Abstract
There is increasing interest in identifying new pathways and candidate genes that confer susceptibility to osteoporosis. There is evidence that adipogenesis and osteogenesis may be related, including a common bone marrow progenitor cell for both adipocytes and osteoblasts. Perilipin 1 (PLIN1) and Perilipin 4 (PLIN4) are members of the PATS family of genes and are involved in lipolysis of intracellular lipid deposits. A previous study reported gender-specific associations between one polymorphism of PLIN1 and bone mineral density (BMD) in a Japanese population. We hypothesized that polymorphisms in PLIN1 and PLIN4 would be associated with bone measures in adult Caucasian participants of the Framingham Osteoporosis Study (FOS). We genotyped 1,206 male and 1,445 female participants of the FOS for four single-nucleotide polymorphism (SNPs) in PLIN1 and seven SNPs in PLIN4 and tested for associations with measures of BMD, bone ultrasound, hip geometry, and height. We found several gender-specific significant associations with the measured traits. The association of PLIN4 SNP rs8887, G>A with height in females trended toward significance after simulation testing (adjusted P = 0.07) and remained significant after simulation testing in the combined-sex model (adjusted P = 0.033). In a large study sample of men and women, we found a significant association between one SNP in PLIN4 and height but not with bone traits, suggesting that PATS family genes are not important in the regulation of bone. Identification of genes that influence human height may lead to a better understanding of the processes involved in growth and development.
Keywords: Perilipin 1, Perilipin 4, Bone mineral density, Bone geometry, Framingham Osteoporosis Study
Osteoporosis is a skeletal disorder characterized by compromised bone strength and subsequent increase in fracture risk. In the United States alone, 10% of adults aged 50 years or more suffer from osteoporosis, and the direct care expenditures for osteoporotic fractures are estimated to be $12–$18 billion annually. The incidence of osteoporosis is expected to increase significantly in the next few years due to extended life expectancies and the aging of the US population [1]. Twin and family studies have implicated genetic factors in up to 75% of the intersubject variance in the regulation of bone mineral density (BMD) and other determinants of osteoporotic fracture risk, such as quantitative ultrasound properties of bone and skeletal geometry [2–5]. Height itself may also be considered a bone-related phenotype since the phenotype is largely explained by the length of the legs and vertebral segments [6–8]. Based on numerous candidate gene-association studies and recent genome-wide association studies, there is reason to believe that previously unidentified pathways and new candidate genes will be described that confer susceptibility to osteoporosis [9–13].
A common bone marrow progenitor cell for both adipocytes and osteoblasts suggests that adipogenesis and osteogenesis may be related [14–16]. Osteoporotic bone has been noted to have an expansion of adipose tissue in the bone marrow at the expense of osteogenic cells, a finding that has been reported in several species including humans and mice [17–19]. Lineage-specific transcription factors that direct differentiation of this common progenitor have been identified, including the runt-related transcription factor 2 (Runx2) for osteoblastogenesis and the peroxisome proliferator-activator gamma (PPARγ) for adipogenesis [20, 21]. An inverse relationship appears to exist between the two lineages, with agents inducing osteoblast differentiation inhibiting adipogenesis and vice versa [21]. Polymorphisms in PPARγ have been shown to affect bone mass in mice and humans [22]. If there are direct effects of adipocytes on other marrow progenitors vital to bone health, there may be associations between genes that regulate fat metabolism and skeletal integrity. Finally, an inverse relation between fat mass and bone density has been observed in both cross-sectional and longitudinal studies in different age groups [23–25]. For these reasons, we examined two genes involved in fat metabolism to investigate their association with a variety of skeletal phenotypes.
Perilipin, whose HUGO Gene Nomenclature Committee name is now Perilipin 1 (PLIN1), encodes a protein that was first identified in the late 1980 s by Londos et al. [26] and named “perilipin” in reference to its physical location surrounding lipid droplets [27]. PLIN4, formerly known as S3-12, which has sequence similarity to PLIN1, was found in a screen for adipocyte-specific proteins, suggesting that it is involved in adipocyte lipid storage. As such, both PLIN1 and PLIN4 are members of the “PATS” family of proteins (PLIN, ADFP, TIP-47, S3-12), a group of structural proteins defined by their protein sequence similarity and association with lipid droplets [28]. These proteins are found almost exclusively in adipocytes and steroidogenic cells and play a vital role in the lipolysis of intracellular lipid deposits [29, 30]. Expression of these genes is elevated in obese animals and humans, with PLIN1 being the most studied of the group [28–33]. Polymorphisms in PLIN1 have been shown to have a female-specific association with obesity in various ethnic groups [31–33].
Yamada et al. [34] studied the PLIN1 1243C>T (rs2304796) single-nucleotide polymorphism (SNP) and its relationship to BMD in a community-dwelling Japanese population. They found a male-specific association, with the TT genotype having greater BMD for the total hip, lumbar spine, femoral neck, and trochanter than the combined CC and CT genotypes. Here, we studied PLIN1 and PLIN4 SNPs and their relation to BMD, broadband ultrasound attenuation (BUA), bone geometric traits, and height in participants of the Framingham Osteoporosis Study (FOS), a population of European ancestry. We hypothesized that SNPs in PLIN4 and PLIN1 would be associated with these bone properties.
Materials and Methods
Study Sample
The FOS is an ancillary study of the Framingham Heart Study. The Framingham Heart Study began in 1948 with the primary goal of prospectively evaluating the multivariable risk factors for cardiovascular disease. The original cohort participants were selected from a systematic sampling of two-thirds of the households of the population of Framingham, MA, at that time. In 1971, the Framing-ham Offspring Cohort Study was initiated with the intent to evaluate the role of genetic factors in the etiology of coronary artery disease and was comprised of 71% of all the eligible adult offspring of couples from the original cohort and offspring’s spouses. The offspring cohort consists of 5,124 subjects, of whom 2,616 are offspring of the 1,164 original cohort spouse pairs, 34 are stepchildren, 898 are offspring of one parent in the original cohort at greater risk of cardiovascular disease, and 1,576 are spouses. After their initial evaluation, these individuals underwent repeat examinations approximately every 4 years and participated in the FOS at either examination cycle 6 or 7 from 1996 to 2001. The design of this study has been described previously [35–38].
