BBSome has a key role in regulating IFT particle assembly at either the base or the tip of the C. reinhardtii cilium.
The function of PLD in C. reinhardtii
The function of PLD in C. reinhardtii is likely to involve phospholipid signaling (Meijer et al., 2002). Consistent with this, we found that the accumulation of PLD in cilia of C. reinhardtii mutants is correlated with major changes in the phospholipid content of the ciliary membrane. It is possible that PLD cycles through the wild-type cilium as part of a system that monitors and responds to the cell’s environment (e.g., to light or to osmotic conditions) via a phospholipid-based signaling pathway. We previously reported that C. reinhardtii bbs mutants are nonphototactic. Phototaxis is dependent on membrane depolarization and Ca2+ influx (Witman, 1993), both of which activate PLD in C. reinhardtii (Munnik et al., 2000). Changes in membrane lipid composition could affect the functioning of membrane channels and hence phototaxis. Consistent with this, we previously observed that bbs4-1 cells with newly formed cilia were able to phototax for over an hour, but lost that ability as the cilia aged and presumably accumulated PLD and acquired an altered membrane lipid composition (Lechtreck et al., 2009).
C. reinhardtii PLD has homologues in mammals and many other ciliated eukaryotes
Our finding that C. reinhardtii PLD is apparently a bona fide ciliary protein that cycles through the cilium raises the question of whether a homologue occurs in the cilia of other organisms. Close homologues of C. reinhardtii PLD are encoded in the genomes of many vertebrates, nonvertebrate metazoans (with C. elegans being a notable exception), and some ciliated protists; they are absent from Arabidopsis thaliana, yeasts, and other nonciliated organisms (Fig. S4, A and B). Such a phylogenetic distribution is often indicative of a centriolar or ciliary function of the gene product. The mammalian homologue of PLD is PLD6 (C. reinhardtii to H. sapiens BLAST E = 8e−37; 40% identity); in a phylogenetic analysis PLD6 groups more closely with C. reinhardtii PLD than with other murine PLDs (Fig. S4 B). In mice, overexpressed PLD6 localizes to mitochondria and is anchored to the mitochondrial surface by a transmembrane domain (Huang et al., 2011). This transmembrane domain is not predicted in C. reinhardtii PLD but, like C. reinhardtii AMPK and CAH6, the protein is predicted to be myristoylated and palmitoylated (Fig. S4 C), likely accounting for its association with the ciliary membrane. PLD6 is widely expressed in human tissues; in mice, expression is especially high in lung and testis, tissues that are rich in cilia, and Pld6 −/− mice are infertile (Huang et al., 2011; Watanabe et al., 2011). It will be of interest to determine if mammalian PLD6 functions also in cilia, and whether BBSome deficiency promotes ciliary accumulation of PLD6. It may be relevant that cilia of Bbs−/− mice accumulate unusual intraciliary vesicles at the ciliary tip (Shah et al., 2008); this might be brought about by an excess of ciliary DAG caused by ciliary accumulation of PLD6. Phospholipid signaling also has been implicated in the functioning of olfactory cilia (Klasen et al., 2010), and malfunction of olfactory cilia has been reported in Bbs8−/− mice (Tadenev et al., 2011).
BBSome deficiency results in secondary defects in ciliary composition
Quantitative analysis of age-sorted cilia revealed that the ciliary protein CAH6 is progressively lost from bbs4-1 cilia. Because CAH6 is initially present in mutant cilia, it is unlikely that BBSome defects inhibit CAH6 ciliary import. Rather, the progressive loss of CAH6 from bbs4-1 cilia is likely to be caused indirectly by BBSome deficiency. Loss of a peripheral membrane protein such as CAH6 could follow from the severe changes in membrane lipid content that occur in bbs4-1 cilia, presumably as a result of increased ciliary PLD activity; the ability of proteins, such as CAH6, which is predicted to be fatty acid modified (Fig. S4 C), to associate with membranes is influenced by the lipid composition of the membrane (Levental et al., 2010). An altered lipid composition of bbs cilia also could contribute in other ways to changes in the pattern of ciliary membrane proteins. Loss of certain GPCRs from cilia has been reported in murine Bbs−/− mutants (Berbari et al., 2008b). In light of our data on CAH6 in bbs mutant cilia of C. reinhardtii, it seems possible that absence of GPCRs from mammalian cilia also could be an indirect consequence of the loss of the BBSome.
