Fluorescent lifetime imaging of an ER-tuned redox-responsive probe revealed an unanticipated stability of ER thiol redox to fluctuations in unfolded protein load, in contrast with sensitivity to lumenal calcium.
Abstract
Interfering with disulfide bond formation impedes protein folding and promotes endoplasmic reticulum (ER) stress. Due to limitations in measurement techniques, the relationships of altered thiol redox and ER stress have been difficult to assess. We report that fluorescent lifetime measurements circumvented the crippling dimness of an ER-tuned fluorescent redox-responsive probe (roGFPiE), faithfully tracking the activity of the major ER-localized protein disulfide isomerase, PDI. In vivo lifetime imaging by time-correlated single-photon counting (TCSPC) recorded subtle changes in ER redox poise induced by exposure of mammalian cells to a reducing environment but revealed an unanticipated stability of redox to fluctuations in unfolded protein load. By contrast, TCSPC of roGFPiE uncovered a hitherto unsuspected reductive shift in the mammalian ER upon loss of luminal calcium, whether induced by pharmacological inhibition of calcium reuptake into the ER or by physiological activation of release channels. These findings recommend fluorescent lifetime imaging as a sensitive method to track ER redox homeostasis in mammalian cells.
Introduction
Whereas the position of disulfides in folded proteins is ruled largely by thermodynamic considerations (Anfinsen, 1973), the rate at which disulfides form is influenced by the concentration and activity of reduced and oxidized protein thiol oxidoreductases in the ER lumen. The mammalian ER contains at least 20 such protein disulfide isomerase (PDI)–like oxidoreductases, which have in common the presence of one or more thioredoxin domains through which they accept and donate disulfides to contingent proteins (Ellgaard and Ruddock, 2005; Hatahet and Ruddock, 2009). Several factors affect the redox poise of this heterogeneous collection of oxidoreductases: Upstream are enzymes that function as ER oxidases; they accept electrons from the reduced oxidoreductases and transfer them to small molecule acceptors. ERO1, conserved from yeast to mammals, oxidizes PDI family members by reducing molecular oxygen to hydrogen peroxide (Tu et al., 2000; Gross et al., 2006) and the ER localized peroxiredoxin 4 (PRDX4) serves a backup, oxidizing PDIs by reducing hydrogen peroxide to water (Tavender et al., 2008; Zito et al., 2010b). Other enzymatic (Kodali and Thorpe, 2010; Nguyen et al., 2011) and nonenzymatic processes (Ruddock, 2012) may also contribute to disulfide bond formation in the ER lumen. The aforementioned oxidizing features of the ER are opposed by the ingress of reduced cysteine residues in newly synthesized secreted proteins and by the pool of reduced glutathione (Hwang et al., 1992; Cuozzo and Kaiser, 1999; Chakravarthi et al., 2006).
ER oxidation is vigorously defended. The genes encoding ERO1 and several PDIs are under positive transcriptional control by the unfolded protein response (Frand and Kaiser, 1998; Pagani et al., 2000; Travers et al., 2000) and allosteric mechanisms are in place to regulate the enzymes (Sevier et al., 2007; Appenzeller-Herzog et al., 2008; Baker et al., 2008). These regulatory features likely account for the recovery of disulfides within minutes of reversing a reductive pulse (Braakman et al., 1992; Appenzeller-Herzog et al., 2010). Mice tolerate severe loss-of-function mutations to the two genes encoding ERO1 isoforms, Ero1L and Ero1Lb (Zito et al., 2010a; Chin et al., 2011), further attesting to the redundancy of pathways for disulfide bond formation. Against this background of robustness are studies reporting strong deviation of redox poise under conditions of unfolded protein stress in the ER. Pharmacological and genetic manipulations that adversely affect protein folding homeostasis in the yeast ER are reported to promote a more reducing environment (Merksamer et al., 2008), whereas in mammals, enforced expression of a difficult to fold secreted protein leads to hyperoxidation, which contributes to failure of secretion (Malhotra et al., 2008).
These studies suggest that deviation from healthy thiol redox poise may contribute to impaired protein folding in the overloaded ER, but offer contradictory indications as to the direction of the deviation. Resolving this conundrum requires tools to accurately measure ER redox poise; a nontrivial problem. It is possible to detect changes in ER redox poise by following the distribution of disulfides and dithiols in sentinel ER resident proteins. Bulk biochemical methods to track the redox status of individual ER proteins exist, but these end-point assays are subject to post-lysis experimental artifacts, lack spatial and temporal resolution, and provide limited information on intra-sample variation. These limitations have stimulated the development of redox-sensitive intravital optical probes that function in the ER lumen. A particularly promising approach has been to exploit the optical perturbation introduced by the presence of a disulfide on the surface of the green fluorescent protein β-barrel (Björnberg et al., 2006; Cannon and Remington, 2008; Meyer and Dick, 2010). Introduction of cysteine residues on the surface of such redox-sensitive GFP (roGFP) results in a fluorescent protein whose excitation properties differ between the dithiol and the disulfide state: The ratio of emitted fluorescence when excited at 390 nm versus 475 nm reports on the ratio of disulfide versus dithiol in the population of probe molecules (Hanson et al., 2004).
The first generation of roGFPs, although useful in tracking redox in the cytoplasm and mitochondrial matrix, proved too reducing for the oxidizing environment of the ER. This challenge was ingeniously met by introducing a single amino acid insertion into the roGFP β-barrel adjacent to cysteine 147, in what were named “roGFP insertion X” (roGFPiX, where X is the inserted amino acid). Destabilization of the optically perturbing 147–204 disulfide in roGFPiX resulted in a less reducing protein whose midpoint potential was thus predicted suitable to detect fluctuations in redox poise in the oxidizing environment of the ER (Lohman and Remington, 2008). One such probe, roGFPiL, was introduced into the ER lumen of mammalian cells and proved sensitive to changing redox changes (van Lith et al., 2011). Unfortunately, the insertion that tunes roGFPiX redox potential to the oxidizing environment in the ER has a negative impact on fluorescent intensity, compromising sensitivity (Lohman and Remington, 2008). One can partially overcome this problem with higher illumination, but at the risk of photo-toxicity and perturbation of the redox environment (Hoebe et al., 2007; van Lith et al., 2011). We have discovered that the dithiol and disulfide states of roGFPiX variants are associated with vastly different fluorescence lifetimes. Thus, real-time fluorescent lifetime imaging microscopy of a suitable variant, roGFPiE, can circumvent the crippling dimness of the probe. Here we report on the application of this technological advance to analyze ER redox poise under diverse physiological and pathophysiological conditions.
Results
roGFPiE: A protein thiol redox probe suited to the ER
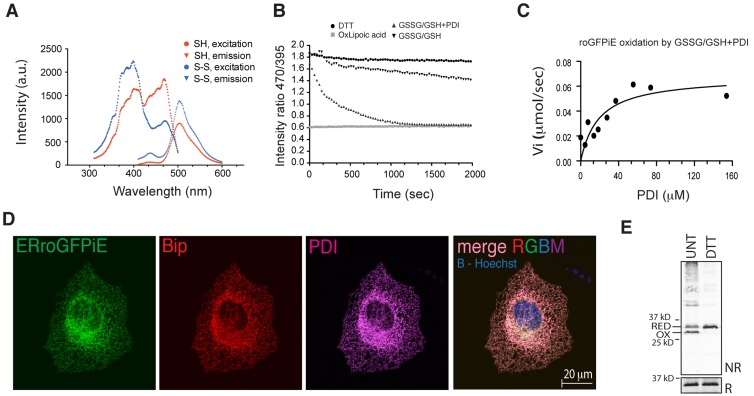
Fluorescent spectroscopy of roGFPiE purified from Escherichia coli showed the expected double peaked excitation spectrum with a prominent peak at 475 nm, after reduction with dithiothreitol (DTT), and its attenuation by oxidation with lipoic acid. The emission spectrum was unaltered by the redox state of the protein (Fig. 1 A). Thus, ratiometric measurements of roGFPiE faithfully reported on the redox status of the probe in vitro, as previously described (Lohman and Remington, 2008).
