Abstract
Dynamic regulation of the actin cytoskeleton is required for diverse cellular processes. Proteins regulating the assembly kinetics of the cytoskeletal biopolymer F-actin are known to impact the architecture of actin cytoskeletal networks in vivo, but the underlying mechanisms are not well understood. Here, we demonstrate that changes to actin assembly kinetics with physiologically relevant proteins profilin and formin (mDia1 and Cdc12) have dramatic consequences on the architecture and gelation kinetics of otherwise biochemically identical cross-linked F-actin networks. Reduced F-actin nucleation rates promote the formation of a sparse network of thick bundles, whereas increased nucleation rates result in a denser network of thinner bundles. Changes to F-actin elongation rates also have marked consequences. At low elongation rates, gelation ceases and a solution of rigid bundles is formed. By contrast, rapid filament elongation accelerates dynamic arrest and promotes gelation with minimal F-actin density. These results are consistent with a recently developed model of how kinetic constraints regulate network architecture and underscore how molecular control of polymer assembly is exploited to modulate cytoskeletal architecture and material properties.
Introduction
The actin cytoskeleton is composed of a diverse assortment of dynamic filament networks and bundles that play crucial roles in a myriad of morphogenic cellular processes including migration, division, endocytosis, and intracellular trafficking (1–10). Spatiotemporal regulation of the actin cytoskeleton occurs through a constant cycle of F-actin polymerization and network assembly followed by actin depolymerization and network disintegration, resulting in turnover of F-actin polymers within cells on the timescale of seconds to minutes. It remains to be determined how actin and actin-binding proteins conspire to self-organize into functional dynamic mechanical modules with distinct kinetics and architectures required for diverse cellular processes.
Actin network architecture can be modulated through actin-binding proteins that regulate filament assembly, length, cross-linking, and dynamics. The role of actin cross-linking proteins in regulating the architecture and mechanics of actin networks has been well established (11–13). In recent years, a myriad of proteins that regulate the rates of actin filament nucleation and elongation have been identified and characterized. Proteins such as profilin, which bind actin monomers, collaborate with different formin isoforms, a protein that interacts with the barbed end of F-actin, to control the nucleation and elongation rates of F-actin assembly (4,14–20). Perturbation of these proteins in vivo significantly impacts the assembly of diverse actin architectures ranging from the contractile ring to stress fibers (7,16,21,22). Previous efforts have focused on macroscopic properties of network formation in the presence of actin-binding proteins, including, but not limited to, growth rate (23), elastic properties (24), and network density (25). However, the underlying mechanisms by which these actin regulatory proteins regulate the local actin network architecture remain poorly understood. Recent work has also shown that spatial regulation of nucleation factors can direct actin bundling (26), although work from our lab and others has shown that the architecture of cross-linked actin networks is regulated by kinetic constraints during assembly (27–29). Together, these results suggest a potentially important role for molecular regulation of actin polymerization kinetics on the architecture of otherwise biochemically identical actin networks.
We sought to understand how altered rates of filament nucleation and elongation driven by physiologically relevant proteins profilin and formin impact the local actin architecture and gelation kinetics of cross-linked F-actin networks. We studied the dynamic assembly and gelation of in vitro actin networks formed in the presence of the cross-linker α-actinin using high-resolution quantitative imaging to characterize the network architecture and changes in thermally driven motions over time. To isolate the contribution of changes in actin filament nucleation rates, the concentrations of G-actin and α-actinin remained fixed, whereas the concentrations of profilin and the formin isoforms, Cdc12 and mDia1, were varied. Our results show that changes in the filament nucleation rate have significant effects on the gelation kinetics and the architecture of the resultant networks. Low nucleation rates promote the slow formation of networks with thick bundles and large mesh sizes, whereas high nucleation rates promote a more rapid assembly of networks composed of thinner bundles and smaller mesh sizes. The filament elongation rate is also found to play an important role. Below a critical filament elongation rate, bundles form by diffusion-mediated aggregation to form very rigid, straight bundles that fail to integrate into a mechanically coherent network. As the elongation rate increases, the network gelation is accelerated and rapid elongation promotes the formation of space-spanning gels with a minimal concentration of polymerized actin. These results establish the important roles actin assembly factors play in controlling actin cytoskeletal architecture by altering kinetic constraints during their assembly. These results have significant implications for the mechanisms by which actin assembly factors control cytoskeletal organization in vivo.
Materials and Methods
Protein preparation
Ca-ATP-actin was purified from chicken skeletal muscle (4). Gel-filtered actin was labeled on Cys-374 with pyrenyl iodoacetamide for pyrene assays (Invitrogen, Carlsbad, CA) (30). Immediately before each polymerization reaction, Ca-ATP-actin was converted to Mg-ATP-actin by adding 0.5 volumes of 0.6 mM EGTA and 0.15 mM MgCl2 for 3 min at 25°C. Extinction coefficients were used to determine protein concentrations of actin and pyrene-actin (4). Chicken smooth muscle α-actinin was purified from chicken gizzard tissue by the protocol in Feramisco and Burridge (31), concentrated to 10 μM and stored at 4°C until use. The concentration of chicken smooth muscle α-actinin was measured using the extinction coefficient estimated using ProtParam (http://us.expasy.org/tools/) and the amino acid composition: A280 = 128,500 M−1 cm−1. Fission yeast and mouse profilin SpPrf and pfn1 were expressed in Escherichia coli and purified as described (32). pET21-Cdc12(882-1390) (FH1FH2) and pET21a-mDia1(552-1255) (FH1FH2) formins were expressed and purified as previously described (4,21).