Our genetic analysis included an unselected subset of 2,651 (1,206 men and 1,445 women) offspring subjects (age range 29–94 years) who had both genotyping and at least one of the measured phenotypes available. Among these 2,651 participants with PLIN1 or PLIN4 genotyping, 2,448 (55.08% female, age 62.06 ± 10.59, range 29–94) had valid BMD measures of the total hip, 2,495 (55.37% female, age 62.14 ± 10.30, range 29–90) had BMD measures of the spine, 2,504 (55.3% female, age 62.1 ± 10.3, range 29–90) had BMD measures of the trochanter, 2,505 (55.4% female, age 62.1 ± 10.38, range 29–90) had BMD measures of the femoral neck, 2,435 (56.07% female, age 62.51 ± 10.86, range 29–94) had BUA, 2,468 (55.4% female, age 62.0 ± 10.2, range 31–90) had femoral neck length measurements, and 1,400 (53.4% female, age 54.5 ± 8.9, range 29–81) had metacarpal length measurements. All participants had height measurements obtained at or several years before the time of the bone exam. Neither the Framingham original nor offspring cohort was selected on the basis of cardiovascular diseases or osteoporosis. This study was approved by the Institutional Review Board for Human Subjects Research of Boston University, Tufts University, and the Hebrew Rehabilitation Center.
Measurement of BMD and Quantitative Ultrasound
Participants underwent bone densitometry by DXA with a Lunar (Madison, WI) DPX-L from 1996 to 2001. The coefficients of variation (CV) in normal subjects for the DPX-L have been previously noted to be 0.9% for the lumbar spine, 1.7% for the femoral neck, and 2.5% for the trochanter [37]. Calcaneal quantitative bone ultrasonography was performed to obtain right calcaneal BUA with a portable device, the Sahara® bone sonometer (Hologic, Waltham, MA), in members of the offspring cohort between 1996 and 2001. Based on duplicate, same-day measurements on 29 subjects, the CV for BUA was 5.3% [38].
Measurement of Bone Geometry
Hip Geometry
DXA scans obtained in 1992–1993 or 1996–1997 (for 31 original cohort subjects who missed the earlier measured DXAs) were taken with an interactive computer program [39]. The program derived a number of proximal femoral structural variables, including femoral neck length (FNL), defined as the distance from the center of the femoral head to the intersection of the neck and shaft axes; the outer diameters at both the femoral neck and the femoral shaft; as well as the neck-shaft angle (see [40] for details). CVs for the different geometric indices were previously reported, ranging from 3.3% (femoral neck outer diameter) to 9.1% (FNL) [12].
Metacarpal Measurements
As part of a study investigating the heritability of osteoarthritis, participants of the FOS who had at least one parent (member of the original cohort) with hand radiographs were radiographed using the same techniques in 1993–1995. Hand radiographs were digitized at 450-dpi resolution using a COBRAscan 612 SL Digitizer (Radiographic Digital Imaging, Compton, CA) and Windows Imaging software (Microsoft, Redmond, WA) to acquire an image with a pixel size of 0.12 mm and 1,024 gray levels. A custom-written script developed for ImageTool software (UTHSCSA, San Antonio, TX) was used to semiautomatically obtain measurements of the second, third, and fourth metacarpals, by outlining the contour of the digitized metacarpal bones of interest on a flat-screen monitor. The algorithm implemented in the software then determined the coordinates of outlined bone, generated a mass profile, and converted the data into the cortical macrostructure. For our analyses we focused on lengths of the second through fourth metacarpals and averaged them as previously described [41]. Finally, considering height to be a bone-related phenotype, it was measured to the nearest one-fourth inch using a stadiometer [35, 37].
Covariables
Information including age, sex, weight, and, for women, estrogen use and menopausal status was obtained at the time of bone density/ultrasound and geometry measurements. Weight was measured using a standardized balance beam scale. Body mass index (BMI, kg/m2) was calculated using height measured as above. Each woman was assigned to one of the two estrogen status groups: (1) premenopausal or postmenopausal women taking estrogen (estrogen-replete) or (2) postmenopausal not on estrogen (estrogen-deplete), where menopause was defined as having no menstrual period for at least 1 year [35, 37].
SNP Selection and Genotyping
The variants chosen for PLIN1—rs2289487, T>C (intron 2), named PLIN1 SNP; rs894160, G>A (intron 6), named PLIN4 SNP; rs2304795, A>G (exon 8), named PLIN5 SNP; and rs1052700, A>T (exon 9), named PLIN6 SNP—were studied based in part on our results from previous publications [32, 42]. To assist selection of SNPs in the PLIN1 locus, we performed a search of the CEU population HapMap PHASE II data for polymorphic alleles with a minor allele frequency (MAF) ≥ 5% at the PLIN1 locus, defined as 5 kb upstream of the predicted start codon and 2 kb downstream of the termination codon. Eleven SNPs were identified within this region in the CEU population of HapMap, but only five had MAFs ≥ 5%. The rs894160 SNP was in linkage disequilibrium (LD) with rs8179043, having r2 = 1, so rs894160 was used as a tag SNP. These four SNPs provide coverage of all common variation within the PLIN1 locus, as defined above.
To identify common SNPs in the human PLIN4 locus, we performed a search of the HapMap database for polymorphic alleles with an MAF ≥ 5%. The PLIN4 locus was defined as 5 kb upstream of the predicted start codon and 2 kb downstream of the termination codon, a region spanning approximately 25.1 kb. Thirteen such SNPs were identified in this region. We chose eight of these SNPs for genotyping: two promoter (rs884164 and rs1609717), one exonic missense (rs7250947), one 3′-UTR (rs8887), and four intronic (rs8102428, rs892158, rs4807598, and rs11673616). Analysis in the CEU population of the Hap-Map database with the Haploview program determined rs892158 (rs7260518 and rs10406797), rs7250947 (rs8102428 and rs884164), rs11673616 (rs4991027), and rs1609717 (rs4807598) to be tag SNPs capturing variants (in parentheses) in LD with r2 >0.8 over 11 of 13 SNPs in the region.
DNA was isolated from blood samples using DNA blood Midi kits (Qiagen, Hilden, Germany) according to the vendor’s recommended protocol. Ready-made 5′ nucleic allelic discrimination assays were available from Applied Biosystems (Foster City, CA) for the PLIN4 SNPs rs8887, rs11673616, rs892158, rs8102428, and rs884164. We used the Applied Biosystems Custom Assay design Web tool to generate functional assays for SNPs rs1609717 and rs7250947 (appliedbiosystems.com). We performed genotyping of PLIN1 and PLIN4 SNPs, using TaqMan assays on the ABIPrism 7900HT Sequence Detection System (Applied Biosystems). Standard laboratory practices were used to ensure the accuracy of the data.