Figure 1.
Optical and physiological sensitivity of roGFPiE to the ER redox environment. (A) Excitation (measured by the emission at 520 nm) and emission (measured by excitation at 450 nm) scans of purified roGFPiE dithiol (SH) or disulfide (S-S; 1 µM) performed at a spectral resolution of 1 nm. (B) Time-dependent changes in the ratio of fluorescence emission (excitation: 470 nm vs. 395 nm) of the fully reduced purified roGFPiE dithiol (1 µM) introduced at t = 0 into a glutathione redox buffer (5:1 GSH/GSSG, 4 mM total) in the absence or presence of 30 µM PDI. The emission ratio of roGFPiE maintained continuously in the presence of the reducing agent DTT (5 mM) or the oxidizing agent lipoic acid (5 mM) is provided as a reference for the dithiol and disulfide forms, respectively. (C) Initial velocity of the oxidation of roGFPiE from the dithiol to the disulfide in the presence of the indicated concentration of PDI, calculated from the linear phases of measurements as in B. (D) Fluorescent photomicrographs of COS7 cells expressing ERroGFPiE, stained with rabbit polyclonal anti-GFP, mouse monoclonal anti-PDI, and chick polyclonal anti-BiP (as ER markers). The nucleus was visualized by Hoechst staining (in blue in the merged right-most panel). (E) Immunoblot of ERroGFPiE expressed in untreated or DTT-treated (5 mM DTT for 10 min) HEK 293T cells, resolved by nonreducing (NR) and reducing (R) SDS-PAGE. Shown are representative experiments reproduced three times (A–D) or twice (E).
Thiol redox reactions tend to kinetic isolation (Kemp et al., 2008). Therefore, an in vivo thiol redox reporter should be coupled to the prevalent enzymatic system of the organelle. We tested the response of the sensor in the presence of PDI, an abundant chaperone mediating client protein oxidation and reduction in the ER. Ratiometric measurements tracked time-dependent changes in roGFPiE redox status, as trace amounts of the fully reduced protein in vitro were exposed to an excess of a redox buffer of oxidized and reduced glutathione. Alone, roGFPiE equilibrated slowly with the glutathione redox buffer, reflecting kinetic isolation, however PDI markedly accelerated probe oxidation (Fig. 1 B) with half maximal acceleration attained at ∼20 µM PDI (Fig. 1 C).
The tight coupling of roGFPiE to changes in PDI redox status justified its use as an in vivo probe. When targeted to the ER of mammalian cells, ERroGFPiE colocalized with the ER chaperone BiP and the disulfide isomerase PDI (Fig. 1 D). COS7 cells, with superior spatial resolution, were used to assess colocalization, but similar observations were made in HEK 293T cells (see Fig. 3), which were used in subsequent experiments. Non-reducing immunoblot confirmed the presence of a mixed population of reduced and oxidized ERroGFPiE, which were completely reduced in cells exposed to DTT (Fig. 1 E).
Figure 3.
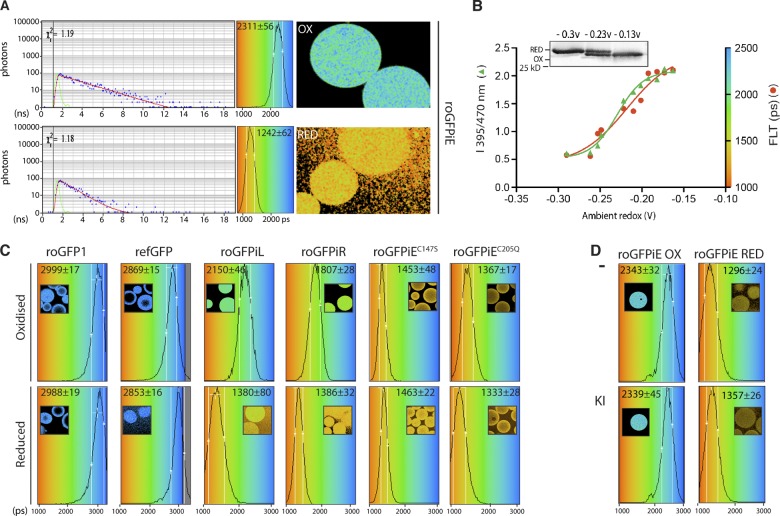
Redox-dependent changes to fluorescence lifetime of ER-localized roGFPiE in live cells. (A) Fluorescent intensity–based (in grayscale) and lifetime images (color-coded to the scale of the histogram on the right) of HEK 293T cells expressing wild-type ERroGFPiE (C147-E147a-C204) and its ER-localized mutant derivatives, with or without 2 mM DTT (10 min). A histogram of the distribution of lifetimes in the population of cells is provided (right), noting the mean ± SD lifetime. The asterisk in panel 6 points to a region of intense autofluorescence with similar spectral properties as roGFPiE, but recognizable by its relatively long fluorescence lifetime. (B) Nonreducing (NR) and reducing (R) immunoblot of the GFP in the cells shown in A. The dithiol (RED) and disulfide (OX) forms are indicated. (C) Schema showing the mean florescence lifetime of the indicated GFP proteins in vivo and in vitro, as measured in the experiments shown in Fig, 2, A and C; panel A here; and in Fig. S2 B. roGFPiE and ERroGFPiE are in bold font. Redox state is indicated by S-S (disulfide) and SH (dithiol). Treatments with DTT, the oxidizing agent 2,2′-dipyridyl disulfide (DPS), or lipoic acid (LA) are noted. A and B are representative experiments reproduced twice.
Despite these favorable features, in vivo ERroGFPiE proved unreliable as a ratiometric ER redox probe: Whereas the excitation spectrum of Sepharose beads coated with roGFPiE tracked its redox status in vitro (Fig. S1 A), nonspecific autofluorescence competed with the probe’s feeble signal in vivo, severely compromising the detection of changes in ER redox in cells using the same microscope setup (Fig. S1, B–D). This motivated the development of an alternative readout that could be tracked more readily in vivo.
Fluorescence lifetime imaging detects changes in ER protein thiol redox
The lifetime of the compromising autofluorescence (3.0 ns) was found to be significantly longer than that reported for GFP (2.4 ns; Esposito et al., 2005; Wallrabe and Periasamy, 2005) and was unresponsive to DTT (Fig. S2 A). Therefore, we hypothesized that this parameter may provide a basis for deconvoluting the specific signal.
Proximity of the C147/C204 disulfide to the modified 2,3-didehydrotyrosine residue of roGFPiE (PDB: 3CD1) suggested that it might influence the lifetime of the fluorophore. Indeed, fluorescence lifetimes of roGFPiE varied from a low of 1242 ± 62 picoseconds, for the fully reduced probe, to a high of 2311 ± 56 ps for the fully oxidized form (Fig. 2 A). Fluorescence lifetimes correlated with changes in excitation ratio in a titration with reduced and oxidized lipoic acid (Fig. 2 B).
Figure 2.