In vitro network formation
Actin networks were formed by mixing nonactin components first: glucose oxidase mix (4.5 mg/mL glucose, 0.5% β-mercaptoethanol, 4.3 mg/mL glucose oxidase, 0.7 mg/mL catalase), F-buffer (10 mM Imidazole pH 7.0, 1 mM MgCl2, 50 mM KCl, 0.2 mM EGTA, 0.5 mM ATP), Ca-G-buffer (2 mM Tris pH 8.0, 0.2 mM ATP, 0.5 mM DTT, 1 mM Sodium Azide, 0.1 mM CaCl2), α-buffer (pH 7.6, 20 mM NaCl, 0.1 mM EDTA, 15 mM β-mercaptoethanol, 20 mM Tris-HCl, 10% Glycerol), and 5% molar ratio of Alexa Fluor 488 phalloidin (Invitrogen)/actin, as well as any α-actinin, profilin, and formin. Monomeric Mg-ATP-actin was then added to start the network assembly reaction. Each sample was mixed by pipetting up and down 3 times, loaded into its 5–10 μL sample chamber, sealed with VALAP (1:1:1 by weight of Vaseline, lanolin, parafin wax) and immediately transferred onto the confocal microscope for imaging. The time from the addition of monomeric actin to the start of imaging was between 45 and 85 s. The reaction time was measured relative to the time point when monomeric actin was added to the sample.
Confocal microscopy and bundle analysis
Sample chambers were constructed to dimensions of ∼22 mm (l) × 1 mm (w) × 100 μm (h). All images were taken 50 μm above the bottom coverslip to minimize any edge effects that could affect bundle formation. Time-lapse images were taken with a 20×, 0.75 numerical aperture plan fluor objective (Nikon, Melville, NY). Spinning disk confocal images were collected with a CoolSNAP HQ2 camera (Photometrics, Tucson, AZ) using a CSU X-1 scanhead from Yokogawa (Sugar Land, TX). We quantified bundle density in each frame with successive line scans in the x and y directions. 63 mm of linear density was analyzed in each frame as described in (27); steady state was determined when the bundle density reached 95% of its final value.
Structure factor calculation
The structure factor (S(q,t)) in Fig. 1 was calculated from the two-dimensional Fourier transform of a 1024 ×1024 pixel region taken from the center of each confocal slice in the time-lapse series of images used in the calculation of Fig. 1. For each transform, a series of 225 radial line scans encompassing 360° were drawn out from the center of the transform and averaged over the radial distance; steady state was determined by when the magnitude attained 95% of the final value. The S(q,t) was evaluated for a value of q = 0.0316 μm−1 as this corresponds to a length scale of ∼32 μm, which is above the mesh size for these networks. As the S(q,t) was calculated from a pixilated image, the highest frequency it can detect occurs at a length scale of 2 pixels, whereas the lowest frequency it can detect occurs at N/2 pixels, where N is the number of pixels in a given dimension of the image. For the images presented here, these limits represent 0.645 and 165 μm, respectively. Thus, the S(q,t) chosen for this work falls well within the limits of this technique.
Figure 1.
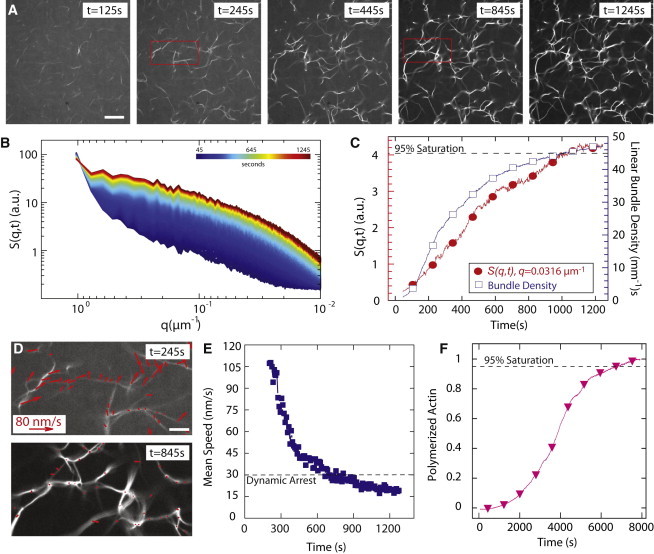
Quantification of assembly kinetics of cross-linked profilin-actin networks. (A) Images of bundles formed by 5 μM actin, 1.2 μM α-actinin, and 15 μM SpPrf. Scale bar = 30 μm. (B) S(q,t) calculated for sample described in (A) for times from 45 to 1245 s. (C) S(q,t) at q = 0.0316 μm−1 and linear bundle density over time. (D) Vector map of bundle displacements obtained in regions identified in (A) at 245 s (top) and 845 s (bottom); scale bar = 5 μm. (E) The mean speed of bundle mobility is plotted as a function of time. Dynamic arrest is defined when mean speed is reduced to 30 nm/s, near our resolution limit. (F) Time course of the spontaneous assembly of 5 μM Mg-ATP-actin monomers (10% pyrene labeled) in the presence of 15 μM SpPrf.
Particle image velocimetry analysis
The motions of actin networks were obtained by quantitative analysis of fiduciary marks within the bundles and analyzed with a previously published particle imaging velocimetry technique (33).
Pyrene assay
Actin assembly was measured from the fluorescence of pyrene-actin with a Safire2 (Tecan, Durham, NC) fluorescent plate reader. Spontaneous assembly assays were performed on samples of 5 μM actin as used in in vitro network formation, except with 10% pyrene-labeled Mg-ATP-actin. A 25 μM mixture of pyrene-labeled and unlabeled Mg-ATP-actin with 100× antifoam 204 (0.005%; Sigma, St. Louis, MO) was added to the upper row of a 96-well nonbinding black plate (Corning, Corning, NY). All other components of the assay were added to the lower well: α-actinin, α-buffer, profilin, formin, glucose oxidase mixture, 10× F-buffer, and Mg-G-buffer. Reactions were started by mixing lower wells with upper wells.
Barbed end concentration calculation
Normalized pyrene curves with data points every 10 s were fit to a line over the surrounding 5 data points at the point where 60% of the monomers in each sample were polymerized. The calculated slope in arbitrary units/s was converted to [actin]/s by a total amount of monomer in the normalized curve (5 μM). The equation used is from (34), where the kbarbed is the elongation rate of the barbed end in sub/s/μM. Elongation rates were taken from published literature (16).