Statistical Analysis
MAF estimation and Hardy–Weinberg equilibrium (HWE) testing were performed using unrelated individuals. We used an exact Chi-squared test statistic (Genetic Package) to compare observed genotype frequencies to those expected under HWE. We excluded one SNP (PLIN4, rs4807598) from analysis because of significant deviation from HWE (P = 0.001).
To test for association between SNPs and height, BMD, geometry, and metacarpal measures, we used sex-specific regression models with height and bone measures as dependent variables and SNPs and covariates as independent variables. To account for within-familial correlation, linear mixed effects (LME) regression was used (lmekin, R-Kinship Package). We used a codominant model (comparing three genotype groups, 2 df test) for five SNPs with ≥ 20% MAF and a dominant model (combining the heterozygotes and minor allele homozygotes) for the remaining six SNPs with <20% MAF. Association analyses between SNPs and height were adjusted for age and estrogen status in women, and those for bone measures were adjusted for age, BMI, height, and estrogen status in women. We also performed sex-combined analyses, additionally adjusting for sex.
We used simulation methods to adjust for the number of statistical tests while accounting for the dependency of tests due to LD among the SNPs and correlation among phenotypes. First, independently of phenotypes and covariates, we simulated SNP genotypes with allele frequencies and LDs similar to the observed genotype data (simqtl, SOLAR, http://www.sfbr.org [38]). Second, we merged the set of phenotypes and covariates for each individual with the simulated genotype, while maintaining the allele frequency, LD information, Mendelian consistency, and correlation among phenotypes and covariates. Then, using the simulated data, we performed LME regression analysis to obtain sex- and gene-specific minimum P values. This procedure was repeated 1,000 times, and for each test the adjusted P value was calculated as the proportion of simulated data sets in which the minimum P value was less than the observed P value. “Adjusted P values” are significant when they are below P <0.05 as these have been adjusted for multiple comparisons using simulation testing.
We calculated the proportion of variance explained by SNP effects for those traits with significant P values. Among significant P values in men, h2 ranged from 0.35% to 0.58%. Among significant P values in women, h2 ranged from 0.26% to 0.87%. We then used QUANTO (http://hydra.usc.edu/gxe/) to compute the power for males (n = 1,206) and females (n = 1,446) using a significance level of 0.05. With h2 of 0.5%, the power is 70% for males and 77% for females.
Results
Subject Characteristics
Table 1 contains the descriptive characteristics of FOS participants included in this analysis by gender with bone density/ultrasound measures and bone geometric traits. Both men and women were of similar age (mean 62 years) and similar BMI measurements (mean 28 kg/m2). Among the 1,445 women, 42% were classified as estrogen status–positive, defined as premenopausal women or postmenopausal women taking estrogen supplementation. In general, compared to women, men had higher BMD/BUA measurements by 0.11–0.18 g/cm2 for the femoral neck, trochanter, spine, and total hip and by 12.5 db/mHz for BUA. In addition, men had greater values for the bone geometric traits compared to women, with larger values of FNL and metacarpal length by 0.88 and 0.58 cm, respectively.
Table 1.
Subject characteristicsa
| Characteristics | Males (n = 1,206)
|
Females (n = 1,446)
|
P | ||
|---|---|---|---|---|---|
| n | Mean ± SD | n | Mean ± SD | ||
| Age (years) | 1,078 | 62.4 ± 10.0 | 1,317 | 61.8 ± 10.3 | 0.1124 |
| BMI (kg/m2) | 1,074 | 28.7 ± 4.5 | 1,313 | 27.5 ± 5.6 | <0.0001 |
| Height (cm) | 1,074 | 174.5 ± 7.1 | 1,313 | 160.5 ± 6.6 | <0.0001 |
| Estrogen status positiveb (%) | N/A | 562/1,316 (42.71%)c | N/A | ||
| Bone density/ultrasound measurements | |||||
| Spine BMD (g/cm2) | 1,063 | 1.33 ± 0.20 | 1,302 | 1.15 ± 0.20 | <0.0001 |
| Femoral neck BMD (g/cm2) | 1,068 | 0.97 ± 0.14 | 1,306 | 0.86 ± 0.15 | <0.0001 |
| Trochanter BMD (g/cm2) | 1,068 | 0.89 ± 0.14 | 1,305 | 0.71 ± 0.14 | <0.0001 |
| Total hip BMD (g/cm2) | 1,050 | 1.04 ± 0.14 | 1,275 | 0.91 ± 0.15 | <0.0001 |
| BUA (db/mHz) | 1,022 | 82.7 ± 18.9 | 1,290 | 70.2 ± 19.2 | <0.0001 |
| Bone geometry measurements | |||||
| Femoral neck length (cm) | 1,053 | 5.97 ± 5.97 | 1,289 | 5.09 ± 0.67 | <0.0001 |
| Metacarpal length (cm) | 613 | 6.71 ± 0.38 | 701 | 6.13 ± 0.34 | <0.0001 |
| Age (years) | 613 | 54.9 ± 8.9 | 701 | 54.3 ± 9.0 | 0.2493 |
| BMI (kg/m2) | 610 | 28.4 ± 4.0 | 697 | 27.0 ± 5.7 | <0.0001 |
| Height (cm) | 610 | 175.0 ± 6.9 | 697 | 161.3 ± 5.8 | <0.0001 |
| Estrogen status positiveb (%) | N/A | 339/698 (48.57%)c | N/A | ||
Values taken for subjects with PLIN1 genotyping; not statistically different from the subgroup of subjects with PLIN4 genotyping
Estrogen status positive includes premenopausal women and postmenopausal women taking estrogen
Prevalence count/total count (percentage)
Single-Nucleotide Polymorphisms
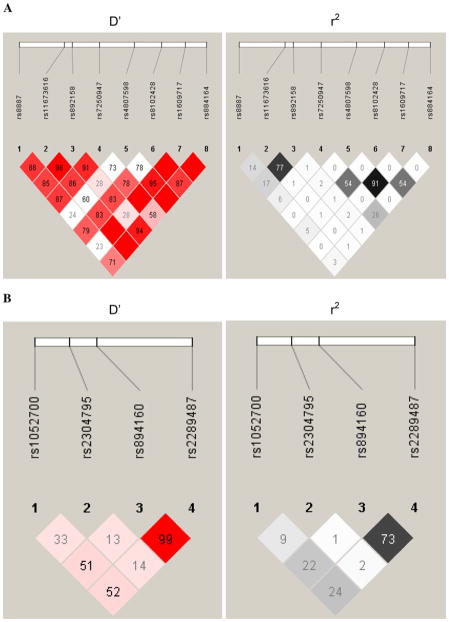
Table 2 contains detailed information regarding the seven polymorphisms in PLIN4 and the four polymorphisms in PLIN1 evaluated in this analysis. One of the PLIN4 SNPs (rs4807598) was not in HWE and was therefore not included in this analysis. Figure 1a shows the LD for each pair of PLIN4 SNP markers using D′ and r2 calculated from estimated haplotype frequencies. Figure 1b shows the PLIN1 LD for each pair of markers. MAFs of the SNPs ranged from 4.7% to 44.7%, with no significant differences in the genotype frequencies by sex.