Fluorescence lifetime of roGFPiE is altered by its oxidation state in vitro. (A) Color-coded fluorescence lifetime images acquired by time-correlated single-photon counting (TCSPC) spectroscopy of Sepharose beads coated with roGFPiE purified from E. coli and tethered via an antibody (right). Where indicated, the beads were exposed to an oxidizing environment of lipoic acid (OX) or a reducing environment of DTT (RED). A time-correlated photon count, from a representative pixel (after a laser excitation pulse, blue dots) and its fit to a mono-exponential decay (red line, from which the value of lifetime is extracted) are shown (left), alongside the instrument response function (green line). reports on goodness of fit. A histogram of the distribution of lifetimes observed in the pixels sampled is shown in the central panel superimposed on the continuous rainbow scale representing fluorescence lifetime values of 1,000–3,000 ps, which is also used to color code the adjacent FLIM images. The mean ± SD lifetime of each type of measurement is noted. Reduction of disulfides in the tethering immunoglobulin triggers dissociation of probe molecules from the bead, accounting for the halo effect observed in the DTT-treated (RED) samples. (B) Ratio of fluorescence emission (excitation 470 nm vs. 395 nm, left axes, green trace) and fluorescence lifetime (in picoseconds, right axes, red trace) of roGFPiE equilibrated with a lipoic acid–based redox buffer of the indicated predicted redox potential (in volts). The inset is an immunoblot of roGFPiE resolved on a nonreducing SDS-PAGE from samples at the indicated redox potential. (C) Histogram of the distribution of lifetimes of oxidized and reduced GFP proteins attached to Sepharose beads. A color-coded lifetime image is shown in the inset (as in A). The mean ± SD of the fluorescence lifetime is also indicated. (D) Histogram of distribution of lifetimes of samples as in C. Where indicated, the buffer contained the collisional quencher potassium iodide (KI, 100 mM). Shown are representative experiments reproduced three times (A) or twice (B–D).
In its oxidized form, the fluorescence lifetime of roGFPiE approached that of conventional EGFP and refGFP (a thiol-lacking revertant mutant roGFP; Fig. 2 C). Together these observations suggested that the fluorophore was perturbed in the reduced state of roGFPiE. Interestingly, the fluorescence lifetimes of roGFP1, the C147/C204-containing parent of roGFPiE that lacks the E147a insertion, was long in both its reduced and oxidized configuration (Fig. 2 C), indicating that the perturbation promoted by reduction of the disulfide requires the insertion at 147a. The identity of the inserted residue seemed less important, as reduced roGFPiE, roGFPiL, and roGFPiR had similar short fluorescence lifetimes. However, in their oxidized configuration roGFPiR and roGFPiL had shorter fluorescence lifetimes than roGFPiE (1807 ± 28 and 2150 ± 46 vs. 2311 ± 56 ps, respectively), endowing the latter with a superior dynamic range (Fig. 2, A and C). Fluorescence lifetime of roGFPiE was unaffected by the presence of iodine (Fig. 2 D), suggesting that the lifetime alterations in roGFPiX variants does not result from exposure of the fluorophore to collisional quenching and predicting a measure of indifference to the ionic environment.
Given that roGFPiE had the greatest dynamic range of fluorescence lifetime in vitro, it was chosen for further in vivo studies. Fluorescence lifetime imaging microscopy (FLIM) of mammalian cells expressing ERroGFPiE showed a conspicuous difference in lifetime before and after exposure to DTT (Fig. 3, A and B). Reduced roGFPiE had similar lifetimes in vitro and in vivo (Fig. 3 C). In unperturbed cells, the lifetime of ERroGFPiE corresponded to that of roGFPiE at equilibrium with a redox buffer just above its midpoint (−0.236 V; Figs. 2 B and 3 C; Lohman and Remington, 2008), an assignment consistent with the distribution of reduced and oxidized probe observed in immunoblot of cell lysates (Fig. 1 E).
As expected, the revertant mutant, ERrefGFP, maintained its long fluorescence lifetime in cells exposed to reducing stress. ERroGFPiE with a C204Q mutation (precluding disulfide formation) had a persistently short fluorescence lifetime. Though unable to form a disulfide, the C147S/ΔE147a double mutant lacking the insertion had persistent long lifetime (Fig. 3 A, samples 7 and 8). Whether expressed in the oxidizing ER or reducing cytosol, conventional EGFP had an indistinguishable long lifetime, as expected (Fig. S2 A). The autofluorescent signal of untransfected cells had a patchy distribution and although foci of high intensity autofluorescence were noted, pixels containing such signal could be rejected based on their unusually long lifetime and lack of response to DTT (Fig. S2 A). Together, these observations confirmed that roGFPiE FLIM tracked protein thiol redox in the ER of living cells and could be unmixed from polluting background autofluorescence.
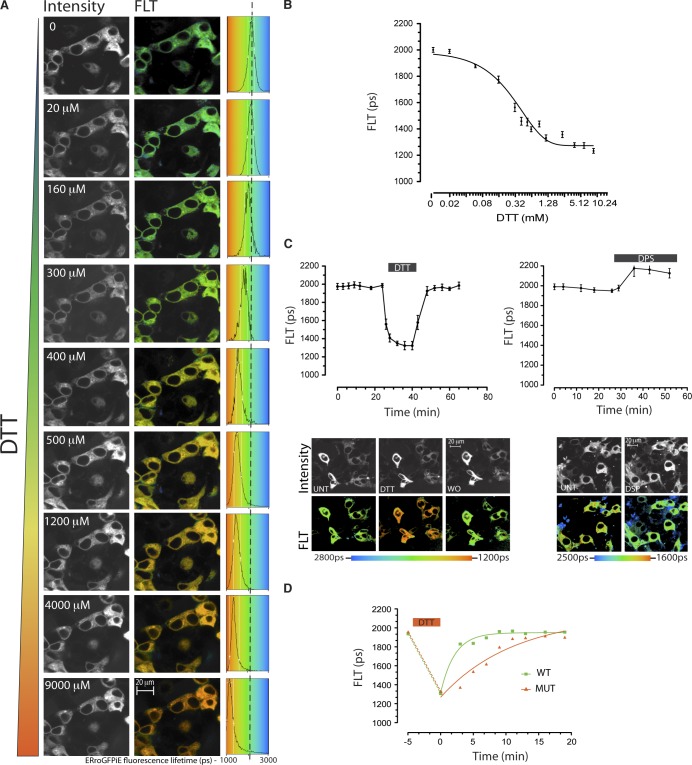
Despite the dimness of the ERroGFPiE signal, fluorescence lifetime imaging had the sensitivity and time resolution needed to detect subtle and rapid changes in ER redox poise in wild-type cells exposed to DTT or to the oxidizing agent 2,2′-dipyridyl disulfide (DPS; Fig. 4, A–C). Unlike the ratiometric method used to detect redox changes in roGFPiL in vivo, which suffered from the confounding effects of a creeping baseline (van Lith et al., 2011), a steady baseline of fluorescence lifetime was observed in cells expressing ERroGFPiE (Fig. 4 C). FLIM of roGFPiE also readily revealed a defect in disulfide formation in cells genetically deficient in ER disulfide oxidases, which readily defend ER thiol redox at baseline but exhibit a subtle kinetic delay in disulfide formation after recovery from a pulse of DTT (Fig. 4 D; Zito et al., 2012).
Figure 4.
Faithful tracking of dynamic changes in ER redox by ERroGFPiE. (A) Fluorescent intensity–based (in grayscale) and lifetime images (color-coded to the scale of the histogram on the right) of HEK 293T cells expressing ERroGFPiE exposed sequentially at 10-min intervals to escalating concentrations of the reducing agent DTT (concentration noted in the inset). A histogram of the distribution of lifetimes in the population of cells is provided (right), noting the mean ± SD lifetime. (B) Plot of the relationship between DTT concentration and probe lifetime derived from the experiment described in A. (C) A trace of time-dependent changes in ERroGFPiE fluorescence lifetime (FLT in picoseconds) in live HEK 293T cells, before, during, and after brief exposure to 2 mM DTT or 1 mM of the oxidizing agent DPS. Fluorescent emission intensity (in black and white) and lifetime (color-coded) images of cells before (UNT), during the DTT pulse, and after its washout (WO) are shown in the bottom panel. (D) As in C, comparison of lifetime changes after a reductive pulse of DTT applied to wild-type (WT) and compound Ero1l-, Ero1lB-, and Prdx4-deficient mutant (MUT) mouse embryonic fibroblasts. Shown are representative experiments reproduced twice.