BBS likely involves progressive deterioration of ciliary function
The observation that ciliary defects resulting from BBSome deficiency increase with time potentially has implications for the pathomechanism of BBS in mammals. Several features of BBS in murine Bbs−/− models or human patients with BBS also increase with time or manifest themselves only years or decades after birth (Beales, 2005). The outer segments of photoreceptor cells in the eye, for example, initially develop and function normally in Bbs2 and Bbs4 knockout mice. About 6 wk postnatally, rhodopsin mislocalization becomes apparent and retinal degeneration begins (Nishimura et al., 2004). Therefore, BBSome defects appear to initiate a cascade of events that increasingly deteriorate ciliary protein composition and function. The time scales in which ciliary defects develop in different organisms and different tissues within an organism may be different. Nevertheless, the mechanism by which cells use the BBSome to control ciliary protein transport and to maintain the balance of ciliary signaling proteins might very well be conserved.
Materials and methods
Strains and culture conditions
Wild-type and mutant strains of C. reinhardtii were mostly maintained as described in Lechtreck et al. (2009). The bbs8-1 strain was generated as described in the legend of Fig. S1. The retrograde IFT mutants fla15 and fla17-1 were aerated without CO2 supplement and maintained at 22°C.
Antibodies and Western blotting
Using primers 5′-CGCGAATTCATGGGTTGCGCCAGCTCC-3′ and 5′-CGCGAATTCTTAGATGGCAAACTTATGGTGCATG-3′, a partial cDNA encoding C. reinhardtii PLD was obtained by PCR and cloned into bacterial expression vectors pGEX and pMAL. Truncated PLD fused to maltose-binding protein was used as an antigen to immunize two rabbits, and a fusion protein of truncated PLD and glutathione S–transferase was used to affinity purify the antisera produced by GenScript. Anti-PLD was diluted 1:2,000 for Western blotting and 1:100 for immunofluorescence microscopy. Polyclonal antibodies to C. reinhardtii CAH6 (1:2,000) and FAP12 (1:1,000) were provided by J.V. Moroney (Louisiana State University, Baton Rouge, LA) and E. Betleja and D.G. Cole (University of Idaho, Moscow, ID), respectively. Other rabbit polyclonal antibodies used in this study were anti-PKD2 L1 (1:1,000; Huang et al., 2007), anti-IFT140 (1:400), anti-IFT742 (1:4,000; Qin et al., 2004), anti-BBS4 (1:800; Lechtreck et al., 2009), anti-DHC1b (1:2,000; Pazour et al., 1999), and anti-D1bLIC (1:1,000; Hou et al., 2004). Mouse monoclonal antibodies used included anti-IC2 (1:100; King and Witman, 1990); anti-IFT139 (1:50), anti-IFT81 (1:250), anti-IFT57 (1:100), and anti-IFT172 (1:50; all Cole et al., 1998); anti-FMG-1 No. 61 (1:1,000; Bloodgood and Salomonsky, 1994); and anti-mastigoneme No. 37 (1:100; Nakamura et al., 1996). Western blots were developed using anti–mouse or anti–rabbit IgG conjugated to horseradish peroxidase (Molecular Probes) and chemiluminescence. An Alpha Innotech FluorChem Q chemiluminescence imaging system was used for documentation and quantification of signals, except for those Western blots including fla15, fla17-1, and fla10-1, which were documented using x-ray film and signals quantified using ImageJ (National Institutes of Health).
