Abstract
Many biological, physical, and social interactions have a particular dependence on where they take place; e.g., in living cells, protein movement between the nucleus and cytoplasm affects cellular responses (i.e., proteins must be present in the nucleus to regulate their target genes). Here we use recent developments from dynamical systems and chemical reaction network theory to identify and characterize the key-role of the spatial organization of eukaryotic cells in cellular information processing. In particular, the existence of distinct compartments plays a pivotal role in whether a system is capable of multistationarity (multiple response states), and is thus directly linked to the amount of information that the signaling molecules can represent in the nucleus. Multistationarity provides a mechanism for switching between different response states in cell signaling systems and enables multiple outcomes for cellular-decision making. We combine different mathematical techniques to provide a heuristic procedure to determine if a system has the capacity for multiple steady states, and find conditions that ensure that multiple steady states cannot occur. Notably, we find that introducing species localization can alter the capacity for multistationarity, and we mathematically demonstrate that shuttling confers flexibility for and greater control of the emergence of an all-or-nothing response of a cell.
Introduction
Cells constantly have to adapt and respond to their environment. In single-celled organisms those cells least well adjusted to their surroundings will tend to contribute less to future generations than cells that are able to assimilate better or more quickly to changing circumstances. In multicellular organisms, aberrant response of individual cells to environmental or physiological cues may result in developmental anomalies or disease. The way in which cells respond to external signals, process them and act upon them, is thus intimately and inextricably linked to an organism’s fate (1,2); and in the long-term, evolution will shape the molecular machinery underlying cellular decision-making processes (3,4).
One central aspect of biological information processing is the mapping of environments onto intracellular states determined by the abundances of the molecular species (proteins, mRNAs, metabolites, etc.) under consideration. In this processing of information one or more environmental variables need to be represented in a way that facilitates the appropriate response. Continuous and discrete representations have been reported, and it is easy to see how an increased number of states will start to mimic the analog nature of continuously varying states; here, of course, we are typically only interested in stable states. A simple on/off switch, for example, is a binary representation or response mechanism; this behavior is particularly interesting if there is a regime of conditions where the system can populate either state. In this case we speak of a bistable switch; outside this regime we only find a single state for the system. More generally, we speak of multistability if more than two stable states are obtainable simultaneously.
The number of states in which a cell can be at any given time is linked to the flexibility in its decision-making (5). If only one state is accessible (and stable) then there is obviously no room for choice (even if such choices are made by random processes) and any cell-to-cell variability will derive from intrinsic or extrinsic sources of noise which will broaden out the population behavior around such stable states. For bi- and multistable systems, however, cell-to-cell variability may to a large extent be explained in terms of different states being occupied by different cells (even though they are genetically identical). From such variation different cell fates may be differentially accessible and hence understanding the causes of multistability in signal transaction will have ramifications across many areas of modern biology, notably stem-cell biology and regenerative medicine.
One canonical class of biological systems exhibiting multistability is the protein kinase cascade, which involves multiple phosphorylation of a substrate (6). Mitogen-activated protein kinase (MAPK) cascades are the most popular exponents of this type of system (7). Depending on the mechanism of (de-)phosphorylation, bistability in such systems can arise (8) that would give such systems the ability to use different levels of phosphorylation of the final substrate, e.g., Erk.
The ultimate function of Erk is to initiate a host of transcriptional responses. To fulfill such a function, activated Erk has to shuttle into the nucleus and a growing body of recent work is paying attention to such spatial aspects of signal transduction (9–11). Here we show that this spatial organization (12) of signal transduction processes plays a pronounced role in increasing the computational space available to cells. Interestingly, the same effect has been observed at the onset of mitosis (13). Very much like the physical address space in a computer processor (14), the biological equivalent is influenced by the (bio-)physical organization of such systems. And here we show how the compartmentalization increases the number of stable states that can become simultaneously accessible, conferring greater flexibility and plasticity to such systems. Importantly, spatial organization can induce multistability into systems that otherwise would be monostable, as well as (sometimes considerably) increase the number of states in systems where the presence of multiple-phosphorylation sites would already give rise to multistable behavior. This work complements the findings in Bhalla (15) by providing a detailed mathematical analysis focused on the Erk shuttling mechanisms.