Stability of ER protein thiol redox poise in the face of changing levels of unfolded protein stress
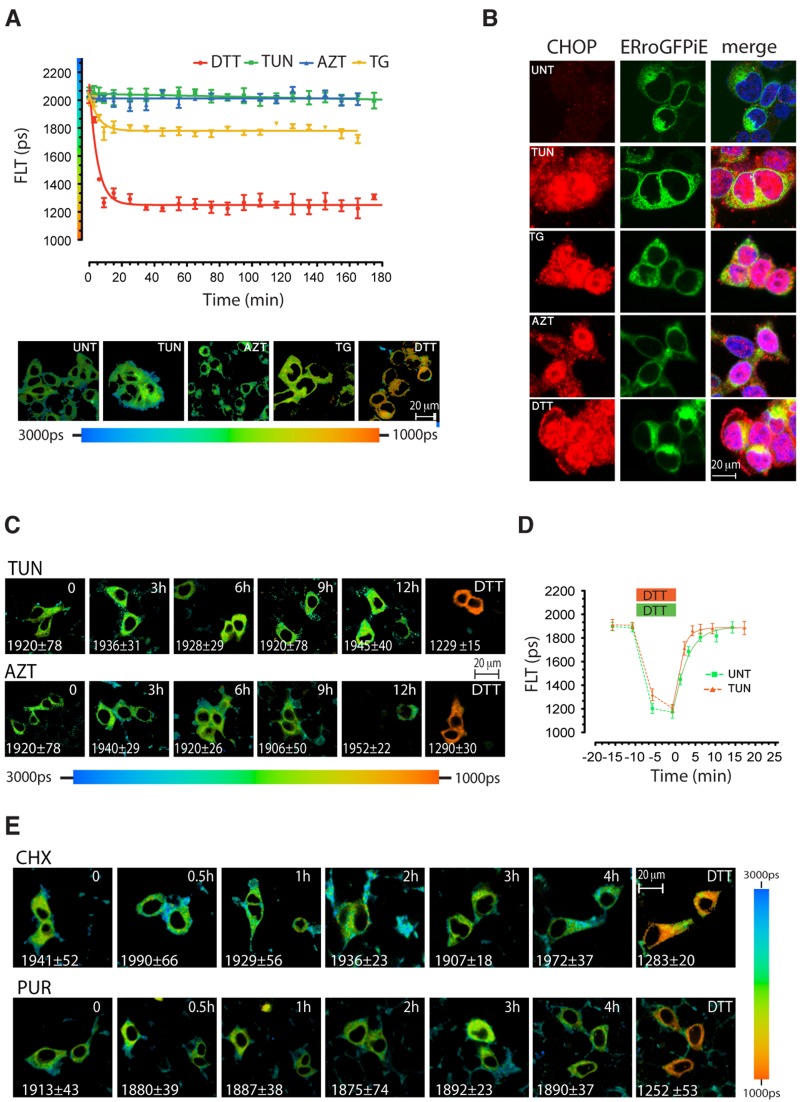
Diverse pathophysiological processes perturb protein folding homeostasis in the ER lumen (Walter and Ron, 2011). However, the effect of the resulting stress on ER thiol redox remains poorly characterized. In budding yeast, an ER-localized roGFP2 had been reported to undergo reduction on exposure to tunicamycin, an inhibitor of N-linked glycosylation that causes unfolded protein stress (Merksamer et al., 2008). However, given the reducing midpoint potential of roGFP2, −0.272 V (Hanson et al., 2004), it is suited to detect catastrophic deviations in redox state, but cannot track physiological variations around the organelles’ midpoint potential. Therefore, ERroGFPiE-expressing 293T cells were exposed to conditions that promote unfolded protein stress in the ER: azetidine (a proline analogue that perturbs protein structure), thapsigargin (an inhibitor of the smooth endoplasmic reticulum calcium [SERCA] uptake pump that inhibits protein folding by depleting ER calcium stores), DTT, or tunicamycin. 3-h exposure to tunicamycin or azetidine had no measureable effect on ERroGFPiE (Fig. 5 A), despite strong induction of ER stress, reflected in the induction and nuclear localization of the ER unfolded protein responsive marker CHOP (Fig. 5 B). ERroGFPiE remained unresponsive for up to 12 h (Fig. 5 C), a point at which death became noticeable in the stressed cells.
Figure 5.
Fluorescence lifetime of ERroGFPiE is unaffected by physiological levels of unfolded protein stress. (A) A trace of time-dependent changes in ERroGFPiE fluorescence lifetime in live HEK 293T cells after exposure to 2 mM DTT, 5 µg/ml tunicamycin (TUN), 3 mM azetidine (AZT), or 2 µM thapsigargin (TG). Color-coded fluorescence lifetime images of cells 3 h into the exposure are shown in the bottom panel. Each data point represents the mean ± SD of fluorescence lifetime measured in ≥10 cells. (B) Fluorescent photomicrographs of cells as in A, immunostained for the ER stress–induced nuclear protein CHOP (left), GFP (middle), and a superposition of the two images with Hoechst nuclei staining (right). (C) Extended time course of color-coded fluorescence lifetime images of ERroGFPiE expressing live HEK 293T cells exposed to tunicamycin or azetidine. (D) A trace of time-dependent changes in ERroGFPiE fluorescence lifetime in untreated (green) and 3-h tunicamycin-treated HEK 293T cells (orange), before, during, and after brief exposure to DTT. (E) Extended time course of color-coded fluorescence lifetime images of ERroGFPiE expressing live HEK 293T cells exposed to 50 µg/ml of the protein synthesis inhibitors cycloheximide (CHX) or 10 µg/ml of puromycin (PUR). Shown are representative experiments reproduced twice.
These observations indicate that prolonged unfolded protein stress does not affect the steady-state thiol redox poise of the ER. To determine if prolonged unfolded protein stress affects the dynamics of disulfide formation, we compared the recovery of oxidized ERroGFPiE after a reductive pulse of DTT in untreated and 3-h tunicamycin-pretreated cells. Both populations rapidly recover their steady-state oxidative poise, a process that was mildly accelerated by tunicamycin pretreatment (Fig. 5 D), presumably due to ERO1 induction. Lowering the burden of unfolded proteins by treating cells with protein synthesis inhibitors was likewise with no measurable effect on ER redox. Neither cycloheximide, which arrests protein synthesis during elongation, freezing nascent chains in the translocon, nor puromycin, which releases the nascent chains from the translocon, affected roGFPiE redox (Fig. 5 E).
Next we turned our attention to the role of the unfolded protein response in maintaining the stability of ER redox. ER redox was defended equally well in mutant mouse cells lacking all IRE1 activity and in mutant cells rescued with an IRE1 transgene (Fig. 6 A; Calfon et al., 2002). Unfortunately, spectral overlap with roGFPiE precluded the use of IRE1 inhibitors (which fluoresce strongly in that range) and confined our analysis to the consequences of long-term disruption of the IRE1 pathway. However, as in mammals the IRE1 pathway plays a crucial role in the long-term process of building ER capacity (Cross et al., 2012); this comparison is likely relevant to any role IRE1 might have in maintaining redox equilibrium.
Figure 6.
Failure of ER redox regulation at extreme unfolded protein stress. (A) Color-coded fluorescence lifetime images of ΔIre1α-deleted mouse cells and deleted cells rescued by an IRE1α transgene (IRE1+) after 4 h exposure to 5 µg/ml tunicamycin or 2 mM DTT. Values represent mean ± SD of fluorescence lifetime measured in ≥20 cells. (B) A trace of time-dependent changes in ERroGFPiE fluorescence lifetime in live COS7 cells that had been exposed to 5 µg/ml tunicamycin (TUN), 10 µM of the selective PERK kinase inhibitor GSK2606414 (PERKi), or both agents. Each data point represents the mean ± SD of fluorescence lifetime measured in ≥10 cells. (C) Color-coded fluorescence lifetime images along with histograms of lifetime frequency distribution of ERroGFPiE-expressing untreated COS7 cells and cells 3 h into the exposure to tunicamycin and PERK inhibitor. Shown are representative experiments reproduced twice.