Mating experiments and fluorescence microscopy
To generate gametes, cells were grown for 10 d on Tris-acetate-phosphate medium plates (Gorman and Levine, 1965) and resuspended in modified medium I without nitrogen (8 ml/plate; M-N; http://www.chlamy.org/SG.html), and the suspension was incubated overnight in constant light with gentle agitation. The next morning, cells were incubated in 20% M-N supplemented with 10 mM Hepes for 3 h. To obtain a series of zygotes of distinct age, gametes were mixed at various consecutive time points. Next, the cells were harvested (3 min at 2,000 rpm, at room temperature); resuspended in 10 mM Hepes, 5 mM MgSO4, 5 mM EGTA, and 25 mM KCl (HMEK); mixed with an equal volume of 6% formaldehyde/0.1% Nonidet P-40 in HMEK; and allowed to settle onto poly-l-lysine–coated multi-well slides for ∼3–6 min. Slides were then submerged into −20°C methanol for 6 min and air dried. All subsequent steps were performed as described in the following paragraph.
For immunofluorescence microscopy of vegetative cells, cells in HMEK were mixed with an equal volume of 6% formaldehyde/0.5% Nonidet P-40 in HMEK, allowed to settle onto coated slides for 8–15 min, washed with PBS, and air dried. The following primary antibodies and dilutions were used: anti-PLD (1:100), anti-IFT139 (1:1), and anti–acetylated α-tubulin (1:2,000, mouse; Sigma-Aldrich). Specimens were incubated overnight at 4°C with primary antibodies in PBS/2% BSA/0.05% Tween20. Secondary antibodies linked to Alexa Fluor 488, 568, or 594 (1:1,000; Invitrogen) were applied for 90–120 min at room temperature. Specimens were mounted with ProlongGold (Invitrogen). Images were acquired using Axiovision software (Carl Zeiss) and an AxioCam MRm camera (Carl Zeiss) on a microscope (Axioskop 2 plus; Carl Zeiss) equipped with a 100× NA 1.4 oil DIC Plan-Apochromat objective (Carl Zeiss) and epifluorescence. Image brightness and contrast were adjusted using Photoshop (CS5.1; Adobe), and figures were assembled using Illustrator (CS5.1; Adobe). Capture times and adjustments were similar for images mounted together. An Axio Imager.Z1 equipped with a 100× NA 1.3 EC Plan-Neofluar objective, an Apotome, an axiocam MRm, and Axiovision software (all from Carl Zeiss) was used to obtain optical sections of the immunostained cell shown in Fig. 1 G. Figures were prepared using Adobe Photoshop and Illustrator.
Isolation of cilia
Flagellar isolation using the dibucaine method and flagellar amputation by pH shock were performed as previously described by Witman (1986) and Lefebvre (1995), respectively. In brief, cells were concentrated and repeatedly washed in 10 mM Hepes, pH 7.4, resuspended in HMS (10 mM Hepes, pH 7.4, 5 mM MgSO4, and 4% wt/vol sucrose), and placed on ice; all subsequent steps were performed at 4°C. 2 ml of dibucaine (25 mM in H2O) were added per 10 ml of sample; cells were diciliated by rapidly drawing them up and down in a pipette. 20 ml of 0.7 mM EGTA in HMS was then added to the suspension. Cell bodies were removed by differential centrifugation (3 min at 1,150 g); the supernatant was collected and the remaining cell bodies were removed by centrifugation (10 min at 1,700 g) through a sucrose cushion (10 ml 25% sucrose in HSM). The supernatant was harvested and cilia were pelleted at 27,000 g for 20 min. For deciliation by pH shock, 0.5 M acetic acid was added to cells in M medium with rapid stirring until the pH reached 4.2; after 50 s the solution was neutralized using 0.25 N KOH. Cells were immediately placed into fresh culture medium and allowed to regenerate cilia in bright light at RT with shaking (60 rpm). Isolated flagella were extracted with 1% NP-40 in HMDEKP (30 mM Hepes, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl, and 1% protease inhibitor cocktail [Sigma-Aldrich]) for 20 min on ice, and axonemes and membrane-plus-matrix were separated by centrifugation (27,000 g for 10 min). Alternatively, cilia were extracted with 1% Triton X-114, which allows subsequent separation of the membrane and matrix fractions by phase partitioning. After removing the axonemes, the supernatant was incubated briefly at 37°C and phases were separated by centrifugation (3,300 g for 10 min at RT). The aqueous phase was treated with 1% Triton X-114, the detergent phase was diluted with HMDEKP, and the phase separation was repeated to yield the final aqueous and detergent phases.