Identifying whether a system exhibits multistable behavior or not, however, is challenging. This type of behavior may be limited to small regions of parameter space, and in high-dimensional spaces it is likely not detectable by simulation or random search of the parameter space; instead other approaches are called for. (Our systems considered here have 20 and 36 parameters, the reaction rate constants, respectively.) Here we base our arguments on a set of generalizable heuristics that conclusively assert or reject a system’s capacity for multistability, and which allow us to identify and delineate multi- and monostable regions in parameter space. When multistability occurs, we thereby obtain corresponding values of the rate constants and the steady states. Further, we delimit regions of the parameter space that contain rate constants that give rise to multistationarity.
Material and Methods
Calculations used to assess multistationarity were made using the software Mathematica (Ver. 7.0; Wolfram Research, Champaign, IL) and the chemical reaction network theory (CRNT) toolbox (16).
Bifurcation diagrams were computed using the software Oscill8 (17) and visualized with the software MATLAB (R2011b; The MathWorks, Natick, MA).
Results
The one-site phosphorylation cycle
We first consider a basic building block of many signal transduction systems that, for spatially homogeneous systems, is guaranteed to be monostable. A one-site phosphorylation cycle—other posttranslational modifications can, of course also be considered—consists of the reversible modification of a substrate S into its phosphorylated form S∗. Phosphorylation and dephosphorylation are enzymatically catalyzed by E (kinase) and F (phosphatase), respectively, through a standard Michaelis-Menten mechanism involving the formation of intermediate complexes X,Y,
| (1) |
| (2) |
where denotes the reaction rate constants. Endowed with mass-action kinetics, this cycle is monostable (18,19).
Now suppose we learn that these enzymatic reactions can occur in both the nucleus and the cytoplasm (Fig. 1), and we therefore include the reactions in the cytoplasm (denoted by the superscript c)
| (3) |
| (4) |
as well as shuttling reactions between cytoplasm and nucleus
| (5) |
for the species Z of the one-site phosphorylation cycle that shuttle into the nucleus at rate kin,Z and out at kout,Z. (We remain in a regime where compartmental models are appropriate and where we do not have to model diffusive motion using reaction-diffusion equations. This is appropriate for all cases where transport across a membrane is rate-limiting.)
Figure 1.
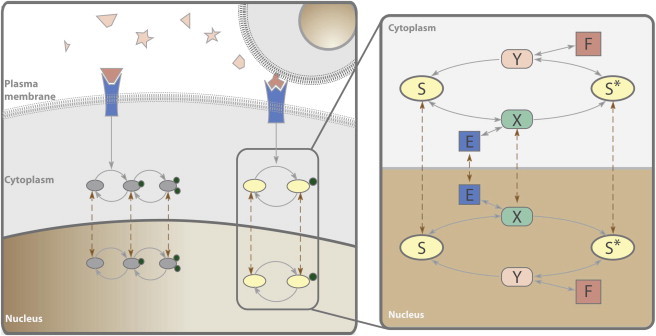
Spatial signaling schematic. (A) Cells require movement of molecular species within the plasma membrane, cytoplasm, nucleus, and more cell locations to turn genes on or off and ultimately induce a response. (B) One-site phosphorylation/dephosphorylation in two compartments, cytoplasm and nucleus. Molecular species: unphosphorylated substrate (S), kinase (E), substrate-kinase complex (X), phosphorylated substrate (S∗), phosphatase (F), phosphatase-phosphorylated substrate complex (Y), and superscript c denotes species in the cytoplasm; all others are in the nuclear compartment. Kinase and substrate total abundances (Etot, Stot) are globally conserved, whereas phosphatase abundances are conserved within each compartment (Ftot, Fctot).
We have analyzed the system for all possible combinations of shuttling species, in total 64 systems. Here we provide a detailed analysis when the species E, X, S, S∗ shuttle (see Fig. 1 B, and the Supporting Material). This system is chosen because it corresponds to a spatial model of a simplified one-site MAPK model system (9,20) that is strictly monostable.
Assuming mass-action kinetics, the species abundances are modeled by a system of ordinary differential equations (ODEs). For the particular system in Fig. 1 B, the ODE system is
| (6) |
Total kinase, phosphatase, and substrate abundances are constant for each compartment or globally, depending on which species are allowed to shuttle between compartments. For instance, in Qiao et al. (6), there are four independent conservation laws (conserved total amounts):
| (7) |
Here we use variation in the amount of active kinase to model how external stimuli are processed, and use the substrate state to capture the effects of stimuli.