PERK-mediated translational repression regulates the moment-to-moment flux of proteins into the ER and is thus best studied by acute pharmacological inhibition, which precludes long-term adaptation in cultured cells (Harding et al., 2012). Neither PERK inhibition with the selective kinase inhibitor GSK2606414 (Axten et al., 2012; Harding et al., 2012) nor tunicamycin affected ERroGFPiE’s redox poise over 15 h of observation (Fig. 6 B). To promote sensitivity to spatial inhomogeneity in redox poise, were it to occur, these measurements were conducted in COS7, with superior spatial resolution. Redox poise was defended throughout the ER in PERK inhibitor–treated cells (Fig. 6 C). Translational control is especially important under conditions of ER stress and PERK deficiency strongly sensitizes cells to tunicamycin (Harding et al., 2000). Interestingly, tunicamycin-treated cells experienced defective ER redox when the PERK pathway was simultaneously compromised (Fig. 6 B, yellow trace; and Fig. 6 C, bottom-most panels).
Calcium store depletion promotes a more reducing ER
The aforementioned observations suggested that ER redox is defended across a broad range of levels of unfolded protein load and breaks down only when the defensive mechanism of translational repression is acutely disabled. It was of interest therefore to note that within minutes of exposure to thapsigargin, ERroGFPiE fluorescence lifetime decreased to a new baseline, corresponding to an apparent reductive shift of 24 mV from −0.209 V to −0.233 V (Fig. 5 A). These observations suggested an unanticipated relationship between perturbed intracellular calcium homeostasis and ER protein thiol redox.
Thapsigargin irreversibly inhibits the SERCA pump; in keeping with this, depletion of ER calcium and the shortening of ERroGFPiE fluorescence lifetime persisted upon thapsigargin washout (Fig. S3, A and B). HEK 293T cells exhibit a wide range of sensitivity to the ER calcium store–depleting effects of thapsigargin, necessitating the use of relatively high concentrations of this SERCA inhibitor (Fig. S3, C and D). To determine if the correlation between depletion of lumenal calcium and a more reducing ER extended to reversible, physiological calcium signaling, we exploited the responsiveness of pancreatic acinar AR42j cells to the gut hormone cholecystokinin (Zhao et al., 1990). As expected, cholecystokinin reversibly depleted lumenal calcium stores, as monitored by the fluorescence lifetime of the FRET-based probe D1ER cameleon (Fig. 7, A and B, left; Palmer et al., 2004). The temporal profile of ER calcium store depletion correlated well with the cholecystokinin-mediated shortening of ERroGFPiE fluorescence lifetime and with its restoration upon washout (Fig. 7, A and B, right). By contrast, ER calcium stores were notably preserved in cells experiencing ER stress and only began to decline when viability was compromised at late time points (Fig. S4), suggesting that stability of ER calcium may contribute to the stability of roGFPiE in stressed cells.
Figure 7.
Calcium depletion promotes a more reducing ER. (A) Time-dependent changes in fluorescence lifetime (FLT) of the redox reporter ERroGFPiE or the ER calcium reporter D1ER cameleon before, during, and after exposure of AR42j cells to a pulse of cholecystokinin (CCK, 2 µM). Shown are temporally superimposed typical measurements reproduced three times in cells expressing either reporter. (B) Fluorescent emission intensity (grayscale) and lifetime (color-coded) images of ERroGFPiE or D1ER cameleon in AR42j cells before, at the peak of CCK action, and after washout of CCK (as in A). (C) Traces of time-dependent changes in cytosolic calcium concentration measured by Indo 1-AM emission ratios before and after exposure to thapsigargin (2 µM). Where indicated, the thapsigargin-induced cytosolic calcium spike was buffered by 50 µM BAPTA-AM. (D) Bar diagram showing mean ± SD (n > 5) fluorescence lifetime of the ER-localized calcium probe, D1ER cameleon, or the redox probe, ERroGFPiE, in cells exposed to thapsigargin, BAPTA-AM, or both (as in C; note that D1ER cameleon’s lifetime is inversely related to ER calcium). (E) A time series of fluorescence lifetime measurements of roGFPiE dithiol (SH), disulfide (S-S), or a mixture of the two that approximates the redox state of ERroGFPiE in untreated cells (S-S/SH in vivo like) exposed in vitro to a solution whose calcium concentration was increased at regular intervals by addition of calcium (gray trace). Note that calcium concentration does not affect fluorescence lifetime of roGFPiE in vitro. Shown are representative experiments reproduced twice.
Thapsigargin has reciprocal effects on lumenal ER calcium (decrease) and cytosolic calcium (increase; Brayden et al., 1989). To deconvolute their influence on fluorescence lifetimes of ERroGFPiE, we buffered cytosolic calcium with the cell-permeant calcium chelator BAPTA-AM. In the presence of BAPTA-AM, the ratiometric cytosolic calcium probe, INDO1-AM, reported nearly complete buffering of the thapsigargin-induced transient increase of cytosolic calcium (Fig. 7 C). ER calcium was depleted by thapsigargin, as expected (Fig. 7 D, left) but, BAPTA-AM did not reverse thapsigargin’s effect on the redox state of ERroGFPiE (Fig. 7 D, right), implicating changes in lumenal calcium. Importantly, the calcium-dependent changes in ERroGFPiE fluorescence lifetime observed in vivo were not reproduced by exposing reduced or oxidized roGFPiE to different calcium concentrations in vitro, indicating that at neither oxidation state is roGFPiE’s lifetime directly sensitive to calcium (Fig. 7 E).
Discussion
The development of ratiometric optical redox probes suited to the oxidizing environment of the ER was a major conceptual advance (Lohman and Remington, 2008; van Lith et al., 2011). However, the feeble fluorescent signal of these roGFPiX probes compromises their utility in vivo, where normal background autofluorescence overlaps their emission spectrum and confounds the intensity-based ratiometric excitation measurement used to estimate the redox state of the probe. Our findings indicate that the insertion of residue 147a, which destabilizes the disulfide and renders the probe less reducing (and thus suited to the ER environment), fortuitously subordinates the fluorescence lifetime of the fluorophore to the redox state of the adjacent cysteines. The dithiol has a considerably shorter fluorescence lifetime than the disulfide, thus the redox state of the probe can be estimated by measuring its fluorescence lifetime in vivo.
Both ratiometrics and fluorescence lifetime imaging provide information that is independent of probe concentration. Lifetime measurements offer advantages in deconvoluting signals emanating from cellular autofluorescence, whose lifetime differs considerably from roGFPiE (as exemplified by Fig. 3 A, panel 6, and Fig. S2 A) and thus provides a measurement gated on the fluorescent properties of the probe. The large number of events acquired by photon counting provides reliable data at weak excitation intensities. This is revealed by the narrowness of the distribution of lifetimes measured in vivo under diverse physiological conditions and favors the detection of subtle differences in the state of the probe, while avoiding photo-toxicity and photo-oxidation.
The binary mode of TCSPC eliminates nonlinear amplification of weak signals, a problem affecting analogue light-detecting devices commonly used in microscopy. In addition, immunity to saturation of the counting mode used to generate the intensity image and the single light path used throughout the FLIM image acquisition both contribute to its superior accuracy. The practical advantages of lifetime imaging over ratiometric measurements are nicely demonstrated by the contrast between the stable baseline of the probe obtained during repeated lifetime measurement in vivo (Fig. 4 C) and the creeping baseline attributed to photo-oxidation caused by the intense illumination required to generate a signal in intensity-based ratiometric measurements (van Lith et al., 2011).
Ratiometric measurement of fluorescent excitation may be acquired by an assortment of configurations. And although these may produce internally consistent data, they are difficult to standardize. Lifetime measurements, by contrast, yield an absolute value that should be comparable across diverse platforms. Notably, our measurement of EGFP fluorescence lifetime, 2,496 ps, is within 3% of values previously reported (Esposito et al., 2005; Wallrabe and Periasamy, 2005).