Results
Assembly kinetics of cross-linked F-actin networks
The polymerization of actin in the presence of actin cross-linking proteins, such as α-actinin, builds space-spanning and mechanically coherent networks of cross-linked and bundled F-actin (12,13). To explore correlations between the kinetics of actin polymerization and network assembly, we have developed tools to characterize changes in their structure and dynamics using high-resolution confocal microscopy (27). We compared these measurements to F-actin polymerization dynamics measured by the pyrene fluorescence of identical samples.
In the representative sample in Fig. 1, 5 μM actin is polymerized in the presence of 1.2 μM α-actinin and 15 μM fission yeast profilin (SpPrf). The reaction is initiated at t = 0 s by the addition of salts, and F-actin is visualized via time-lapse confocal microscopy with a low concentration of Alexa-488 phalloidin. Filaments or small bundles first become visible around 125 s and successively become thicker and more numerous (Fig. 1 A, Movie S1 in the Supporting Material). Evolution of the network architecture is assessed by calculation of the static S(q,t). Because this measurement is made in Fourier-space, it is sensitive to periodicities in structure, and is particularly adept at sensing alignment in noncrystalline collections of objects, such as liquid crystals and the polymer networks presented here. In this case, the actin bundle is the repetitive structure, and the S(q,t) evaluates its organization in the network. The measurement of the magnitude of the S(q,t) is thus proportional to the relative organization of the bundles over a given length scale in the image. As the network forms between 45 and 1245 s (Fig. 1 B), the S(q,t) evolves until reaching steady state once the network is in place. By examining the magnitude of S(q,t) at a given value of q = 0.0316 μm−1, we find that the structure stops changing at a time near 1000 s (Fig. 1 C). The most prominent feature during this period is the increasing number of bundles (Fig. 1, A and C). The linear bundle density, calculated by identification of intensity peaks in line scans taken from successive confocal images (27), increases from 3.4 bundles/mm at t = 100 s to 42.6 bundles/mm at t = 800 s, showing a time evolution strikingly similar to S(t) (Fig. 1 C).
A third characterization of gelation is the arrest of thermally driven movements of actin bundles, which can be measured by use of image processing algorithms to track bundle movement (Fig. 1 D) (33). Initially, bundles are freely mobile with a mean instantaneous speed of 105 nm/s (Fig. 1, D and E). As bundles grow and become interconnected, the mean speed decreases to <10 nm/s. We set a threshold of 30 nm/s, which approaches our measurement resolution, to determine a timescale at which dynamics are arrested. Dynamic arrest occurs at a time of ∼600 s, indicating that the network organization continues to evolve after an integrated cross-linked network is formed.
Networks assembled from actin and α-actinin have been shown to reach steady-state organization on timescales much quicker than the rate of actin polymerization (27). We confirmed that this is still the case in the presence of profilin by a pyrene fluorescence assay containing 5 μM actin and 15 μM SpPrf (Fig. 1 F). The presence of profilin significantly increases the lag phase and only ∼1.2% of actin was polymerized by 1000 s, our approximate gelation time. Actin polymerization was completed by 6745 s. When the network architecture is determined at 1245 s, remaining actin accumulates into the background meshwork or thickens existing bundles. Thus, the kinetics of network assembly is significantly faster than the actin polymerization kinetics.
These tools allow us to assess the role of actin assembly factors in regulating the kinetics and architecture of actin network assembly. We used the well-characterized mouse formin mDia1 and fission yeast formin Cdc12 in various combinations with mouse profilin pfn1 and fission yeast profilin SpPrf. Table 1 summarizes known consequences of these actin assembly factors on the relative filament concentration at steady state, and the actin filament elongation rate.
Table 1.
The relative number of actin filaments and filament elongation rate of actin filaments in the absence or presence of various combinations of formin and profilin isoforms used in this work
| Actin alone | Rel. # of filaments |
Elongation rate |
|
|---|---|---|---|
| 100% (42) | 11.3 sub/(μM·s) (16) | ||
| +0 μM SpPrf profilin | +100 nM Cdc12 formin | ∼4000% (42) | 0.1 sub/(μM·s) (16) |
| +3:1 S. pombe profilin: actin | +100 nM Cdc12 formin | ∼200% (42) | 13.3 sub/(μM·s) (16) |
| +3:1 mouse profilin: actin | +100 nM mDia1 formin | ∼20% (43) | 48 sub/(μM·s) (16) |
The data were obtained from literature values, referenced in the table.
Profilin slows network assembly, forming a network of sparse, thick bundles
Profilin interacts with actin monomers to inhibit F-actin nucleation, although not significantly altering the filament elongation rate (Table 1) (17). As the concentration of pombe profilin SpPrf is increased from 0 to 25 μM, the rate of F-actin nucleation decreases and thus increases the time needed to complete actin polymerization as assessed by pyrene fluorescence assays (Fig. 2 A) (14). For saturating levels of profilin, the polymerization profile of actin has an extended lag phase and an increased time to steady state (35–37). The time at which 95% of the actin is polymerized, F-actin polymerization time, increases from 2000 to 6800 s over a range of profilin concentrations from 0 to 25 μM (Fig. 2 B).
Figure 2.