We analyze the steady states of the ODEs to determine if the system can exhibit multiple steady states (multistationarity) or not. This analysis employs a suite of different mathematical techniques, including the Jacobian injectivity criterion and recent developments from chemical reaction network theory (CRNT) (16,21–23). Started in the 1970s by Feinberg and Horn (24), Feinberg (25), and Horn and Jackson (26), CRNT draws relationships between network structure and qualitative dynamical properties. The role of the Jacobian injectivity criterion is to preclude multistationarity and to derive conditions on the rate constants that ensure multistationarity cannot occur. A review of these techniques is given in the next section.
Theoretical approaches to multistationarity
We will consider polynomial functions that are derived from Eqs. 6 and 7. The variables of the functions will be the species concentrations. Let
be a differentiable function. The Jacobian, Jx(f), of f at a point x = (x1,…, xn) in is the n × n matrix with entry (i, j) being ∂fi/∂xj. If f is a polynomial function then all entries of the matrix Jx(f) are polynomials in x and consequently, the determinant of Jx(f) is a polynomial in x too. The Jacobian injectivity criterion states that if all components fi have total degree at most two, and the determinant of the Jacobian does not vanish in a convex domain , then f is an injective function in Ω (21,22).
The positive steady states of a system of biochemical reactions are given as the positive solutions to a system of polynomial equations fκ,i(x) = 0, i = 1,…, n, where n is the number of species (variables) in the system. The coefficients of polynomials fκ,i depend on the form of the conservation laws and the rate constants κ = {kr}, where kr is the rate constant of reaction r. Here it is explained with reference to the system in Fig. 1 B. There are n = 12 variables and
There is one function for each conservation law, obtained as, e.g., fκ,i(x) = −Ftot + [F] + [Y]; see Ferrell and Bhatt (7). The remaining equations are obtained as a set of nonredundant steady-state equations from Qiao et al. (6). That is to say, if l (here, l = 4) is the number of independent conservation laws, we choose n – l (here, n – l = 8) independent equations from Qiao et al. (6). For example, if we choose the equation for , that is, fκ,i(x) = kon,F [S∗][F] – koff,F [Y] – kcat,F [Y], we cannot choose the equation for , as (for details, see the Supporting Material).
If the function fκ = (fκ,1,…, fκ,n) is injective over the positive real numbers then there can at most be one positive solution to fκ(x) = (0,…, 0), and consequently, multistationarity cannot occur. In our case, the polynomials fκ,i are either quadratic or linear in x, and hence the Jacobian injectivity criterion can be applied. The determinant of the Jacobian, Jx(f), of fκ is a polynomial in x with coefficients depending on the rate constants. Each term in the polynomial is of the form , where a(κ) is a coefficient and mj is 0, 1, or 2. If the coefficients of the determinant of the Jacobian are all positive or all negative for a specific choice of rate constants, then the determinant cannot vanish when x is positive. Hence, it follows from the Jacobian injectivity criterion that multiple positive steady states cannot occur. Importantly the coefficients a(κ) themselves are polynomials in the rate constants κ = {kr}. Thus, by analyzing these coefficients we can find polynomial inequalities on the rate constants that, if satisfied, ensure that the system cannot have multiple steady states. In other words, we can find conditions on the rate constants that are necessary for multistationarity to occur.
Failure of the Jacobian injectivity criterion is not sufficient to conclude that multistationarity occurs. To investigate whether multistationarity occurs when the Jacobian injectivity criterion fails, we make use of the algorithms implemented in the CRNT toolbox (16). For some systems modeled with mass-action kinetics (as is the case here), the toolbox can conclusively determine if multistationarity can or cannot occur. If the system admits multiple steady states, we obtain a unique set of rate constants for which there exists a pair of positive steady states (fulfilling the conservation laws with the same total amounts). However, the rate constants that the toolbox outputs cannot be constrained or controlled in any way. That is to say, it is not possible to restrict the search to certain regions that are considered biologically realistic. To overcome this difficulty we manually vary the rate constants provided by the toolbox to obtain satisfactory rate constants that exhibit multistationarity.