Table 2.
PLIN1 and PLIN4 SNPs genotyped in this study
| Gene | SNP rs number | Positiona | Location | Alleles | MAF (%) | HWE P | |
|---|---|---|---|---|---|---|---|
| 1 | PLIN4 | rs8887 | 4453201 | Downstream | G >A | 44.7 | 0.92 |
| 2 | PLIN4 | rs11673616 | 4457915 | Intron 4 | A >G | 13.2 | 0.68 |
| 3 | PLIN4 | rs892158 | 4458716 | Intron 4 | G >A | 16.2 | 0.38 |
| 4 | PLIN4 | rs7250947 | 4461530 | Exon 3 | G >A | 9.5 | 0.40 |
| b | PLIN4 | rs4807598 | 4465135 | Intron 2 | T >C | 4.7 | 0.001 |
| 5 | PLIN4 | rs8102428 | 4467982 | Intron 1 | A >G | 10.5 | 0.80 |
| 6 | PLIN4 | rs1609717 | 4470450 | Promoter | A >G | 4.7 | 0.05 |
| 7 | PLIN4 | rs884164 | 4472625 | Downstream | A >G | 7.6 | 0.61 |
| 8 | PLIN1 | rs1052700 | 88009314 | 3′-UTR | T >A | 34.2 | 0.64 |
| 9 | PLIN1 | rs2304795 | 88011267 | Pro371Pro | T >C | 38 | 0.25 |
| 10 | PLIN1 | rs894160 | 88012827 | Intron 6 | C >T | 29.8 | 0.69 |
| 11 | PLIN1 | rs2289487 | 88018100 | Intron 2 | T >C | 36.8 | 0.76 |
Position relative to NCBI coordinates (viewed 08/20/08)
This SNP was not in Hardy–Weinberg equilibrium and is not included in this analysis
Fig. 1.
a LD for each pair of PLIN4 SNP markers using D′ and r2 calculated from estimated haplotype frequencies. b LD for each pair of PLIN1 SNP markers using D′ and r2 calculated from estimated haplotype frequencies
Individual SNP Associations with BMD/BUA and Geometric Traits
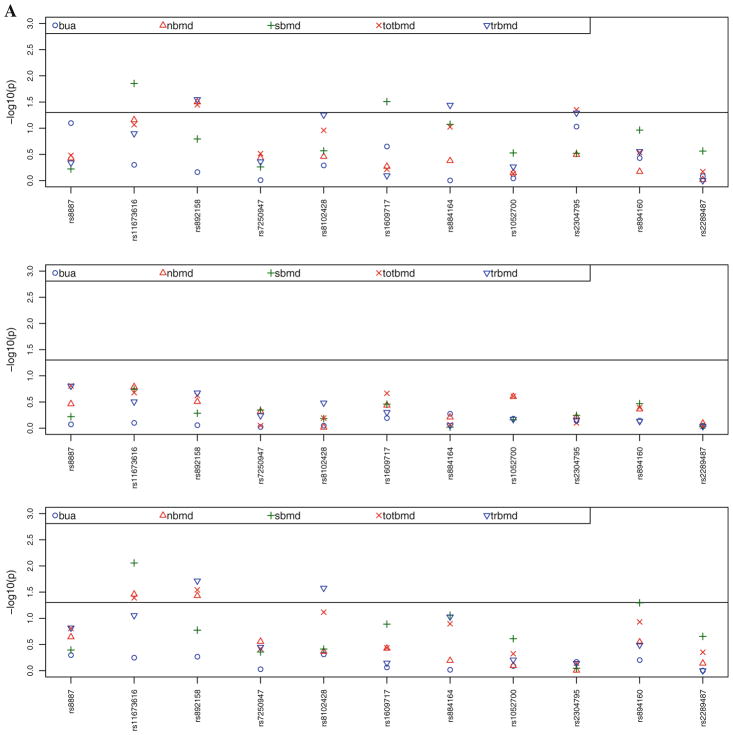
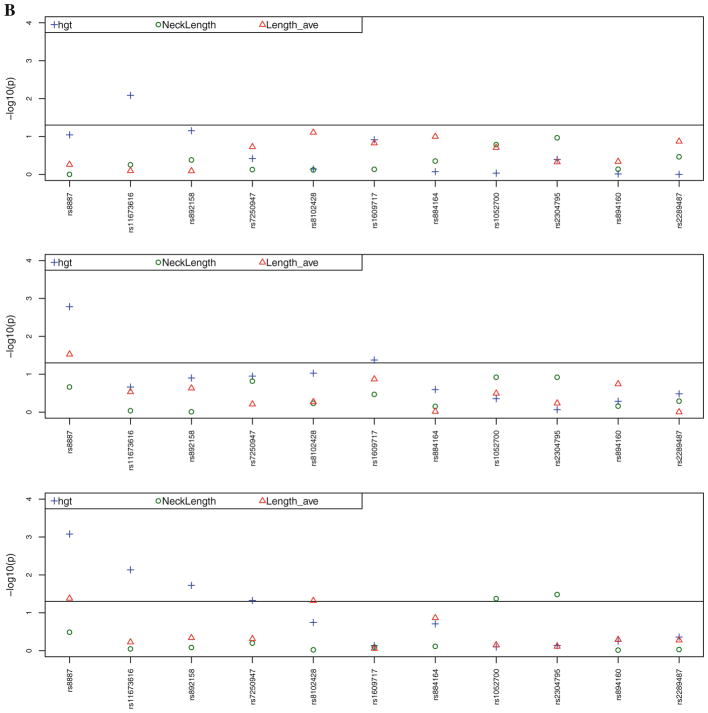
Figure 2a contains the multivariate adjusted, sex-specific (age, BMI, height, and estrogen status in women) P values for the associations between the seven SNPs in the PLIN4 gene and the four SNPs in the PLIN1 gene with BMD for the total hip, femoral neck, trochanter, and spine and BUA. Figure 2b contains the multivariate adjusted (age, BMI, height, and estrogen status in women) P values for the associations between SNPs in the PLIN4 and PLIN1 genes and height, FNL, and metacarpal length. Table 3 displays only the significant (P <0.05) gender-specific associations and the magnitude of these associations in men and women between the PLIN4 and PLIN1 SNPs and BMD/BUA and geometric traits, including the data for nonsignificant findings in the other gender. Our analysis revealed that eight of the 11 SNPs were associated at a nominal significance level with one or more of the measurements of bone density/ultrasound or geometry after adjustment for covariates.