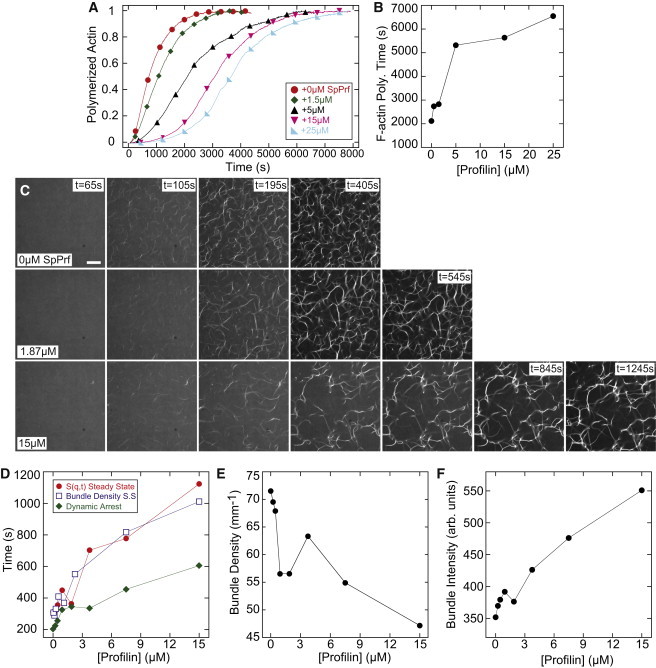
Profilin slows network assembly and promotes the formation of a network of sparse, thick bundles. (A) Time course of the spontaneous assembly of 5 μM Mg-ATP-actin monomers (10% pyrene labeled) in the presence of the indicated concentrations of SpPrf. (B) The 95% F-actin polymerization time from (A) as a function of SpPrf concentration. (C) Representative images of fluorescent (Alexa 488) phalloidin labeling of F-actin in networks forming via a spontaneous assembly of 5 μM Mg-ATP-actin, and 1.2 μM α-actinin, in the presence of varying concentrations of SpPrf. Polymerization is initiated at 0 s; scale bar = 30 μm. (D) The time to steady-state bundle density (open squares), S(q,t) steady state (solid circles), and dynamic arrest (solid diamonds) as a function of SpPrf concentration. (E) Steady-state linear bundle density as a function of SpPrf concentration. (F) Average steady-state bundle intensity as a function of SpPrf concentration.
To investigate the effect of profilin on steady-state network structure we spontaneously assembled networks of 5 μM actin, 1.2 μM α-actinin, and varying concentrations of SpPrf. At 0 μM SpPrf, individual filaments and bundles appear within 100 s of initiating polymerization. Dynamic arrest occurs at 200 s, and the steady-state structure is formed by 311 s (Fig. 2, C and D, Movie S1). The steady-state architecture contains a network of bundles with a high density (70 mm−1, Fig. 2 E), and low bundle intensity (350 units, Fig. 2 F). As the concentration of profilin increases, the time of structure formation and gelation increases concomitantly (Fig. 2, C–F). These changes in network assembly kinetics correlate with changes in the steady-state architecture, with the higher concentrations of profilin forming fewer, but thicker bundles. We observed qualitatively similar changes to network assembly kinetics and structure with mouse profilin pfn1 (Fig. S1, Movie S2). Thus, inhibiting filament nucleation with profilin has significant consequences on the network structure and assembly kinetics.
At all profilin concentrations, the actin assembly time was at least fivefold longer than network formation (Fig. 2, B and D). For samples containing 7.5 μM SpPrf, the steady-state bundle density is established at 800 s, but it takes >5000 s for 95% of the actin in the sample to polymerize. During this period of ∼4000 s, bundles are locked in place relative to one another and de novo bundle assembly is prohibited. As polymerization continues, the thickness of existing bundles increases (Fig. S2), presumably by recruiting new filaments or by the elongation of existing filaments. Thus, limiting the F-actin nucleation rate promotes the formation of a smaller number of thicker bundles and increases the amount of time required to reach a steady state.
Formin Cdc12 accelerates dynamics and increases bundle density of cross-linked actin networks in the presence of profilin
In the presence of saturating SpPrf concentrations, the fission yeast formin Cdc12 accelerates filament nucleation but does not significantly affect filament elongation rates (Table 1) (16). As Cdc12 is added to profilin-actin samples, the nucleation rate has been shown to increase proportionally with Cdc12 concentration (20,38). To assess this effect on actin assembly dynamics, we first examined actin assembly via pyrene fluorescence by spontaneously assembling samples of 5 μM G-actin, 15 μM SpPrf, and varying concentrations of Cdc12. Increasing the concentration of Cdc12 from 1 to 250 nM dramatically accelerates actin polymerization (Fig. 3 A), decreasing the time to 95% F-actin assembly from 5655 s to 975 s, with the effect of Cdc12 saturating at around 100 nM (Fig. 3 B).
Figure 3.
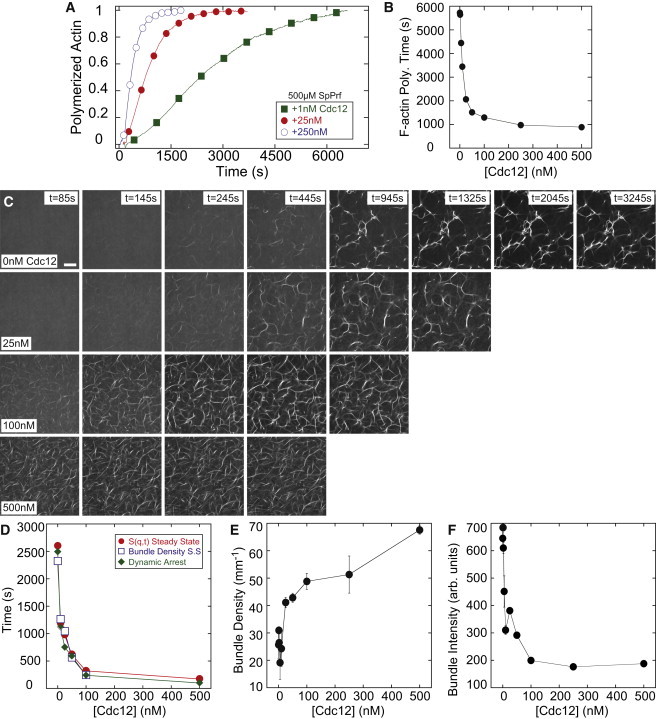
Cdc12 formin accelerates dynamics and increases the bundle density in profilin-actin networks. (A) Time course of the spontaneous assembly of 5 μM Mg-ATP-actin monomers (10% pyrene labeled) in the presence of 15 μM SpPrf and the indicated concentrations of Cdc12. (B) The 95% F-actin polymerization time from (A) as a function of Cdc12 concentration. (C) Representative images of fluorescent (Alexa 488) phalloidin labeling of F-actin in networks forming via a spontaneous assembling of 5 μM Mg-ATP-actin, 1.2 μM α-actinin, and 15 μM SpPrf in the presence of varying concentrations of Cdc12. Polymerization is initiated at t = 0 s; scale bar = 30 μm. (D) The time to steady-state bundle density (open squares), steady-state S(q,t) (solid circles), and dynamic arrest (solid diamonds) as a function of Cdc12 concentration. (E) Steady-state linear bundle density as a function of Cdc12 concentration. (F) Average bundle intensity as a function of Cdc12 concentration.