The photophysical basis of the dramatic difference in fluorescence lifetime of the disulfide and dithiol forms of roGFPiE remains to be determined. The GFP fluorophore is well protected by the encasing β-barrel (Remington, 2011), and fluorescence of reduced roGFPiE is not quenched by halogens. Thus, it seems unlikely that a structural alteration in the wall of the GFP β-barrel exposes the fluorophore to collisional quenching when the C147/C204 disulfide has been reduced. More likely, the shorter lifetime is intrinsic to the disposition of the fluorophore in relation to other residues of the 147a insertion-containing GFP β-barrel, a perturbation that is reversed by formation of the C147/C204 disulfide.
Autonomy of fluorescence lifetime is an important feature in restricting the responsiveness of roGFPiE to changes in the redox environment. ER redox poise is a convenient heuristic term applied to a nonequilibrium situation; no single sentinel disulfide can serve as a general redox probe for the compartment (Kemp et al., 2008). Nonetheless, the dramatic acceleration by PDI of the equilibration of roGFPiE with a glutathione redox buffer, observed here, suggests that roGFPiE is a good surrogate for clients of PDI. As these include reduced, unfolded, nascent secreted proteins, roGFPiE is likely to report on a physiologically relevant aspect of the redox of PDI substrates.
Applying this refined tool to in vivo measurements in living cells unveiled remarkable stability of the ER redox setpoint in the face of considerable variation in unfolded protein load. Neither tunicamycin nor azetidine, which impede folding and induce high levels of ER stress, affected redox poise. Nor was ER redox measurably affected by shutting off the influx of unfolded proteins into the ER. An extreme imbalance between client protein influx and the capacity of the cellular machinery, imposed by disabling the PERK-mediated translational limb of the unfolded protein response in cells exposed to the glycosylation inhibitor tunicamycin, led to an inappropriately reducing ER. But these extreme circumstances showcase the stability of ER redox under more physiological circumstances.
Our findings seemingly conflict with earlier observations made in yeast, which suggest a failure of disulfide bond formation in ER-stressed yeast (Merksamer et al., 2008). These differences may reflect phylogenetic distance, but it may also be worth noting that the yeast study was conducted with roGFP2, which is tuned to the cytosol’s redox potential and would likely miss subtle perturbations like the ones we measured upon calcium release or combined exposure to tunicamycin and PERK kinase inhibitor. Thus, the shift in roGFP2 redox status noted in ER-stressed yeast more likely reflects a different, dramatic process, such as catastrophic permeabilization of the ER or accumulation of cytosolic roGFP2 due a stress-induced translocation defect in the yeast (Rubio et al., 2011).
In mammalian cells the ER unfolded protein response activates a gene expression program that defends against accumulation of reactive oxygen species and whose compromise exposes ER-stressed cells to accumulation of endogenous peroxides (Harding et al., 2003). But the stability of ER thiol redox observed here suggests that the accompanying global increase in reactive species reflects something other than ER hyperoxidation and that the restoration of secretion of difficult-to-fold proteins by electron donors (antioxidants; Malhotra et al., 2008) is unlikely to involve their direct action at the level of the ER. There are other hints that in mammalian cells redox in the ER and cytosol may be quite isolated. Partial loss of ERO1 function, which lowers levels of endogenous peroxides in ER-stressed worms (Marciniak et al., 2004), had no similar protective effect in mouse pancreatic β cells (Zito et al., 2010a). These considerations should prompt a reexamination of the site of action of the pro-oxidant strands of the ER unfolded protein response, mediated by the transcription factor CHOP (McCullough et al., 2001).
This refined method for tracking ER redox revealed that depletion of ER calcium results in a more reduced ERroGFPiE at steady state. This was observed in both thapsigargin-treated cells and in a more physiological setting of regulated ER calcium release, induced by an agonist of the membrane cholecystokinin receptor. Calcium depletion promptly leads to unfolded protein stress in the ER lumen (Prostko et al., 1992), but a role for ER protein misfolding in the shift of roGFPiE to a more reduced poise is rendered unlikely by the observation that tunicamycin and azetidine, which severely perturb ER protein folding homeostasis, have no such effect.
The underlying molecular mechanism linking changes in ER calcium stores to ER redox remain to be worked out. It is nonetheless tempting to speculate on the physiological significance of the shift to a less oxidizing ER after calcium release. Reduction of a lumenal disulfide involving cysteines 2496 or 2505 by the ER-localized ERp44 attenuates calcium release by the R1 isoform of the inositol phosphate-3 receptor (Higo et al., 2005), and thiol oxidation has been shown to attenuate the reuptake of calcium into the ER via the SERCA pump (Scherer and Deamer, 1986). Conversely, compromise of ERO1 activity limits activity-dependent calcium release in cardiomyocytes (Chin et al., 2011). This dialectic of thiol redox and calcium flux across the ER membrane is consistent with a rectifying feedback mechanism that opposes the consequences of ER calcium depletion on thiol redox poise and a role for the reducing poise elicited by calcium release as a negative feedback loop to conserve lumenal calcium or limit cytosolic calcium signaling.
Materials and methods
Plasmid construction and mutagenesis
The GFP-based probe variants were produced by introducing point mutations into roGFPiE (a gift from S.J. Remington, University of Oregon, Eugene, OR; Lohman and Remington, 2008) in the pQE10 vector for bacterial expressible variants and in ERroGFPiE in pCDNA3.1-His vector (created using roGFPiE-pQE10 as a source) for mammalian ER targeted variants, directed to the ER by a cleavable signal peptide and C-terminal KDEL ER retrieval signal, based on the FLAGM1 tag in the plasmid pFLAG-CMV1 (Sigma-Aldrich).
Transfections, immunoblotting, and immunofluorescence
HEK 293T, COS7, and AR42j cells were obtained from the American Type Culture Collection (Manassas, VA). Ire1α-deleted embryonic mouse cells (ΔIre1) derived from outbred 9.5-d embryos homozygous for a targeted deletion of the exon encoding the transmembrane domain and lacking all IRE1 activity, and their retrovirally rescued counterpart (IRE1+), were described previously (Calfon et al., 2002), as were the outbred mouse embryonic fibroblasts derived from 13.5-d embryos with combined homozygosity for mutant alleles of PRDX4 (that delete the signal peptide containing exon), and gene-trap disruptions of Ero1l and Ero1lb encoding both mouse isoforms of ERO1 (Zito et al., 2012). Cells were cultured and imaged in DMEM (Sigma-Aldrich), supplemented with 10% fetal calf serum. Transfections were performed using the Neon Transfection System (Invitrogen).
Transfected HEK293T cells from confluent 100-mm plates were washed in phosphate-buffered saline (PBS) with 20 mM N-ethylmaleimide and lysed in 0.5% Triton X-100, 150 mM NaCl, 20 mM Hepes, pH 7.4, 10 mM CaCl2, 20 mM N-ethylmaleimide, and protease inhibitors. Proteins were resolved by 12% SDS-PAGE and blotted with an anti-GFP serum raised in rabbit. CHOP immunoreactivity was detected by immunofluorescence of 2% paraformaldehyde-fixed cells using a primary rabbit polyclonal serum to mouse CHOP (diluted 1:500) as described previously (Ron and Habener, 1992). Goat anti–rabbit DyLight488- and anti–mouse DyLight543-conjugated IgG were used as secondary antibodies diluted 1:500 (Jackson ImmunoResearch Laboratories, Inc.). To localize roGFPiE the protein was immunostained in 2% paraformaldehyde-fixed cells using the aforementioned anti-GFP serum alongside a commercial mouse monoclonal anti-PDI antibody (1D3; Assay Designs) and a polyclonal antibody raised in hen against full-length bacterially expressed hamster BiP. Goat anti–rabbit DyLight488, anti–mouse DyLight633, and anti–chicken Cy3-conjugated IgG were used as secondary antibodies diluted 1:500 (Jackson ImmunoResearch Laboratories, Inc.).