Fig. 2.
a Adjusted P values for the associations between SNPs in the PLIN4 and PLIN1 genes and BMD for the total hip (totbmd), femoral neck (nbmd), trochanter (trbmd), spine (sbmd), and right calcaneal broadband ultrasound attenuation (bua) (adjusted for age, BMI, height, and estrogen status in women). SNPs rs8887, rs11673616, rs892158, rs7250947, rs8102428, rs1609717, and rs884164 are from PLIN4 and rs1052700, rs2304795, rs894160, and rs2289487 are from PLIN1. First panel represents the results for males, second panel for females, and third panel the combined-gender results. b Adjusted P values for the associations between SNPs in the PLIN4 and PLIN1 genes and height (hgt, adjusted for age and estrogen status in women), femoral neck length (NeckLength), and metacarpal length (Length_-ave) (adjusted for age, BMI, height, and estrogen status in women). First panel represents the results for males, second panel for females, and third panel the combined-gender results
Table 3.
Gender-specific associations of selected PLIN1 and PLIN4 SNPs and bone density/ultrasound and geometric traits
| Men | Genotype | Adjusted mean | P | Women | Genotype | Adjusted mean | P |
|---|---|---|---|---|---|---|---|
| Geometry | Geometry | ||||||
| SNP 1: Metacarpal length | GG | 6.70 | 0.550 | SNP 1: Metacarpal length | GG | 6.10 | 0.030 |
| GA | 6.72 | GA | 6.12 | ||||
| AA | 6.72 | AA | 6.18 | ||||
| SNP 1: Height | GG | 68.97 | 0.090 | SNP 1: Height | GG | 63.28 | 0.002 |
| GA | 68.79 | GA | 63.47 | ||||
| AA | 68.46 | AA | 62.87 | ||||
| SNP 2: Height | AA | 68.90 | 0.008 | SNP 2: Height | AA | 63.34 | 0.218 |
| AG and GG | 68.40 | AG and GG | 63.15 | ||||
| SNP 6: Height | AA | 68.74 | 0.122 | SNP 6: Height | AA | 63.34 | 0.042 |
| AG and | 69.18 | AG and GG | 62.89 | ||||
| Bone density/ultrasound | Bone density/ultrasound | ||||||
| SNP 2: Spine BMD | AA | 1.34 | 0.014 | SNP 2: Spine BMD | AA | 1.16 | 0.181 |
| AG and GG | 1.30 | AG and GG | 1.14 | ||||
| SNP 3: Neck BMD | GG | 0.98 | 0.032 | SNP 3: Neck BMD | GG | 0.86 | 0.312 |
| GA and AA | 0.96 | GA and AA | 0.86 | ||||
| SNP 3: Trochanter BMD | GG | 0.89 | 0.028 | SNP 3: Trochanter BMD | GG | 0.72 | 0.211 |
| GA and AA | 0.87 | GA and AA | 0.71 | ||||
| SNP 3: Total hip BMD | GG | 1.05 | 0.036 | SNP 3: Total hip BMD | GG | 0.91 | 0.238 |
| GA and AA | 1.03 | GA and AA | 0.90 | ||||
| SNP 6: Spine BMD | AA | 1.32 | 0.031 | SNP 6: Spine BMD | AA | 1.15 | 0.348 |
| AG and GG | 1.37 | AG and GG | 1.17 | ||||
| SNP 7: Trochanter BMD | AA | 0.88 | 0.036 | SNP 7: Trochanter BMD | AA | 0.71 | 0.889 |
| AG and GG | 0.91 | AG and GG | 0.71 | ||||
| SNP 9: Total hip BMD | TT | 1.05 | 0.044 | SNP 9: Total hip BMD | TT | 0.91 | 0.794 |
| TC | 1.04 | TC | 0.91 | ||||
| CC | 1.02 | CC | 0.91 |
Results multivariate adjusted, only results with P <0.05 or the corresponding data for the other gender when one gender is significant
P values in bold are significant
The combined-gender results showed significant associations at a nominal level between PLIN4 SNP 1 (rs8887, G>A) with height and metacarpal length; PLIN4 SNP 2 (rs11673616, A>G) with height and BMD of the spine, femoral neck, and total hip; PLIN4 SNP 3 (rs892158, G>A) with height and BMD of the femoral neck, trochanter, and total hip; PLIN4 SNP 4 (rs7250947, G>A) with height; and PLIN4 SNP 5 (rs8102428, A>G) with metacarpal length and BMD of the trochanter. PLIN1 SNPs were not associated with any of the phenotypes examined in the combined-gender analysis.
Among men, we found significant associations at a nominal level between PLIN4 SNP 2 (rs11673616, A>G) with height and BMD of the spine; PLIN4 SNP 3 (rs892158, G>A) with BMD of the femoral neck, trochanter, and total hip; PLIN4 SNP 6 (rs1609717, A>G) with BMD of the spine; PLIN4 SNP 7 (rs884164, A>G) with BMD of the trochanter; and PLIN1 SNP 9 (rs2304795, A>G) with BMD of the total hip. In women, we noted significant associations between PLIN4 SNP 1 (rs8887, G>A) with metacarpal length and height and SNP 6 (rs1609717, A>G) with height. None of the studied PLIN4 SNPs was significantly associated with the measured BMD/BUA traits in women.
Accounting for the multiple testing, the association of SNP 1 with height remained significant in the combined-sex results with adjusted P = 0.033. None of the gender-specific associations remained significant for either PLIN1 or PLIN4, although the association of PLIN4 SNP 1 (rs8887, G>A) with height in females was borderline significant with adjusted P = 0.07.