In this work, a steady state is considered stable if it is asymptotically stable, that is, if the real part of the eigenvalues of the Jacobian of the system at the steady state are all negative. Asymptotic stability ensures that if the initial state of the system is sufficiently close to the steady state then it will eventually be attracted toward the steady state.
Analytic conditions for multistationarity
Armed with these tools we establish that multistationarity cannot occur if only one species of the one-phosphorylation cycle shuttles; if two species shuttle, then only the combinations {X, S∗} or {S, Y} can induce multistationarity for certain total amounts and rates. The full list of sets of species that can induce multistationarity are shown in Table 1. It is interesting that multistationarity can be introduced and destroyed by including more shuttling species. For example, if X, Y shuttle then the system shows multistationarity, but if, in addition, E shuttles it is monostable. If S∗ also shuttles, that is, X, Y, E, S∗ shuttle, then the system again shows multistationarity. Thus, it is very susceptible to the precise shuttling species. The fact that the model includes the formation of at least one of the intermediate complexes X, Y (and hence some form of sequestration) is critical for the creation of multistationarity.
Table 1.
One-site phosphorylation system
| No. | No multistationarity | Multistationarity |
|---|---|---|
| 1 | All | None |
| 2 | {S, S∗}{E, Y}{F, X}{S∗, E}{S, F}{S, X}{S∗, Y}{E, X}{F, Y}{S, E}{S∗, F} | {E, F} {X, Y} {S∗, X} (S, Y} |
| 3 | {X, E, F}{Y, E, F}{X, Y, E}{X, Y, F}{S, F, X}{S∗, E, Y}{S, E, F}{S∗, F, E} | {S, E, X}{ S∗, F, Y}{S, E, Y}{ S∗, E, X}{S∗, F, X}{S, E, S∗}{ S∗, F, S}{S, S∗, X}{S, X, Y}{ S∗, Y, X}{S, F, Y}{S, S∗, Y} |
| 4 | {Y, X, E, F} | {S, S∗, X, F}{S, S∗, Y, E}{S, E, X, Y}{S∗, F, X, Y}{S, F, X, Y}{S∗, E, X, Y}{S, S∗, X, Y}{S∗, S, E, F}{S, E, F, X}{S∗, E, F, Y}{S, E, F, Y}{S∗, E, F, X}{S, S∗, X, E}{S, S∗, Y, F} |
| 5, 6 | None | All |
For all possible sets of shuttling species, it is indicated whether or not the system has the capacity for multiple steady states.
In particular, multistationarity occurs if the species E, S, X, S∗ is allowed to shuttle (Fig. 1 B). Applying the Jacobian injectivity criterion, we conclude that if the shuttling rates fulfill
| (8) |
then multistationarity cannot occur even for the spatial model, whatever the total amounts and reaction constants may be within each cycle (see the Supporting Material). The shuttling rates are paired for species E, X and species S, S∗ and therefore a necessary condition for multistationarity is that either X moves in or out of the nucleus slower than the kinase, E; or alternatively that S shuttles more slowly than its active form, S∗.
We next home-in on a set of biologically plausible rate constants and total abundances for which the system has three steady states, two of which are stable. We find that at low and high stimulus doses (i.e., total kinase level Etot) there is only one stable steady state of the phosphorylated nuclear substrate, S∗, whereas for intermediate stimulus doses two stable steady states coexist (Fig. 2, A–C). As the stimulus level changes (due to kinase production or degradation), the state of S∗ may switch either to a highly or lowly phosphorylated (activity) steady state based on the system’s memory: switching between states can occur in the form of a hysteresis loop (see black and red arrows in Fig. 2 A).
Figure 2.