These changes in actin assembly have pronounced effects on the kinetics and steady-state architectures of networks formed with 1.2 μM α-actinin (Fig. 3 C, Movie S3). As the concentration of Cdc12 increases, the time at which filaments and bundles first appear decreases, from 245 s for 0 nM to <85 s (the first image acquired) for ≥100 nM. Increasing the concentration of Cdc12 from 0 to 100 nM dramatically reduces the time to form the steady-state architecture, assessed both by S(t) and the bundle density, as well as the dynamic arrest time (Fig. 3 D). The reduced network assembly time correlates with increased bundle density (Fig. 3 E) and reduced bundle thickness (Fig. 3 F). Effects saturate at Cdc12 concentrations higher than 100 nM (Fig. 3, C–F).
The results described in Figs. 2 and 3 indicate that the rate of filament nucleation plays a crucial role in the kinetics and steady-state architecture of actin networks. Cdc12 increases filament nucleation by a sufficient amount to recover the steady-state architecture formed by spontaneous assembly of actin and α-actinin alone, in the absence of SpPrf and Cdc12. A high filament nucleation rate promotes the formation of bundles at early times and the rapid integration of these bundles into a mechanically coherent network of thin actin bundles with high density. A low filament nucleation rate reduces the number of bundles, as well as the rate at which these bundles become arrested into networks.
A critical filament elongation rate is necessary for network gelation
To investigate the effect of filament elongation rates on network formation, we examined the assembly of F-actin and α-actinin structures in the presence of a saturating Cdc12 concentration (500 nM) and varying concentrations of SpPrf from 0 to 15 μM. Bulk assembly assays reflect a rapid assembly of F-actin over this concentration range, with 95% of F-actin being assembled within 200–600 s (Fig. 4 A). The elongation rate of Cdc12 bound filaments has been shown to increase 100-fold from 0.1 to 13.3 sub/(μM·s) from 0 to 5 μM SpPrf (16); these results are illustrated in Fig. 4 B. Because this increase in elongation rate incorporates more actin monomer per filament, the number of filaments nucleated over the course of a reaction decreases by 100-fold (Fig. 4 B) to yield the pyrene assembly curves in Fig. 4 A. Thus, by titrating SpPrf in the presence of saturating Cdc12, the elongation rate is dramatically altered.
Figure 4.
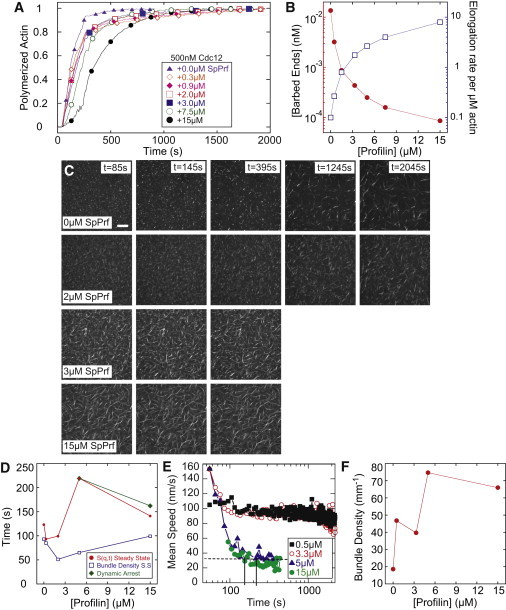
Network gelation is inhibited at low filament elongation rates. (A) Time course of the spontaneous assembly of 5 μM Mg-ATP-actin monomers (10% pyrene labeled) in the presence of 500 nM Cdc12 and the indicated concentrations of SpPrf. (B) Calculated barbed end concentration from (A) and elongation rate per formin-bound filament as a function of SpPrf concentration. (C) Representative images of fluorescent (Alexa 488) phalloidin labeling of F-actin in networks forming via a spontaneous assembling of 5 μM Mg-ATP-actin, 1.2 μM α-actinin, and 500 nM Cdc12 in the presence of varying concentrations of SpPrf. Polymerization is initiated at 0 s; scale bar = 30 μm. (D) The time to steady-state bundle density (open squares), structure factor S(q,t) (solid circles), and dynamic arrest (solid diamonds) for the samples described above as a function of SpPrf concentration. (E) The mean instantaneous speed as a function of time for bundles formed with 0.5 μM Cdc12 (black square), 3.3 μM Cdc12 (open circles, 5 μM Cdc12 (solid triangles), and 15 μM Cdc12 (solid circles). Only samples with 5 and 15 μM Cdc12 reached the threshold for dynamic arrest (horizontal dashed line). (F) Steady-state linear bundle density for samples with 500 nM Cdc12 as a function of SpPrf concentration.