Protein purification and enzymatic assays
For in-vitro assays, human PDI (PDIA1 18–508; a gift of C. Thorpe, University of Delaware, Newark, DE) and roGFP variants were expressed in the E. coli BL21 (D3) strain, purified with Ni-NTA affinity chromatography, dialyzed into the reaction buffer, reduced by incubation with 20 mM of DTT, and then buffer exchanged on a PD-10 gel filtration column (GE Healthcare; Blais et al., 2010). Fluorescent lifetime imaging of purified roGFP variants was conducted on samples of fluorescent protein immobilized on protein A–Sepharose beads with rabbit polyclonal anti-GFP serum. Redox titrations and redox potential calculations were performed by first exposing bacterially expressed roGFPiE to a reducing concentration of DTT (20 mM), removing the DTT by gel filtration, and then equilibrating the reduced protein with different redox buffers containing defined concentrations of oxidized and reduced glutathione, as described previously (Lohman and Remington, 2008).
For kinetic assays, reduced PDI (5–150 µM) was equilibrated in 100 mM Hepes, pH 7.4, and 150 mM NaCl buffer containing an excess of a combination of reduced and oxidized glutathione (4 mM; Sigma-Aldrich) for 1 h at room temperature, then added simultaneously to all the samples in the experiment. The ratio of fluorescence emission at 520 nm after sequential excitation at 395 and 470 nm was measured using EnSpire Multimode Plate Readers (PerkinElmer).
FLIM and intensity-based confocal microscopy and image analysis
A laser-scanning confocal system (510 Meta; Carl Zeiss) with a Plan-Apochromat 63× oil immersion lens (NA 1.4) was used to acquire intensity-based fluorescence microscopy images of cells fixed with 2% paraformaldehyde PBS. Images were analyzed using ImageJ with JACoP and RatioPlus plugins (National Institutes of Health) to assess colocalization and ratiometrics.
FLIM experiments were performed on a modified version of a previously described laser-scanning multiparametric imaging system (Frank et al., 2007), with a 63× oil immersion lens (NA 1.4) coupled to a microscope incubator, maintaining standard tissue culture conditions (Okolab), using a pulsed (sub-10 ps, 40 MHz) supercontinuum (430–2,000 nm) light source (SC 450; Fianium Ltd.). Desired excitation wavelength (typically 470 nm for FLIM of GFP variants and 440 nm for D1ER cameleon FRET) was tuned using an acousto-optic tunable filter (AOTFnC-VIS; AA Opto-Electronic). Desired emission was collected using 510/42 and 470–490 bandpass filters for GFP variants and D1ER cameleon accordingly and detected by a fast photomultiplier tube (PMC-100; Becker & Hickl GmbH). Lifetimes for each pixel were recorded using TCSPC circuitry (SPC-830; Becker & Hickl GmbH), keeping count rates below 1% of the laser repetition rate to prevent pulse pile-up. Images were acquired over 20–60 s, with a typical flow rate of 5 × 104 photons sec−1, while detector speed saturation (pile-up effect) is not observed below 106 photons sec−1 in this instrument. The data were processed using SPCImage (Becker & Hickl GmbH) fitting the time-correlated photon count data obtained for each pixel of the image to a monoexponential decay function, yielding a value for lifetime on the picosecond scale.
After filtering out autofluorescence (by excluding pixels with a fluorescence lifetime that was out of range of the roGFP probes, i.e., longer than 2,800 ps), mean fluorescence lifetime of single cells or beads was established. The value obtained represented the redox as sensed by roGFPiE in vivo/in vitro or FRET efficiency of D1ER cameleon (inversely proportional to Ca2+ concentration). Each data point is constituted by the average and SD of measurements from at least 10 cells or beads. Unless indicated otherwise, the images shown are of a representative experiment reproduced at least twice.
Calcium imaging
In vivo assessments of cytosolic Ca2+ concentration were performed on cells loaded with 20 µM Indo 1-AM for 30 min (Sigma-Aldrich), excited at 350 nm and measuring the ratio of emission intensities at 405 vs. 485 nm. The FRET-based, ER-optimized D1ER cameleon (Palmer et al., 2004) was used for ER calcium imaging by FLIM (exploiting fluorescence lifetime shortening effect of FRET on the emission of the cyan fluorescent protein segment of D1ER cameleon). Cytosolic Ca2+ buffering was achieved by exposing cells to the acetoxymethyl ester of 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid, BAPTA-AM (50 µM; Sigma-Aldrich).
Online supplemental material
Fig. S1 shows insensitivity of ratiometric fluorescent emission spectroscopy of roGFPiE to changes in ER redox of live cells. Fig. S2 shows that fluorescent lifetime of conventional GFP is not altered by changes to the redox environment. Fig. S3 shows irreversible ER calcium depletion and altered ER redox poise in thapsigargin-treated cells. Fig. S4 shows that unfolded protein stress does not promote lumenal calcium depletion. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211155/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201211155.dv.
Supplementary Material
Acknowledgments
We thank S.J. Remington for invaluable advice and for sharing reagents, A. Palmer and R. Tsein for the gift of D1ER cameleon, C. Thorpe for the PDI expression plasmid, and A. Zyryanova for assistance in plasmid construction.
This work is supported by a Wellcome Trust Principal Research Fellowship (no. 084812/Z/08/Z) and by an EU FP7 grant (no. 277713, BetaBat) to D. Ron and by grants from the Wellcome Trust/MRC, the Engineering and Physical Sciences Research Council, and Alzheimer’s Research UK to C.F. Kaminski.
Footnotes
Abbreviations used in this paper:
- DTT
- dithiothreitol
- FLIM
- fluorescence lifetime imaging
- PDI
- protein disulfide isomerase
- roGFP
- redox-sensitive green fluorescent protein
- SERCA
- smooth endoplasmic reticulum calcium
- TCSPC
- time-correlated single-photon counting
- UPR
- ER unfolded protein response
References
- Anfinsen C.B. 1973. Principles that govern the folding of protein chains. Science. 181:223–230 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Riemer J., Christensen B., Sørensen E.S., Ellgaard L. 2008. A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J. 27:2977–2987 10.1038/emboj.2008.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Riemer J., Zito E., Chin K.T., Ron D., Spiess M., Ellgaard L. 2010. Disulphide production by Ero1α-PDI relay is rapid and effectively regulated. EMBO J. 29:3318–3329 10.1038/emboj.2010.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axten J.M., Medina J.R., Feng Y., Shu A., Romeril S.P., Grant S.W., Li W.H., Heerding D.A., Minthorn E., Mencken T., et al. 2012. Discovery of 7-methyl-5-(1-[3-(trifluoromethyl)phenyl]acetyl-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 55:7193–7207 10.1021/jm300713s [DOI] [PubMed] [Google Scholar]
- Baker K.M., Chakravarthi S., Langton K.P., Sheppard A.M., Lu H., Bulleid N.J. 2008. Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 27:2988–2997 10.1038/emboj.2008.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnberg O., Ostergaard H., Winther J.R. 2006. Measuring intracellular redox conditions using GFP-based sensors. Antioxid. Redox Signal. 8:354–361 10.1089/ars.2006.8.354 [DOI] [PubMed] [Google Scholar]
- Blais J.D., Chin K.T., Zito E., Zhang Y., Heldman N., Harding H.P., Fass D., Thorpe C., Ron D. 2010. A small molecule inhibitor of endoplasmic reticulum oxidation 1 (ERO1) with selectively reversible thiol reactivity. J. Biol. Chem. 285:20993–21003 10.1074/jbc.M110.126599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Helenius J., Helenius A. 1992. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 11:1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayden D.J., Hanley M.R., Thastrup O., Cuthbert A.W. 1989. Thapsigargin, a new calcium-dependent epithelial anion secretagogue. Br. J. Pharmacol. 98:809–816 10.1111/j.1476-5381.1989.tb14609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Cannon M.B., Remington S.J. 2008. Redox-sensitive green fluorescent protein: probes for dynamic intracellular redox responses. A review. Methods Mol. Biol. 476:51–65 [DOI] [PubMed] [Google Scholar]