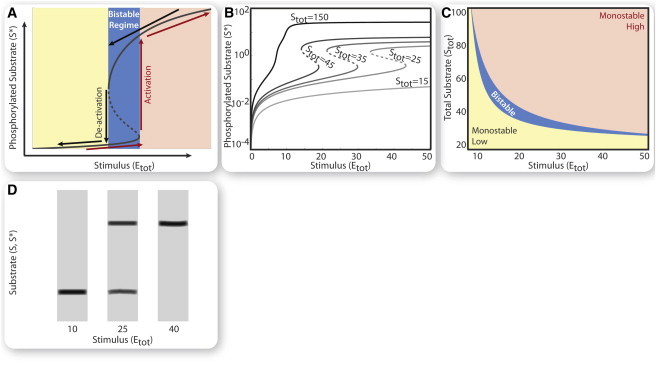
Bistability in one-cycle localization model. Rate constants and total amounts are given in the Supporting Material. (A) Steady-state curve of phosphorylated nuclear substrate (S∗) shows bistability and hysteresis as a function of stimulus (total kinase, Etot). Stable states (solid lines); unstable state (dashed lines). Activation (phosphorylation) and deactivation (dephosphorylation) switches discontinuously from one stable branch to another at different stimulus thresholds, corresponding to the boundary values of the bistable regime (blue region). (B) Steady-state curves of S∗ as a function of stimulus (Etot) at varying amounts of total substrate (Stot). (C) Steady-state diagram identifying the regions of parameter space supporting monostability (yellow, orange) or bistability (blue) as a function of the total amounts of kinase (Etot) and substrate (Stot). (D) Theoretical Western blot of bistable (two bands) or monostable (one band) behavior depending on dose of stimulus.
Shuttling plays a pronounced role in modulating the number of discrete states of the nuclear substrate concentration S∗ that can be realized. An increase of the shuttling rate constant causes the system to change either from a high to a low stable state (for kout,E, kout,X, kin,S, kin,S∗) or vice versa (for kin,E, kin,X, kout,S, kout,S∗), with an unstable steady state in between (Fig. 3 A) and saddle-node bifurcations. Depending on which rate constants are altered, the resulting switches can be reversible (Fig. 3 A, kout,E, kin,X) or irreversible (Fig. 3 A, kin,S∗). The response curves of the rate constants kin,E, kin,S∗, kout,X, kout,S can be tuned by altering, e.g., the total kinase levels, to alter the size of multistable regimes (Fig. 3 B) or to turn reversible into irreversible switches. For example, if the shuttling rate constant kin,S∗ is small, then the nuclear S∗ can exist in either a high or low state (bistable region); however, following an increase of the shuttling rate constant across a threshold, the system switches to a monostable low state and cannot switch back to the high state nor reenter the bistable regime. Similarly the rate constants kout,S, kin,E provide irreversible switches favoring the high state. As the shuttling rate constants are controlled by a variety of other processes, this endows spatially structured systems with a high level of flexibility and increased information-processing ability compared to spatially homogeneous systems: for example, any violation of the inequalities (the expressions in Eq. 8) may result in induction of multiple stable states.
Figure 3.
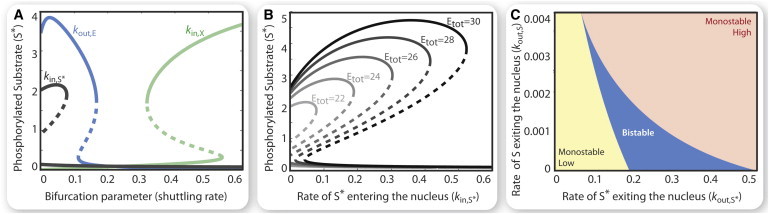
Effects of shuttling in the one-site localization model. Rate constants and total amounts as in Fig. 2. (A) Three steady-state curve behaviors of phosphorylated nuclear substrate (S∗) as shuttling is varied: high to low reversible bistability (blue, kout,E), low to high reversible bistability (green, kin,X), high to low irreversible bistability (black, kin,S∗). (B) Steady-state curves of phosphorylated nuclear substrate (S∗) as a function of the rate constant of S∗ shuttling into the nucleus (kin,S∗) at varying amounts of stimulus (Etot). (C) Steady-state diagram identifying the regions of parameter space supporting monostability (yellow, orange) or bistability (blue) as a function of kout,S and kout,S∗.