For temperature-shift experiments with fla10-1, cells were concentrated and incubated in Modified Medium I (Witman, 1986) with gentle shaking (60 rpm) either at room temperature (∼22°C) or at 32°C in an incubator. For time course experiments, equal aliquots of concentrated fla10-1 cells were transferred at various time points from 22 to 32°C and incubated for the time period indicated. Then, cilia from all samples were isolated simultaneously using warm (32°C) 10 mM Hepes to wash cells maintained at the restrictive temperature. All experiments using the dhc1b-2 and other IFT mutants were performed at or below 22°C.
Lipidomic and metabolomic analyses of cilia
We used two distinct approaches for the lipidomic analysis of cilia, liquid chromatography coupled to mass spectrometry and an approach based solely on mass spectrometry. In brief, cilia were extracted with methanol/chloroform and lipid extracts were separated by gradient elution normal phase HPLC and the analytes detected by positive atmospheric pressure chemical ionization. For metabolomic analysis by gas chromatography coupled to mass spectrometry, samples were derivatized to their trimethylsilyl derivatives. Chromatography was on a 30-M DB-5MS column. Full scan electron ionization mass spectrometry was performed using a Quattro II mass spectrometer (Waters). The accuracy of quantitation is ±10%.
For quantitative analysis of phospholipids, ∼100 µg of cilia (wet weight) from each sample was subjected to lipid extraction. In brief, the sample was dissolved in 200 µl of liquid chromatography grade water. Samples were spiked with 10 µl of a mixture of internal standards containing 20 pmol DAG 17:0–17:0, 24 pmol PA 17:0–17:0, 52 pmol PE 17:0–17:0, 7.5 pmol phosphatidylglycerol (PG) 17:0–17:0, 43 pmol phosphatidylserine (PS) 17:0–17:0, 40 pmol phosphatidylcholine (PC) 18:3–18:3, 54 pmol phosphatidylinositol (PI) 17:0–17:0, 20 pmol ceramide (Cer) 18:1;2/17:0;0, 40 pmol sphingomyelin (SM) 18:1;2/17:0, 20 pmol galactosylceramide (GalCer) 18:1;2/12:0, and 20 pmol lactosylceramide (LacCer) 18:1;2/12:0. 265 µl of methanol was added to the mixture and agitated for 10 min for homogenization. Then, 730 µl of chloroform were added and the mixture was agitated for an additional 1 h. The lower organic phase was collected and evaporated in a Speedvac concentrator at room temperature. Lipid extracts were dissolved in 100 µl of 0.1% methylamine in methanol and subjected to quantitative lipid analysis on a hybrid QSTAR Pulsar i quadrupole time-of-flight mass spectrometer (MDS Sciex). Samples were infused with a TriVersa NanoMate robotic nanoflow ion source (Advion Biosciences, Inc.). DAG, PA, PS, PE, PI, and PG species were quantified by negative ion mode multiple precursor ion scanning analysis (Ejsing et al., 2006).
Miscellaneous
For mass spectrometric identification of CAH6, bands of interest were excised from silver-stained gels and digested overnight with proteomics-grade trypsin, and the eluted peptides were analyzed using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Kratos Axima QIT; Shimadzu Instruments). Peptides were analyzed in positive ion mode in mid–mass range (700–3,000 D; Lechtreck et al., 2009).
CLUSTALW at the default settings was used to align multiple PLD sequences and for tree construction.