The assembly of these networks in the presence of 1.2 μM α-actinin was examined by confocal microscopy. In all samples dense F-actin puncta were observed at 85 s after initiation of polymerization, consistent with rapid F-actin assembly (Fig. 4 C, Movie S4). Additionally, the bundle density reached saturating values within the first 200 s of each reaction (Fig. 4 D). Samples with elongation rates of ≤2.7 subunit/(μM·s) (≤3.3 μM SpPrf) continued to maintain high filament mobility well beyond the timescale of actin polymerization (Fig. 4 E). In these samples, bundles appeared to form by the annealing of shorter bundles (Fig. S3, Movie S5). During this extended period of high mobility, the bundles that form remained isolated from one another and were very straight. Very few branching points were observed. By contrast, in samples with elongation rates >4 subunit/(μM·s) (≥5 μM SpPrf), the arrest of dynamics occurred within 200 s, well before 95% actin polymerization. These networks were significantly branched and contained a higher density of bundles (Fig. 4, C and F). As such, there appears to be a critical elongation rate of ∼2.7 subunit/(μM·s), or 5 μM SpPrf, that is necessary to form a coherent, space-spanning network. As the overall filament length C is expected to scale with the elongation rate, these results may reflect a critical filament length and filament bending required for high angle cross-linking in network formation. Elongated structures that emerge from short filaments necessitate bundling, which increases the bending rigidity and reduces bending fluctuations that appear to prohibit high angle cross-linking.
Rapid filament elongation by mDia1 promotes network assembly at low F-actin concentrations
The mouse formin mDia1 promotes actin assembly in the presence of mouse profilin Pfn1 by increasing both filament nucleation and elongation rates (Table 1). At the ratios of profilin and actin used here, the elongation rate of mDia1 associated filaments increases to 48 sub/(μM·s) (16), resulting in dramatically enhanced actin assembly dynamics as the concentration of mDia1 is increased from 0 to 100 nM in samples containing 5 μM actin and 15 μM Pfn1 (Fig. 5 A). The time to assemble 95% of actin polymer is reduced from 8370 s to 1570 s as the concentration of mDia1 is increased from 0 to 100 nM (Fig. 5 B). Increased filament nucleation also corresponds to a fivefold increase in density of filament ends (Fig. 5 B).
Figure 5.
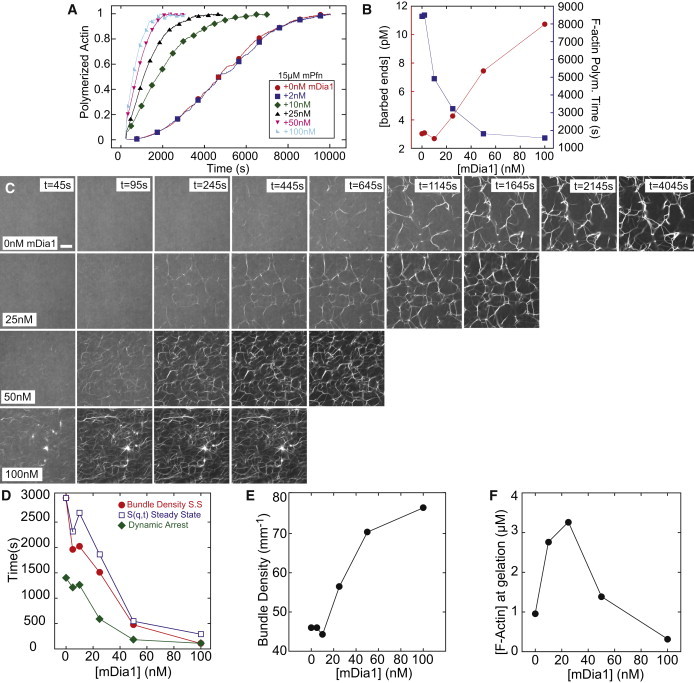
Rapid filament elongation by mDia1 formin promotes fast network assembly at low F-actin concentrations. (A) Time course of the spontaneous assembly of 5 μM Mg-ATP-actin monomers (10% pyrene labeled) in the presence of 15 μM pfn1 and the indicated concentrations of mDia1. (B) Calculated barbed end concentration (left, red circles) and the 95% F-actin polymerization time (right, blue squares) from (A) as a function of mDia1 concentration. (C) Representative images of fluorescent (Alexa 488) phalloidin-labeling of F-actin in networks forming via a spontaneous assembly of 5 μM Mg-ATP-actin, 1.2 μM α-actinin, and 15 μM pfn1 in the presence of varying concentrations of mDia1. Polymerization is initiated at t = 0 s; scale bar = 30 μm. (D) The time to steady-state bundle density (solid circles), S(q,t) steady state (open squares), and dynamic arrest (solid diamonds) as a function of mDia1 concentration. (E) Steady-state linear bundle density as a function of mDia1 concentration. (F) Concentration of actin polymer at gelation time as a function of mDia1 concentration.
Visualizing the network assembly revealed an accelerated filament and bundle appearance with increasing mDia1 concentrations (Fig. 5 C, Movie S6). Increasing the concentration of mDia1 from 0 to 100 nM reduced the time at which the structure factor and bundle density steady state was reached from ∼3000 to ∼200 s, and reduced the dynamic arrest time from 1215 to 115 s (Fig. 5 D). The linear bundle density increases and average bundle thickness decreases as the network assembles over a shorter period (Fig. 5, C–E), consistent with our results from Figs. 2 and 3.
As the concentration of mDia1 increases to 100 nM, a dynamically arrested network is formed in ∼100 s. We compared how much actin polymer is used to form networks at varying concentrations of mDia1 (Fig. 5 F). At 0 nM mDia1, ∼1 μM F-actin was required to form a mechanically coherent network. This amount increases to nearly 3 μM as mDia1 is increased to 25 nM. As mDia1 is increased further, the gelation time decreases faster than the actin polymerization rate increases, resulting in a steep reduction in the amount of F-actin present at the time of gelation. 100 nM mDia1 generates an integrated network with only 0.2 μM F-actin, or 4% of the monomeric pool available. This represents a very efficient network formation by mDia1, not only forming quicker than any other sample, but also using only a minimal amount of polymer.
Discussion
Formins have emerged as prominent regulators of actin filament assembly and are crucial for building diverse networks and bundles used in cell migration, division, and intracellular transport (4,7,39,40). Formin isoforms vary widely in their nucleation efficiency and elongation rates, and may therefore play important roles in forming distinct cytoskeletal structures in vivo. Here, we show that the F-actin nucleation and elongation rates regulated by formin and profilin play a crucial role in the kinetics and steady-state architecture of networks formed from identical concentrations of actin and the cross-linking protein α-actinin.