MAPK two-site phosphorylation
Having established that species shuttling introduces multistability in a system that otherwise is monostable, we next explore the influence of shuttling in a system that can already exhibit multistability. Specifically, we consider nuclear localization in a two-site phosphorylation cycle (Fig. 4 A), such as the layers of canonical MAPK cascades, and its impact on the cell’s ability to establish distinct stable states. Multistability, known to exist in these systems (27,28) with up to three stable states (29), has been discussed without reference to any of the spatial models of MAPK signaling (9,12,20,30,31). Here, to differentiate between biochemical dependent and shuttling-dependent multistability, we consider biochemical parameter sets that preclude multistability in the absence of localization (see the Supporting Material). Again spatial structure and shuttling between compartments can induce bistability; e.g., fixing the shuttling species to be E, X1, X2, S0, S1, S2, bistability is introduced in the system (see the Supporting Material). Provided that the rate constants in at least one of the cycles are in the range of multistability of the two-site phosphorylation cycle, we observe that shuttling creates up to four stable states and three unstable states (Fig. 4 B). Steady-state analysis on a choice of biologically plausible shuttling rates indicates that with shuttling the two-site phosphorylation cycle can undergo hysteresis effects with a large region of multistability (32 ≤ Etot ≤ 455), most of which is bistable. The doubly-phosphorylated substrate in the nucleus (S2) appears in a low/high monostable state for extremely low/high levels of kinase Etot (red and black arrows in Fig. 4 B).
Figure 4.
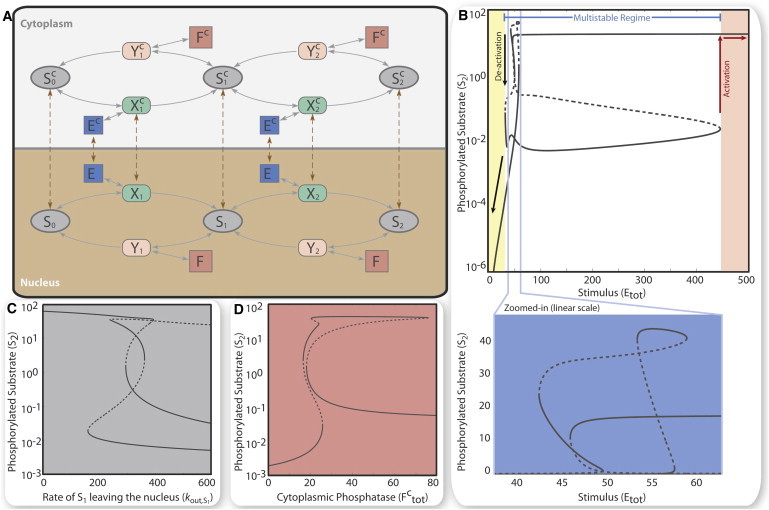
Two-site phosphorylation localization model. Rate constants and total amounts provided in the Supporting Material. (A) Schematic of two-site phosphorylation and dephosphorylation in two compartments, cytoplasm and nucleus. (B) Steady-state curve of phosphorylated nuclear substrate (S2) shows multistability and hysteresis as a function of stimulus (total kinase, Etot). Stable states (solid lines); unstable states (dashed lines). Activation (phosphorylation) and deactivation (dephosphorylation) switches discontinuously from one stable branch to another at different stimulus thresholds, corresponding to the boundary values of the multistable regime (blue interval). Extreme values of stimulus restrict the system to monostability (log scale). Closer inspection reveals up to four stable states simultaneously (blue figure, zoomed, linear scale). (C and D) Steady-state curve of phosphorylated nuclear substrate (S2) as a function of the rate constant of S1 shuttling out of the nucleus (gray) or as a function of the total cytoplasmic phosphatase (Fctot) (coral).
The extended region of multistability with four stable states may provide an explanation for the versatility of MAPK signaling systems and their widespread use as relays in many signal transduction networks. As expected, the steady states of the system can be regulated through reversible switches governed by shuttling parameters and other total amounts (Fig. 4, C and D). But in some cases, stable states may be so close together as to be virtually indistinguishable under some physiological conditions; the switching between three and four stable states in a small region (see zoomed box in Fig. 4 B) may be an example of this. This, too, can be modulated, however, by regulation of the shuttling process, or by adjusting total substrate abundances.
Discussion
Fidelity of information processing and the computational capacity, i.e., the ability to map environmental states onto discernible internal states (in particular of proteins active in the nucleus), are thus profoundly affected by the spatial structure of eukaryotic cells. In a biological context, a high and stable state of nuclear substrate (S∗) can marshal robust responses to environmental cues. To add further flexibility to such information processing, the shuttling speed of a substrate may further depend on the nature of the stimulus; biological examples abound, and, for example, stimulated NIH 3T3 cells shuttle MAPK into the nucleus three times faster than starved cells (32). By simultaneously controlling the stimulus dose and shuttling speed, a nuanced transition to reversible bistability permits hysteresis, and thus introduces the possibility for switching between low and high stable states (Fig. 3 B).