- Chakravarthi S., Jessop C.E., Bulleid N.J. 2006. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 7:271–275 10.1038/sj.embor.7400645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K.T., Kang G., Qu J., Gardner L.B., Coetzee W.A., Zito E., Fishman G.I., Ron D. 2011. The sarcoplasmic reticulum luminal thiol oxidase ERO1 regulates cardiomyocyte excitation-coupled calcium release and response to hemodynamic load. FASEB J. 25:2583–2591 10.1096/fj.11-184622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross B.C., Bond P.J., Sadowski P.G., Jha B.K., Zak J., Goodman J.M., Silverman R.H., Neubert T.A., Baxendale I.R., Ron D., Harding H.P. 2012. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. USA. 109:E869–E878 10.1073/pnas.1115623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuozzo J.W., Kaiser C.A. 1999. Competition between glutathione and protein thiols for disulphide-bond formation. Nat. Cell Biol. 1:130–135 10.1038/11047 [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Ruddock L.W. 2005. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6:28–32 10.1038/sj.embor.7400311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito A., Oggier T., Gerritsen H., Lustenberger F., Wouters F. 2005. All-solid-state lock-in imaging for wide-field fluorescence lifetime sensing. Opt. Express. 13:9812–9821 10.1364/OPEX.13.009812 [DOI] [PubMed] [Google Scholar]
- Frand A.R., Kaiser C.A. 1998. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1:161–170 10.1016/S1097-2765(00)80017-9 [DOI] [PubMed] [Google Scholar]
- Frank J.H., Elder A.D., Swartling J., Venkitaraman A.R., Jeyasekharan A.D., Kaminski C.F. 2007. A white light confocal microscope for spectrally resolved multidimensional imaging. J. Microsc. 227:203–215 10.1111/j.1365-2818.2007.01803.x [DOI] [PubMed] [Google Scholar]
- Gross E., Sevier C.S., Heldman N., Vitu E., Bentzur M., Kaiser C.A., Thorpe C., Fass D. 2006. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA. 103:299–304 10.1073/pnas.0506448103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., Remington S.J. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279:13044–13053 10.1074/jbc.M312846200 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zyryanova A.F., Ron D. 2012. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase, PERK. J. Biol. Chem. 287:44338–44344 10.1074/jbc.M112.428987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatahet F., Ruddock L.W. 2009. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 11:2807–2850 10.1089/ars.2009.2466 [DOI] [PubMed] [Google Scholar]
- Higo T., Hattori M., Nakamura T., Natsume T., Michikawa T., Mikoshiba K. 2005. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell. 120:85–98 10.1016/j.cell.2004.11.048 [DOI] [PubMed] [Google Scholar]
- Hoebe R.A., Van Oven C.H., Gadella T.W., Jr, Dhonukshe P.B., Van Noorden C.J., Manders E.M. 2007. Controlled light-exposure microscopy reduces photobleaching and phototoxicity in fluorescence live-cell imaging. Nat. Biotechnol. 25:249–253 10.1038/nbt1278 [DOI] [PubMed] [Google Scholar]
- Hwang C., Sinskey A.J., Lodish H.F. 1992. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 257:1496–1502 10.1126/science.1523409 [DOI] [PubMed] [Google Scholar]
- Kemp M., Go Y.M., Jones D.P. 2008. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic. Biol. Med. 44:921–937 10.1016/j.freeradbiomed.2007.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodali V.K., Thorpe C. 2010. Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid. Redox Signal. 13:1217–1230 10.1089/ars.2010.3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman J.R., Remington S.J. 2008. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry. 47:8678–8688 10.1021/bi800498g [DOI] [PubMed] [Google Scholar]
- Malhotra J.D., Miao H., Zhang K., Wolfson A., Pennathur S., Pipe S.W., Kaufman R.J. 2008. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl. Acad. Sci. USA. 105:18525–18530 10.1073/pnas.0809677105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066–3077 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough K.D., Martindale J.L., Klotz L.O., Aw T.Y., Holbrook N.J. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21:1249–1259 10.1128/MCB.21.4.1249-1259.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer P.I., Trusina A., Papa F.R. 2008. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 135:933–947 10.1016/j.cell.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A.J., Dick T.P. 2010. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 13:621–650 10.1089/ars.2009.2948 [DOI] [PubMed] [Google Scholar]
- Nguyen V.D., Saaranen M.J., Karala A.R., Lappi A.K., Wang L., Raykhel I.B., Alanen H.I., Salo K.E., Wang C.C., Ruddock L.W. 2011. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 406:503–515 10.1016/j.jmb.2010.12.039 [DOI] [PubMed] [Google Scholar]
- Pagani M., Fabbri M., Benedetti C., Fassio A., Pilati S., Bulleid N.J., Cabibbo A., Sitia R. 2000. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J. Biol. Chem. 275:23685–23692 10.1074/jbc.M003061200 [DOI] [PubMed] [Google Scholar]
- Palmer A.E., Jin C., Reed J.C., Tsien R.Y. 2004. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc. Natl. Acad. Sci. USA. 101:17404–17409 10.1073/pnas.0408030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prostko C.R., Brostrom M.A., Malara E.M., Brostrom C.O. 1992. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J. Biol. Chem. 267:16751–16754 [PubMed] [Google Scholar]
- Remington S.J. 2011. Green fluorescent protein: a perspective. Protein Sci. 20:1509–1519 10.1002/pro.684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Habener J.F. 1992. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 6:439–453 10.1101/gad.6.3.439 [DOI] [PubMed] [Google Scholar]
- Rubio C., Pincus D., Korennykh A., Schuck S., El-Samad H., Walter P. 2011. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J. Cell Biol. 193:171–184 10.1083/jcb.201007077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddock L.W. 2012. Low-molecular-weight oxidants involved in disulfide bond formation. Antioxid. Redox Signal. 16:1129–1138 10.1089/ars.2011.4481 [DOI] [PubMed] [Google Scholar]
- Scherer N.M., Deamer D.W. 1986. Oxidative stress impairs the function of sarcoplasmic reticulum by oxidation of sulfhydryl groups in the Ca2+-ATPase. Arch. Biochem. Biophys. 246:589–601 10.1016/0003-9861(86)90314-0 [DOI] [PubMed] [Google Scholar]
- Sevier C.S., Qu H., Heldman N., Gross E., Fass D., Kaiser C.A. 2007. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 129:333–344 10.1016/j.cell.2007.02.039 [DOI] [PubMed] [Google Scholar]
- Tavender T.J., Sheppard A.M., Bulleid N.J. 2008. Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem. J. 411:191–199 10.1042/BJ20071428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Tu B.P., Ho-Schleyer S.C., Travers K.J., Weissman J.S. 2000. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 290:1571–1574 10.1126/science.290.5496.1571 [DOI] [PubMed] [Google Scholar]
- van Lith M., Tiwari S., Pediani J., Milligan G., Bulleid N.J. 2011. Real-time monitoring of redox changes in the mammalian endoplasmic reticulum. J. Cell Sci. 124:2349–2356 10.1242/jcs.085530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H., Periasamy A. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16:19–27 10.1016/j.copbio.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Walter P., Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Zhao H., Loessberg P.A., Sachs G., Muallem S. 1990. Regulation of intracellular Ca2+ oscillation in AR42J cells. J. Biol. Chem. 265:20856–20862 [PubMed] [Google Scholar]
- Zito E., Chin K.T., Blais J., Harding H.P., Ron D. 2010a. ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 188:821–832 10.1083/jcb.200911086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., Melo E.P., Yang Y., Wahlander A., Neubert T.A., Ron D. 2010b. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell. 40:787–797 10.1016/j.molcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., Hansen H.G., Yeo G.S., Fujii J., Ron D. 2012. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scurvy in mice. Mol. Cell. 48:39–51 10.1016/j.molcel.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.