We found that nucleation potency plays an important role in controlling the network architecture and assembly kinetics (Fig. 6 A). At low filament nucleation rates, sparse filaments are freely mobile for long periods of time that permit their alignment into bundles. Gelation occurs once sufficient elongation of filaments/bundles results in interconnection by cross-linking and/or steric entanglements. As the nucleation rate increases, the average distance between filaments/bundles is reduced and, for a constant elongation rate, reduces the time for dynamic arrest to occur (Figs. 2 and 3, Fig. 6 A). As the F-actin nucleation rate changes, the amount of actin polymer needed to form a dynamically arrested network changes (see Fig. 6 C). In cases where polymerization proceeds past gelation, actin filaments continue to elongate and likely track along bundles to thicken the dynamically arrested bundles, but do not significantly alter the overall network architecture.
Figure 6.
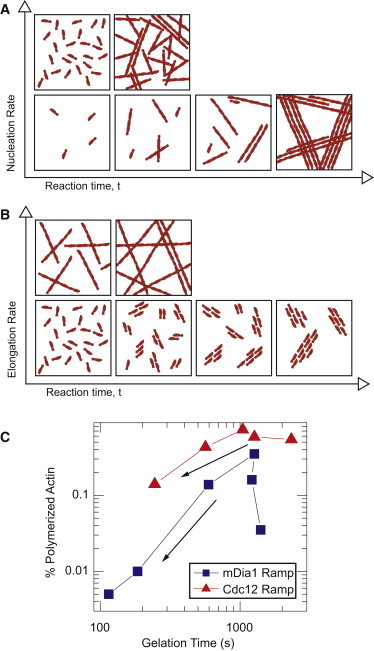
Network architecture is impacted by actin assembly kinetics. (A) Diagram of the effects of increasing nucleation rates on structure as a function of time. At low nucleation rates, networks form over a long period of time and at steady state take on well-spaced bundled structures. At accelerated nucleation rates, more filaments are present at early times, and rapidly form a network composed of lower intensity, but higher density, bundles. (B) Diagram of the effects of increasing elongation rates on network architecture as a function of time. At high elongation rates, actin is able to rapidly form an integrated network with very few actin filaments by reducing the time at which filaments overlap each other. At low elongation rates, a network is unable to form before actin is fully polymerized and forms thick and rigid bundles unable to form an interconnected gel. (C) Plot of the percent of actin monomers polymerized at gelation time across samples used in Figs. 3 and 5. The arrow indicates increasing concentrations of formin. Blue squares, correspond to 0, 5, 10, 25, 50, and 100 nM concentrations of mDia1. Red triangles correspond to concentrations 0, 50, 100, 250, and 500 nM of Cdc12.
Our results also identify an important role for filament elongation rates (Fig. 6 B). For very slow elongation rates (<3 subunits/(μM·s), a high density of very short filaments is formed. We speculate that a short filament length permits filaments and bundles to remain highly mobile for long periods. Local mobility promotes aggregation into thick and long bundles over long time periods and results in a very slow rate of bundle elongation, as compared to that facilitated by F-actin elongation. The increased bending rigidity of these bundles appears to diminish cross-linking into a space-spanning network. As filament elongation rates increase, the tendency to form entangled and cross-linked networks of bundles increases (Fig. 4 and Fig. 6 B). Longer filaments appear to permit high angle cross-linking, perhaps due to large bending fluctuations of individual filaments. As filament elongation increases, the time permissive to bundle formation decreases and the tendency to form dense networks of thin bundles is amplified. Moreover, we find that rapid elongation is able to speed up the gelation time and form networks at vanishingly low polymer densities (Fig. 6 C). Thus, nucleators such as formin mDia1 rapidly elongate filaments and still promote fast gelation, even if their nucleation potency is poor.
Our results shed light into the role of actin assembly factors in forming distinct actin architectures, ranging from isolated bundles to dense meshworks. The formation of rigid, isolated bundles is promoted by the rapid nucleation of very short and slowly elongating filaments that associate into bundles. Space-spanning networks require a critical elongation rate to filaments that are sufficiently long to become interconnected through steric entanglements or cross-links. Enhancing the rate of nucleation reduces the gelation time by speeding up filament assembly (Fig. 6 C). Despite its reduced nucleation potency, mDia1 permits the rapid gelation of networks with a minimal amount (0.5%) of actin polymer used (Fig. 6 C). We speculate that this enhanced gelation efficiency is due to its rapid elongation rate.
The implications for our results on the regulation of in vivo cytoskeletal architecture should be made carefully, as significant differences exist. Our in vitro studies are limited to concentrations of actin (5 μM) that are significantly lower than those found in the cytoplasm (∼50 μM). A lower filament density facilitates quantitative analysis of network architecture with light microscopy and increases the timescale of network formation. The higher concentrations of actin found in the cytoplasm will accelerate filament elongation and network gelation times as well as decrease the average distance between filaments. Although quantitative differences in the gelation time and network architectures surely exist when comparing the results presented here to in vivo systems, we expect the qualitative results to hold. Namely, relative changes in filament elongation rate and density mediated by changes in formin isoform or concentration will drive changes in the gelation rate and extent of bundling in actin networks. It will be interesting to see if such phenomena can be explored in the assembly of cytoskeletal structures in vivo. Additionally, nucleation in our in vitro assay occurred by freely diffusing formins and actin monomers, whereas some nucleation factors localize to membranes or discrete puncta in vivo (41). Such spatial constraints will qualitatively alter the physics controlling network and bundle architecture. Finally, further research is required to connect how changes in the local architecture of bundles are connected to the macroscopic rheological properties of these types of networks. It will be especially interesting to investigate how these structural changes impact the mechanical behavior of cross-linked actin networks and influence the nature of their force transmission, as this could reveal new mechanisms for the regulation of various cellular processes.
Acknowledgments
We are extremely grateful to Martin Lenz for helpful discussions, Patrick McCall for carefully reading the manuscript, and G. Danuser (Harvard) for use of quantitative Fluorescence Speckle Microscopy software developed in his lab.