Two stable states mean that the system can carry information to the value of ∼1 bit, log2(2) = 1 bit; this is in line with existing results of cellular information processing (1). The information that is carried by multistable states is not merely a function of the number of stable states; the separation of these states (and the extent of their respective bases of attraction) also affect the amount of information that is carried in such systems. In principle, however, multistability should tend to confer greater flexibility to the cell’s signal processing and its response to the environment. To this end, it has been argued that multistationarity serves as a mechanism for cellular differentiation and the emergence of different tissues (33). Our results here indicate that the compartmental structure of the eukaryotic cell can confer an increase in the number of available stable states that the cell can use, e.g., to represent different cell fates.
Trafficking is therefore more intimately related to cellular computation than is typically acknowledged, and differences between cell lines in the shuttling rate constants of different substrates may be hard-wired (e.g., in NIH 3T3 mouse cells, phospho-MAPK can accumulate in the nucleus (32), but nuclear accumulation does not occur in PC12 cells (34)). Alternatively, the rich set of mechanisms affecting both phosphorylation (exemplary perhaps also for other posttranslational modifications) and trafficking give cells the flexibility to change their dynamical regime, e.g., from monostable to multistable, on the fly and in response to further environmental and physiological cues. The spatial/compartmental organization of cells thus drives crucial aspects of their information processing capacity; notably the number and robustness of states that can be stably represented is higher (or at least as high) for spatially structured systems compared to homogeneous systems (1). This provides a further rationale for the evolution of cellular compartments (35) but also begs the question as to how bacteria and archaea can increase their computational capacities. Here, we believe, microenvironments generated by molecular crowding (36) confer some of the same advantages. Crowding, such as around membrane-associated histidine kinases of two-component signaling systems, can isolate signal transduction components spatially from one another, which would have similar (although likely weaker) impact on the computational address space as cellular compartments have in eukaryotes.
Acknowledgments
The authors gratefully acknowledge funding from The Leverhulme Trust. MPHS is a Royal Society Wolfson Research Merit Award holder. EF is supported by the fellowship “Beatriu de Pino's” from the Generalitat de Catalunya and the research project MTM2009-14163-C02-01 from Spain. CW is supported by the Danish Research Council and the Lundbeck Foundation, Denmark.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Contributor Information
Carsten Wiuf, Email: wiuf@math.ku.dk.
Michael P.H. Stumpf, Email: m.stumpf@imperial.ac.uk.
Supporting Material
References
- 1.Cheong R., Rhee A., Levchenko A. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toyoshima Y., Kakuda H., Kuroda S. Sensitivity control through attenuation of signal transfer efficiency by negative regulation of cellular signaling. Nat Commun. 2012;3:743. doi: 10.1038/ncomms1745. [DOI] [PubMed] [Google Scholar]
- 3.Balázsi G., van Oudenaarden A., Collins J.J. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearlman S.M., Serber Z., Ferrell J.E., Jr. A mechanism for the evolution of phosphorylation sites. Cell. 2011;147:934–946. doi: 10.1016/j.cell.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozbudak E.M., Thattai M., van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 6.Qiao L., Nachbar R.B., Shvartsman S.Y. Bistability and oscillations in the Huang-Ferrell model of MAPK signaling. PLOS Comput. Biol. 2007;3:1819–1826. doi: 10.1371/journal.pcbi.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrell J.E., Jr., Bhatt R.R. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 1997;272:19008–19016. doi: 10.1074/jbc.272.30.19008. [DOI] [PubMed] [Google Scholar]
- 8.Thomson M., Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujioka A., Terai K., Matsuda M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 10.Lidke D.S., Huang F., Lenormand P. ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. J. Biol. Chem. 2010;285:3092–3102. doi: 10.1074/jbc.M109.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington H.A., Komorowski M., Stumpf M.P. Mathematical modeling reveals the functional implications of the different nuclear shuttling rates of Erk1 and Erk2. Phys. Biol. 2012;9:036001. doi: 10.1088/1478-3975/9/3/036001. [DOI] [PubMed] [Google Scholar]
- 12.Kholodenko B.N., Hancock J.F., Kolch W. Signaling ballet in space and time. Nat. Rev. Mol. Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos S.D.M., Wollman R., Ferrell J.E., Jr. Spatial positive feedback at the onset of mitosis. Cell. 2012;149:1500–1513. doi: 10.1016/j.cell.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feynman R.P. Perseus Books; Jackson, TN: 1996. Feynman Lectures on Computation. [Google Scholar]
- 15.Bhalla U.S. Trafficking motifs as the basis for two-compartment signaling systems to form multiple stable states. Biophys. J. 2011;101:21–32. doi: 10.1016/j.bpj.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison, P., M. Feinberg, and H. Ji. 2011. Chemical reaction network toolbox. http://www.chbmeng.ohio-state.edu/ feinberg/crntwin/.