Online supplemental material
Fig. S1 A shows a molecular map of bbs8-1; the sequence of CAH6 is depicted in Fig. S1 B. Fig. S2 analyzes ciliary length and IFT in dhc1b-2 and the bbs mutant strains and shows protein content of wild-type and bbs mutant cilia and cell bodies. Fig. S3 shows ciliary numbers and length for fla10-1 and dhc1b-2 and compares ciliary regeneration in wild type and bbs4-1. An alignment and a phylogenetic analysis of various PLDs is shown in Fig. S4, A and B, respectively. Fig. S4 C shows the predicted fatty acid modification sites for PLD, AMPK, and CAH6. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207139/DC1.
Supplementary Material
Acknowledgments
We thank Drs. D. Cole, J. Moroney, R. Bloodgood, and J. Rosenbaum for providing antibodies used in this study; Ms. K. Wren for expert help with analyzing fla10-1 cilia; and Dr. J. Leszyk for protein mass spectrometry. We are grateful to Dr. G. Pazour for help with experiments that ultimately could not be included here.
This research was supported by the National Institutes of Health (grant GM030626 to G. Witman), the Grousbeck Family Foundation (to G. Witman), and the Robert W. Booth Endowment (to G. Witman).
Footnotes
Abbreviations used in this paper:
- AMPK
- AMP-regulated protein kinase
- BBS
- Bardet-Biedl syndrome
- CAH6
- carbonic anhydrase 6
- DIC
- differential interference contrast
- GPCR
- G protein–coupled receptor
- IFT
- intraflagellar transport
- MCHR1
- melanin-concentrating hormone receptor 1
- PA
- phosphatidic acid
- PE
- phosphatidylethanolamine
- PLD
- phospholipase D
- SSTR3
- somatostatin receptor type 3
References
- Beales P.L. 2005. Lifting the lid on Pandora’s box: the Bardet-Biedl syndrome. Curr. Opin. Genet. Dev. 15:315–323. 10.1016/j.gde.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Berbari N.F., Johnson A.D., Lewis J.S., Askwith C.C., Mykytyn K.. 2008a. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 19:1540–1547. 10.1091/mbc.E07-09-0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N.F., Lewis J.S., Bishop G.A., Askwith C.C., Mykytyn K.. 2008b. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA. 105:4242–4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W.. 2002. An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 153:401–412. 10.1078/14344610260450136 [DOI] [PubMed] [Google Scholar]
- Blacque O.E., Leroux M.R.. 2006. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell. Mol. Life Sci. 63:2145–2161. 10.1007/s00018-006-6180-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Reardon M.J., Li C., McCarthy J., Mahjoub M.R., Ansley S.J., Badano J.L., Mah A.K., Beales P.L., Davidson W.S., et al. 2004. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 18:1630–1642. 10.1101/gad.1194004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Li C., Inglis P.N., Esmail M.A., Ou G., Mah A.K., Baillie D.L., Scholey J.M., Leroux M.R.. 2006. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell. 17:5053–5062. 10.1091/mbc.E06-06-0571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood R.A., Salomonsky N.L.. 1994. The transmembrane signaling pathway involved in directed movements of Chlamydomonas flagellar membrane glycoproteins involves the dephosphorylation of a 60-kD phosphoprotein that binds to the major flagellar membrane glycoprotein. J. Cell Biol. 127:803–811. 10.1083/jcb.127.3.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.M., Dipetrillo C.G., Smith E.F., Witman G.B.. 2012. A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell Sci. 125:3904–3913. 10.1242/jcs.107151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Snell W.J.. 2009. SnapShot: Intraflagellar transport. Cell. 137:784–784.e1. 10.1016/j.cell.2009.04.053 [DOI] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II–dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domire J.S., Green J.A., Lee K.G., Johnson A.D., Askwith C.C., Mykytyn K.. 2011. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. 68:2951–2960. 10.1007/s00018-010-0603-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing C.S., Duchoslav E., Sampaio J., Simons K., Bonner R., Thiele C., Ekroos K., Shevchenko A.. 2006. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 78:6202–6214. 10.1021/ac060545x [DOI] [PubMed] [Google Scholar]