This research was supported by a Burroughs Wellcome Career Award, Packard Fellowship, and National Institutes of Health (NIH) Director’s Pioneer Award (DP10D00354) to M.L.G., NIH RO1GM079265 to D.R.K. and the University of Chicago Materials Research and Science Consortium. T.T.F. was partially supported by the Howard Hughes Medical Institute and the National Institutes of Health Grant T32 GM007183.
Supporting Material
References
- 1.Galletta B.J., Cooper J.A. Actin and endocytosis: mechanisms and phylogeny. Curr. Opin. Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaksonen M., Toret C.P., Drubin D.G. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 3.Ridley A.J., Schwartz M.A., Horwitz A.R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 4.Kovar D.R., Kuhn J.R., Pollard T.D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vavylonis D., Wu J.Q., Pollard T.D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 6.Wu J.Q., Kuhn J.R., Pollard T.D. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 7.Yang C., Czech L., Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke M., Spudich J.A. Nonmuscle contractile proteins: the role of actin and myosin in cell motility and shape determination. Annu. Rev. Biochem. 1977;46:797–822. doi: 10.1146/annurev.bi.46.070177.004053. [DOI] [PubMed] [Google Scholar]
- 9.Stossel T.P. Contractile proteins in cell structure and function. Annu. Rev. Med. 1978;29:427–457. doi: 10.1146/annurev.me.29.020178.002235. [DOI] [PubMed] [Google Scholar]
- 10.Pollard T.D. Cytoskeletal functions of cytoplasmic contractile proteins. J. Supramol. Struct. 1976;5:317–334. doi: 10.1002/jss.400050306. [DOI] [PubMed] [Google Scholar]
- 11.Gardel M.L., Shin J.H., Weitz D.A. Elastic behavior of cross-linked and bundled actin networks. Science. 2004;304:1301–1305. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 12.Wachsstock D.H., Schwartz W.H., Pollard T.D. Affinity of alpha-actinin for actin determines the structure and mechanical properties of actin filament gels. Biophys. J. 1993;65:205–214. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wachsstock D.H., Schwarz W.H., Pollard T.D. Cross-linker dynamics determine the mechanical properties of actin gels. Biophys. J. 1994;66:801–809. doi: 10.1016/s0006-3495(94)80856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul A.S., Pollard T.D. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott B.J., Neidt E.M., Kovar D.R. The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell. 2011;22:3826–3839. doi: 10.1091/mbc.E11-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovar D.R., Harris E.S., Pollard T.D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 17.Carlsson L., Nyström L.E., Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 18.Moseley J.B., Sagot I., Goode B.L. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goode B.L., Eck M.J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 20.Pruyne D., Evangelista M., Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 21.Rizvi S.A., Neidt E.M., Kovar D.R. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin S.G., Chang F. Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 23.Loisel T.P., Boujemaa R., Carlier M.F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 24.Pujol T., du Roure O., Heuvingh J. Impact of branching on the elasticity of actin networks. Proc. Natl. Acad. Sci. USA. 2012;109:10364–10369. doi: 10.1073/pnas.1121238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernheim-Groswasser A., Wiesner S., Sykes C. The dynamics of actin-based motility depend on surface parameters. Nature. 2002;417:308–311. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- 26.Reymann A.C., Martiel J.L., Théry M. Nucleation geometry governs ordered actin networks structures. Nat. Mater. 2010;9:827–832. doi: 10.1038/nmat2855. [DOI] [PubMed] [Google Scholar]
- 27.Falzone T.T., Lenz M., Gardel M.L. Assembly kinetics determine the architecture of α-actinin cross-linked F-actin networks. Nat. Commun. 2012;3:861. doi: 10.1038/ncomms1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmoller K.M., Lieleg O., Bausch A.R. Structural and viscoelastic properties of actin/filamin networks: cross-linked versus bundled networks. Biophys. J. 2009;97:83–89. doi: 10.1016/j.bpj.2009.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieleg O., Kayser J., Bausch A.R. Slow dynamics and internal stress relaxation in bundled cytoskeletal networks. Nat. Mater. 2011;10:236–242. doi: 10.1038/nmat2939. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn J.R., Pollard T.D. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feramisco J.R., Burridge K. A rapid purification of alpha-actinin, filamin, and a 130,000-dalton protein from smooth muscle. J. Biol. Chem. 1980;255:1194–1199. [PubMed] [Google Scholar]
- 32.Lu J., Pollard T.D. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji L., Danuser G. Tracking quasi-stationary flow of weak fluorescent signals by adaptive multi-frame correlation. J. Microsc. 2005;220:150–167. doi: 10.1111/j.1365-2818.2005.01522.x. [DOI] [PubMed] [Google Scholar]
- 34.Higgs H.N., Blanchoin L., Pollard T.D. Influence of the C-terminus of Wiskott-Aldrich Syndrome Protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 35.Pollard T.D., Aebi U., Tseng P. Cold Spring Harbor Laboratory Press; New York: 1982. Actin Structure, Polymerization, and Gelation. [DOI] [PubMed] [Google Scholar]
- 36.Pollard T.D., Aebi U., Tseng P.C. Actin and myosin function in acanthamoeba. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982;299:237–245. doi: 10.1098/rstb.1982.0129. [DOI] [PubMed] [Google Scholar]
- 37.Kasai M., Asakura S., Oosawa F. The cooperative nature of G-F transformation of actin. Biochim. Biophys. Acta. 1962;57:22–31. doi: 10.1016/0006-3002(62)91073-9. [DOI] [PubMed] [Google Scholar]
- 38.Sagot I., Rodal A.A., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 39.Hotulainen P., Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard T.D., Cooper J.A. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovar D.R., Sirotkin V., Lord M. Three’s company: the fission yeast actin cytoskeleton. Trends Cell Biol. 2011;21:177–187. doi: 10.1016/j.tcb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neidt E.M., Skau C.T., Kovar D.R. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J. Biol. Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F., Higgs H.N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