- 17.Conrad, E. 2008. OSCILL8. http://oscill8.sourceforge.net/doc/.
- 18.Goldbeter A., Koshland D.E., Jr. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldbeter A., Koshland D.E., Jr. Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects. J. Biol. Chem. 1984;259:14441–14447. [PubMed] [Google Scholar]
- 20.Radhakrishnan K., Edwards J.S., Oliver J.M. Sensitivity analysis predicts that the ERK-pMEK interaction regulates ERK nuclear translocation. IET Syst. Biol. 2009;3:329–341. doi: 10.1049/iet-syb.2009.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass H., Connell E., Wright D. The Jacobian conjecture: reduction of degree and formal expansion of the inverse. Bull. Am. Math. Soc. 1982;7:287–330. [Google Scholar]
- 22.Pantea C., Koeppl H., Craciun G. Global injectivity and multiple equilibria in uni- and bi-molecular reaction networks. Discr. Contin. Dynam. Syst. B. 2012;17:6. [Google Scholar]
- 23.Feliu E., Wiuf C. Preclusion of switch behavior in reaction networks with mass-action kinetics. arxiv. 2012 1109.5149. [Google Scholar]
- 24.Feinberg M., Horn F. Chemical mechanism structure and the coincidence of the stoichiometric and kinetic subspaces. Arch. Ration. Mech. Anal. 1977;66:83–97. [Google Scholar]
- 25.Feinberg M. Notes of lectures given at the Mathematics Research Center of the University of Wisconsin, in 1979. University of Wisconsin; Madison, WI: 1979. Lectures on chemical reaction networks. [Google Scholar]
- 26.Horn F., Jackson R. General mass action kinetics. Arch. Ration. Mech. Anal. 1972;47:81–116. [Google Scholar]
- 27.Markevich N.I., Hoek J.B., Kholodenko B.N. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J. Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feliu E., Wiuf C. Enzyme-sharing as a cause of multi-stationarity in signaling systems. J. R. Soc. Interface. 2012;9:1224–1232. doi: 10.1098/rsif.2011.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Sontag E.D. On the number of steady states in a multiple futile cycle. J. Math. Biol. 2008;57:29–52. doi: 10.1007/s00285-007-0145-z. [DOI] [PubMed] [Google Scholar]
- 30.Shankaran H., Ippolito D.L., Wiley H.S. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kholodenko B.N., Birtwistle M.R. Four-dimensional dynamics of MAPK information processing systems. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:28–44. doi: 10.1002/wsbm.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa M., Marchi M., Ratto G.M. Dynamic regulation of ERK2 nuclear translocation and mobility in living cells. J. Cell Sci. 2006;119:4952–4963. doi: 10.1242/jcs.03272. [DOI] [PubMed] [Google Scholar]
- 33.Kauffman S.A. Oxford University Press; New York: 1993. The Origins of Order. [Google Scholar]
- 34.von Kriegsheim A., Baiocchi D., Kolch W. Cell fate decisions are specified by the dynamic ERK interactome. Nat. Cell Biol. 2009;11:1458–1464. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maynard Smith J., Szathmáry E. Oxford University Press; New York: 1997. The Major Transitions in Evolution. [Google Scholar]
- 36.McGuffee S.R., Elcock A.H. Diffusion, crowding and protein stability in a dynamic molecular model of the bacterial cytoplasm. PLOS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