- Firestone A.J., Weinger J.S., Maldonado M., Barlan K., Langston L.D., O’Donnell M., Gelfand V.I., Kapoor T.M., Chen J.K.. 2012. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 484:125–129. 10.1038/nature10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D., de Montaigu A., Galván A., Fernández E.. 2005. Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal. Biochem. 340:330–335. 10.1016/j.ab.2005.01.031 [DOI] [PubMed] [Google Scholar]
- Gorman D.S., Levine R.P.. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA. 54:1665–1669. 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Pazour G.J., Witman G.B.. 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 15:4382–4394. 10.1091/mbc.E04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Diener D.R., Mitchell A., Pazour G.J., Witman G.B., Rosenbaum J.L.. 2007. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 179:501–514. 10.1083/jcb.200704069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Gao Q., Peng X., Choi S.Y., Sarma K., Ren H., Morris A.J., Frohman M.A.. 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell. 20:376–387. 10.1016/j.devcel.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunnicutt G.R., Snell W.J.. 1991. Rapid and slow mechanisms for loss of cell adhesiveness during fertilization in Chlamydomonas. Dev. Biol. 147:216–224. 10.1016/S0012-1606(05)80019-3 [DOI] [PubMed] [Google Scholar]
- Iomini C., Li L., Esparza J.M., Dutcher S.K.. 2009. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics. 183:885–896. 10.1534/genetics.109.101915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Nachury M.V.. 2009. The BBSome. Curr. Biol. 19:R472–R473. 10.1016/j.cub.2009.04.015 [DOI] [PubMed] [Google Scholar]
- Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., Nachury M.V.. 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 141:1208–1219. 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee H.L., Dishinger J.F., Blasius T.L., Liu C.J., Margolis B., Verhey K.J.. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14:431–437. 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kato M., Beachy P.A.. 2009. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA. 106:21666–21671. 10.1073/pnas.0912180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.M., Witman G.B.. 1990. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J. Biol. Chem. 265:19807–19811. [PubMed] [Google Scholar]
- Klasen K., Corey E.A., Kuck F., Wetzel C.H., Hatt H., Ache B.W.. 2010. Odorant-stimulated phosphoinositide signaling in mammalian olfactory receptor neurons. Cell. Signal. 22:150–157. 10.1016/j.cellsig.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L.. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 90:5519–5523. 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Beech P.L., Rosenbaum J.L.. 1995. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131:1517–1527. 10.1083/jcb.131.6.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., Witman G.B.. 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187:1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P.A. 1995. Flagellar amputation and regeneration in Chlamydomonas. Methods Cell Biol. 47:3–7. 10.1016/S0091-679X(08)60782-7 [DOI] [PubMed] [Google Scholar]
- Levental I., Grzybek M., Simons K.. 2010. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 49:6305–6316. 10.1021/bi100882y [DOI] [PubMed] [Google Scholar]
- Meijer H.J., ter Riet B., van Himbergen J.A., Musgrave A., Munnik T.. 2002. KCl activates phospholipase D at two different concentration ranges: distinguishing between hyperosmotic stress and membrane depolarization. Plant J. 31:51–59. 10.1046/j.1365-313X.2002.01336.x [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S.X., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., Jackson P.K.. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24:2180–2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Meijer H.J., Ter Riet B., Hirt H., Frank W., Bartels D., Musgrave A.. 2000. Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22:147–154. 10.1046/j.1365-313x.2000.00725.x [DOI] [PubMed] [Google Scholar]
- Mykytyn K., Mullins R.F., Andrews M., Chiang A.P., Swiderski R.E., Yang B., Braun T., Casavant T., Stone E.M., Sheffield V.C.. 2004. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. USA. 101:8664–8669. 10.1073/pnas.0402354101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., Jackson P.K.. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Seeley E.S., Jin H.. 2010. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 26:59–87. 10.1146/annurev.cellbio.042308.113337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Tanaka G., Maeda T., Kamiya R., Matsunaga T., Nikaido O.. 1996. Assembly and function of Chlamydomonas flagellar mastigonemes as probed with a monoclonal antibody. J. Cell Sci. 109:57–62. [DOI] [PubMed] [Google Scholar]