Combined SNP Associations across PLIN1 and PLIN4 with BMD/BUA and Geometric Traits
SNPs that were shown to have significant individual association(s) with the studied BMD, BUA, and bone geometric traits as shown in Table 3 were selected for additional analyses to evaluate the combined effect of these SNPs (SNP 1, SNP 2, SNP 3, SNP 6, SNP 7, and SNP 9) on the selected traits. Since SNP 2 and SNP 3 have high LD (r2 = 0.77), we also evaluated a model excluding SNP 3 (model 2) and a model excluding SNP 2 (model 3). In these models, we adjusted for age, BMI, height, and estrogen status in women. To evaluate the combined effect on height, we adjusted for age and estrogen status (in women only).
-
Model 1
SNP1 + SNP2 + SNP3 + SNP6 + SNP7 + SNP9 + age + BMI + height + estrogen status
-
Model 2
SNP1 + SNP2 + SNP6 + SNP7 + SNP9 + age + BMI + height + estrogen status
-
Model 3
SNP1 + SNP3 + SNP6 + SNP7 + SNP9 + age + BMI + height + estrogen status
Among men, we found a significant combined SNP effect on spine BMD in model 2 (P = 0.0375) and model 3 (P = 0.0485), where SNP 2 and SNP 6 have significant individual P values. Trochanter BMD had a significant combined SNP effect in model 1 (P = 0.0363) and model 3 (P = 0.0397,) where SNP 3 and SNP 7 have significant individual P values. Total hip BMD had a significant combined SNP effect in model 1 (P = 0.0420) and model 3 (P = 0.0312), where SNP 3 and SNP 9 have significant individual P values.
Among women, we found a significant combined SNP effect on height in all three models, model 1 (P = 0.028), model 2 (P = 0.0155), and model 3 (P = 0.0147), where SNP 1 and SNP 6 have significant individual P values.
Discussion
We evaluated several polymorphisms in the PLIN1 and PLIN4 genes for associations with BMD/BUA, geometric traits, and height in this large subset from the FOS.
In this cohort of Caucasians mostly from European ancestry, we detected several significant (P <0.05) gender-specific and combined-sex associations in the measured traits, including metacarpal length and height. There were several significant associations with BMD found in men only. Using simulation to adjust for multiple testing, the association of SNP 1 (rs8887, G>A) with height in females trended toward significance with adjusted P = 0.07, and in the combined-sex model this association remained significant (P = 0.03).
To date, there are only two references in the literature that investigated PLIN1 and bone density. Yamada et al. [34] studied a cohort of 1,122 men and 1,112 women, ages 40–79, as part of the Japanese National Institute for Longevity Sciences-Longitudinal Study of Aging. These investigators studied only a single polymorphism in PLIN1, 1243C>T (rs2304796), and their analysis revealed a relationship in men only between the TT genotype and greater BMD for the total hip, lumbar spine, femoral neck, and trochanter compared to the combined CC and CT genotypes. They found no relationship between this polymorphism and BMD in Japanese women, regardless of estrogen status. The PLIN1 SNP studied by Yamada et al. does not appear to be present in the Caucasian population, and it is therefore not possible to establish if this SNP is in LD with any of the SNPs evaluated in this analysis. A recent genome-wide analysis of the FOS population [12] found one SNP in the PLIN1 gene, rs8179043 in intron 6 (MAF = 0.30), to be associated with lumbar spine BMD in men only, with a P value of 0.01. This SNP is in strong LD with rs894160 (SNP 10 in this analysis) with r2 ~ 0.97; however, no significant associations were found in our study with this SNP. Differences in the size and characteristics of the population evaluated in our analysis and this genome-wide analysis may account for the null result seen in our study.
Our study has several strengths, including having a large sample size. In addition, this is the first analysis to study a potential relationship between PLIN1 and PLIN4 with bone phenotypes in a Caucasian population. SNP selection covered the PLIN1 gene adequately to enable us to detect associations, and we included PLIN4, a related gene that has not been previously studied. We found several gender-specific associations between various SNPs and the measured traits, consistent with the previous findings by Yamada et al. [34] as well as previous reports of gender-specific linkage of osteoporosis-related phenotypes [3, 39, 40]. After adjustment for multiple testing, we found a trend toward significance in the association between one of the studied PLIN4 polymorphisms and height in females and a significant result in the sex-combined sample. However, we were unable to detect a significant association with this SNP and metacarpal length, which might have been expected if height were reflecting bone growth. Previously, height was shown to be a bone-related phenotype since the phenotype is largely explained by the length of the extremities and vertebral segment, which are a function of bone length [6–8]. Therefore, unsurprisingly, genome-wide association studies of adult height identified several bone-and cartilage-related genes including GDF5 (growth differentiation factor 5), a cartilage-derived morphogenetic protein [7, 8]. Sanna et al. [7] stated that “identifying genetic variants that influence human height will advance our understanding of skeletal growth and development.”
Limitations of this study include the fact that the phenotypes used in our analyses are proxies representing the end product of complex biological processes, rather than the endophenotypes, most proximal to the effects of genes. Other factors, including unknown and unmeasured environmental as well as stochastic factors, may contribute to both the bone and anthropometric traits studied, especially in a sample that is beyond the age of peak bone density and long bone growth. Also, areal BMD combines the trabecular and cortical compartments, making it impossible to distinguish between potentially more important trabecular effects of genes related to marrow fat handling. Future studies should repeat our analyses using volumetric trabecular bone density phenotypes.
There is growing evidence of a close relationship between lipid metabolism and bone remodeling. Osteoblasts and adipocytes share a common progenitor cell in the bone marrow, and an inverse relationship exists between the two lineages, with agents inducing adipogenesis inhibiting osteoblast differentiation and therefore promoting bone loss [23–25]. Thiazolidinediones, inducers of PPARγ and subsequent induction of the mesenchymal stem cell into adipocytes, have been associated with bone loss and osteoporosis [43]. Osteocalcin is a protein produced by osteoblasts that acts as a regulator of bone formation, and osteocalcin-null mice have been found to be obese [44]. There may be profound associations between genes that regulate fat metabolism and skeletal integrity, and future research is necessary to investigate this relationship further, perhaps focusing on other genes involved in stem cell differentiation, rather than fat handling.
The findings of this study should be regarded as hypothesis-generating and indicate that assessment in other populations is needed. Future investigation is necessary to differentiate the role that proteins important in adipocyte metabolism, including PLIN and PLIN4 of the PAT family of proteins, have with regard to bone properties and subsequently fracture risk.