- Nishimura D.Y., Fath M., Mullins R.F., Searby C., Andrews M., Davis R., Andorf J.L., Mykytyn K., Swiderski R.E., Yang B., et al. 2004. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. USA. 101:16588–16593. 10.1073/pnas.0405496101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina P.J., Anderson K.V.. 2008. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev. Dyn. 237:2030–2038. 10.1002/dvdy.21551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., Scholey J.M.. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587. 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Witman G.B.. 1999. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144:473–481. 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L.B., Christensen S.T.. 2012. Regulating intraflagellar transport. Nat. Cell Biol. 14:904–906. 10.1038/ncb2569 [DOI] [PubMed] [Google Scholar]
- Pedersen L.B., Rosenbaum J.L.. 2008. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 85:23–61. 10.1016/S0070-2153(08)00802-8 [DOI] [PubMed] [Google Scholar]
- Piperno G., Siuda E., Henderson S., Segil M., Vaananen H., Sassaroli M.. 1998. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143:1591–1601. 10.1083/jcb.143.6.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L.. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L., Witman G.B.. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Seo S., Baye L.M., Schulz N.P., Beck J.S., Zhang Q., Slusarski D.C., Sheffield V.C.. 2010. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl. Acad. Sci. USA. 107:1488–1493. 10.1073/pnas.0910268107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., Sheffield V.C.. 2011. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 7:e1002358. 10.1371/journal.pgen.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.S., Farmen S.L., Moninger T.O., Businga T.R., Andrews M.P., Bugge K., Searby C.C., Nishimura D., Brogden K.A., Kline J.N., et al. 2008. Loss of Bardet-Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc. Natl. Acad. Sci. USA. 105:3380–3385. 10.1073/pnas.0712327105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield V.C. 2010. The blind leading the obese: the molecular pathophysiology of a human obesity syndrome. Trans. Am. Clin. Climatol. Assoc. 121:172–181, discussion :181–182. [PMC free article] [PubMed] [Google Scholar]
- Tadenev A.L., Kulaga H.M., May-Simera H.L., Kelley M.W., Katsanis N., Reed R.R.. 2011. Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc. Natl. Acad. Sci. USA. 108:10320–10325. 10.1073/pnas.1016531108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Chuma S., Yamamoto Y., Kuramochi-Miyagawa S., Totoki Y., Toyoda A., Hoki Y., Fujiyama A., Shibata T., Sado T., et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell. 20:364–375. 10.1016/j.devcel.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Zhang Y., Li Y., Zhang Q., Ling K., Hu J.. 2012. The BBSome controls IFT assembly and turnaround in cilia. Nat. Cell Biol. 14:950–957. 10.1038/ncb2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G.B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280–290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Witman G.B. 1993. Chlamydomonas phototaxis. Trends Cell Biol. 3:403–408. 10.1016/0962-8924(93)90091-E [DOI] [PubMed] [Google Scholar]
- Witman G.B. 2012. Dynein and intraflagellar transport. In Dyneins: Structure, Biology and Disease. King S.M., editor. Elsevier, New York. 395–421. [Google Scholar]
- Zaghloul N.A., Katsanis N.. 2009. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J. Clin. Invest. 119:428–437. 10.1172/JCI37041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Nishimura D., Seo S., Vogel T., Morgan D.A., Searby C., Bugge K., Stone E.M., Rahmouni K., Sheffield V.C.. 2011. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc. Natl. Acad. Sci. USA. 108:20678–20683. 10.1073/pnas.1113220108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Seo S., Bugge K., Stone E.M., Sheffield V.C.. 2012. BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum. Mol. Genet. 21:1945–1953. 10.1093/hmg/dds004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.