Acknowledgments
This work is from the Framingham Heart Study of the National Heart, Lung, and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The Framingham Heart Study core examinations were supported by the National Heart, Lung, and Blood Institute’s (contract N01-HC-25195). Measurements of phenotypes were funded by the National Institute of Arthritis, Musculoskeletal and Skin Diseases and the National Institute on Aging (grants R01 AR/AG 41398 and R01 AR050066). Genetic analyses were supported by NIH grants HL54776 and DK075030 and by the US Department of Agriculture Research Service (contracts 53-K06–5-10 and 58–1950-9–001). We gratefully acknowledge the Framingham Study members who participated in this study as well as the study coordinators, who contributed to the success of this work.
Footnotes
The authors have stated that they have no conflict of interest.
Disclosures: Dr. Douglas P. Kiel discloses a consultant/advisory role for Wyeth, Merck, Amgen, Novartis, Procter and Gamble, and Lilly and funding from Pfizer, Merck, Amgen, Novartis, and Hologic unrelated to the subject matter of this paper.
Contributor Information
Natalie E. Cusano, Email: nc2433@columbia.edu, Department of Medicine, Beth Israel Deaconess Medical Center, Boston, MA, USA. Division of Endocrinology, Department of Medicine, College of Physicians and Surgeons, Columbia University, 630 West 168th Street, PH 8 W-864, New York, NY 10032, USA
Douglas P. Kiel, Department of Medicine, Beth Israel Deaconess Medical Center, Boston, MA, USA. Hebrew SeniorLife Institute for Aging Research and Harvard Medical School, Boston, MA, USA
Serkalem Demissie, Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA. National Heart, Lung and Blood Institute Framingham Heart Study, Framingham, MA, USA.
David Karasik, Hebrew SeniorLife Institute for Aging Research and Harvard Medical School, Boston, MA, USA.
L. Adrienne Cupples, Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA. National Heart, Lung and Blood Institute Framingham Heart Study, Framingham, MA, USA.
Dolores Corella, Jean Mayer-US Department of Agriculture Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA. Centro de Investigación Biomédica en Red Fisiopatología de la Obesidad y Nutrición, University of Valencia, Valencia, Spain.
Qiong Gao, Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA.
Kris Richardson, Jean Mayer-US Department of Agriculture Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA.
Nikos Yiannakouris, Jean Mayer-US Department of Agriculture Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA.
Jose M. Ordovas, Jean Mayer-US Department of Agriculture Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA. Department of Epidemiology and Population Genetics, Centro Nacional Investigación Cardiovasculares (CNIC), Madrid, Spain
References
- 1.Department of Health and Human Services, Office of the Surgeon General. Bone health and osteoporosis: a report of the surgeon general. DHHS; Rockland, MD: 2004. [PubMed] [Google Scholar]
- 2.Shen H, Recker RR, Deng HW. Molecular and genetic mechanisms of osteoporosis: implication for treatment. Curr Mol Med. 2003;3:737–757. doi: 10.2174/1566524033479375. [DOI] [PubMed] [Google Scholar]
- 3.Ioannidis JP, Ng MY, Sham PC, Zintzaras E, Lewis CM, Deng HW, Econs MJ, Karasik D, Devoto M, Kammerer CM, Spector T, Andrew T, Cupples LA, Duncan EL, Foroud T, Kiel DP, Koller D, Langdahl B, Mitchell BD, Peacock M, Recker R, Shen H, Sol-Church K, Spotila LD, Uitterlinden AG, Wilson SG, Kung AW, Ralston SH. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J Bone Miner Res. 2007;22:173–183. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flicker L, Faulkner KG, Hopper JL, Green RM, Kaymacki B, Nowson CA, Young D, Wark JD. Determinants of hip axis length in women aged 10–89 years: a twin study. Bone. 1996;18:41–45. doi: 10.1016/8756-3282(95)00418-1. [DOI] [PubMed] [Google Scholar]
- 5.Chinappen-Horsley U, Blake GM, Fogelman I, Kato B, Ahmadi KR, Spector TD. Quantitative trait loci for bone lengths on chromosome 5 using dual energy X-Ray absorptiometry imaging in the Twins UK cohort. PLoS ONE. 2008;3:e1752. doi: 10.1371/journal.pone.0001752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, Shen H, Timpson N, Lettre G, Usala G, Chines PS, Stringham HM, Scott LJ, Dei M, Lai S, Albai G, Crisponi L, Naitza S, Doheny KF, Pugh EW, Ben-Shlomo Y, Ebrahim S, Lawlor DA, Bergman RN, Watanabe RM, Uda M, Tuomilehto J, Coresh J, Hirschhorn JN, Shuldiner AR, Schlessinger D, Collins FS, Davey Smith G, Boerwinkle E, Cao A, Boehnke M, Abecasis GR, Mohlke KL. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5(4):e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari SL, Karasik D, Liu J, Karamohamed S, Herbert AG, Cupples LA, Kiel DP. Interactions of interleukin-6 promoter polymorphisms with dietary and lifestyle factors and their association with bone mass in men and women from the Framingham Osteoporosis Study. J Bone Miner Res. 2004;19:552–559. doi: 10.1359/JBMR.040103. [DOI] [PubMed] [Google Scholar]
- 10.Grant SF, Reid DM, Blake G, Herd R, Fogelman I, Ralston SH. Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I alpha 1 gene. Nat Genet. 1996;14:203–205. doi: 10.1038/ng1096-203. [DOI] [PubMed] [Google Scholar]
- 11.Ralston SH. Genetic control of susceptibility to osteoporosis. J Clin Endocrinol Metab. 2002;87:2460–2466. doi: 10.1210/jcem.87.6.8621. [DOI] [PubMed] [Google Scholar]
- 12.Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riggs BL, Nguyen TV, Melton LJ, 3rd, Morrison NA, O’Fallon WM, Kelly PJ, Egan KS, Sambrook PN, Muhs JM, Eisman JA. The contribution of vitamin D receptor gene alleles to the determination of bone mineral density in normal and osteoporotic women. J Bone Miner Res. 1995;10:991–996. doi: 10.1002/jbmr.5650100622. [DOI] [PubMed] [Google Scholar]
- 14.Meunier P, Aaron J, Edouard C, Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- 15.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 16.Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 17.Burkhardt R, Kettner G, Bohm W, Schmidmeier M, Schlag R, Frisch B, Mallmann B, Eisenmenger W, Gilg T. Changes in trabecular bone, hematopoiesis and bone marrow vessels in aplastic anemia, primary osteoporosis, and old age: a comparative histomorphometric study. Bone. 1987;8:157–164. doi: 10.1016/8756-3282(87)90015-9. [DOI] [PubMed] [Google Scholar]
- 18.Bergman RJ, Gazit D, Kahn AJ, Gruber H, McDougall S, Hahn TJ. Age-related changes in osteogenic stem cells in mice. J Bone Miner Res. 1996;11:568–577. doi: 10.1002/jbmr.5650110504. [DOI] [PubMed] [Google Scholar]
- 19.Gimble JM. The function of adipocytes in the bone marrow stroma. New Biol. 1990;2(4):304–312. [PubMed] [Google Scholar]
- 20.Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 21.Gimble JM, Zvonic S, Floyd ZE, Kassem M, Nuttall ME. Playing with bone and fat. J Cell Biochem. 2006;98:251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- 22.Ackert-Bicknell CL, Demissie S, Marín de Evsikova C, Hsu YH, DeMambro VE, Karasik D, Cupples LA, Ordovas JM, Tucker KL, Cho K, Canalis E, Paigen B, Churchill GA, Forejt J, Beamer WG, Ferrari S, Bouxsein ML, Kiel DP, Rosen CJ. PPARG by dietary fat interaction influences bone mass in mice and humans. J Bone Miner Res. 2008;23:1398–1408. doi: 10.1359/JBMR.080419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao LJ, Liu YJ, Liu PY, Hamilton J, Recker RR, Deng HW. Relationship of obesity with osteoporosis. J Clin Endocrinol Metab. 2007;92:1640–1646. doi: 10.1210/jc.2006-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards JB, Valdes AM, Burling K, Perks UC, Spector TD. Serum adiponectin and bone mineral density in women. J Clin Endocrinol Metab. 2007;92:1517–1523. doi: 10.1210/jc.2006-2097. [DOI] [PubMed] [Google Scholar]
- 25.Dimitri P, Wales JK, Bishop N. Fat and bone in children: differential effects of obesity on bone size and mass according to fracture history. J Bone Miner Res. 2010;25:527–536. doi: 10.1359/jbmr.090823. [DOI] [PubMed] [Google Scholar]
- 26.Londos C, Sztalryd C, Tansey JT, Kimmel AR. Role of PAT proteins in lipid metabolism. Biochimie. 2005;87:45–49. doi: 10.1016/j.biochi.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 28.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. 2000;275:38486–38493. doi: 10.1074/jbc.M007322200. [DOI] [PubMed] [Google Scholar]
- 30.Tansey JT, Huml AM, Vogt R, Davis KE, Jones JM, Fraser KA, Brasaemle DL, Kimmel AR, Londos C. Functional studies on native and mutated forms of perilipins. A role in protein kinase A–mediated lipolysis of triacylglycerols. J Biol Chem. 2003;278:8401–8406. doi: 10.1074/jbc.M211005200. [DOI] [PubMed] [Google Scholar]
- 31.Kern PA, Di Gregorio G, Lu T, Rassouli N, Ranganathan G. Perilipin expression in human adipose tissue is elevated with obesity. J Clin Endocrinol Metab. 2004;89:1352–1358. doi: 10.1210/jc.2003-031388. [DOI] [PubMed] [Google Scholar]
- 32.Qi L, Shen H, Larson I, Schaefer EJ, Greenberg AS, Tregouet DA, Corella D, Ordovas JM. Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obes Res. 2004;12:1758–1765. doi: 10.1038/oby.2004.218. [DOI] [PubMed] [Google Scholar]
- 33.Corella D, Qi L, Sorli JV, Godoy D, Portoles O, Coltell O, Greenberg AS, Ordovas JM. Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. J Clin Endocrinol Metab. 2005;90:5121–5126. doi: 10.1210/jc.2005-0576. [DOI] [PubMed] [Google Scholar]
- 34.Yamada Y, Ando F, Shimokata H. Association of polymorphisms in forkhead box C2 and perilipin genes with bone mineral density in community-dwelling Japanese individuals. Int J Mol Med. 2006;18:119–127. [PubMed] [Google Scholar]
- 35.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 36.Cupples LA, Arruda HT, Benjamin EJ, D’Agostino RB, Sr, Demissie S, DeStefano AL, Dupuis J, Falls KM, Fox CS, Gottlieb DJ, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Kathiresan S, Kiel DP, Laramie JM, Larson MG, Levy D, Liu CY, Lunetta KL, Mailman MD, Manning AK, Meigs JB, Murabito JM, Newton-Cheh C, O’Connor GT, O’Donnell CJ, Pandey M, Seshadri S, Vasan RS, Wang ZY, Wilk JB, Wolf PA, Yang Q, Atwood LD. The Framingham Heart Study 100 K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannan MT, Felson DT, Dawson-Hughes B, Tucker KL, Cupples LA, Wilson PW, Kiel DP. Risk factors for longitudinal bone loss in elderly men and women: the Framingham Osteoporosis Study. J Bone Miner Res. 2000;15:710–720. doi: 10.1359/jbmr.2000.15.4.710. [DOI] [PubMed] [Google Scholar]
- 38.McLean RR, Hannan MT, Epstein BE, Bouxsein ML, Cupples LA, Murabito J, Kiel DP. Elderly cohort study subjects unable to return for follow-up have lower bone mass than those who can return. Am J Epidemiol. 2000;151:689–692. doi: 10.1093/oxfordjournals.aje.a010263. [DOI] [PubMed] [Google Scholar]
- 39.Hannan MT, Tucker KL, Dawson-Hughes B, Cupples LA, Felson DT, Kiel DP. Effect of dietary protein on bone loss in elderly men and women: the Framingham Osteoporosis Study. J Bone Miner Res. 2000;15:2504–2512. doi: 10.1359/jbmr.2000.15.12.2504. [DOI] [PubMed] [Google Scholar]
- 40.Yates LB, Karasik D, Beck TJ, Cupples LA, Kiel DP. Hip structural geometry in old and old–old age: similarities and differences between men and women. Bone. 2007;41:722–732. doi: 10.1016/j.bone.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karasik D, Shimabuku NA, Zhou Y, Zhang Y, Cupples LA, Kiel DP, Demissie S. A genome wide linkage scan of metacarpal size and geometry in the Framingham Study. Am J Hum Biol. 2008;20:663–670. doi: 10.1002/ajhb.20791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi L, Corella D, Sorli JV, Portoles O, Shen H, Coltell O, Godoy D, Greenberg AS, Ordovas JM. Genetic variation at the perlipin (PLIN) locus is associated with obesity-related phenotypes in white women. Clin Genet. 2004;66:299–310. doi: 10.1111/j.1399-0004.2004.00309.x. [DOI] [PubMed] [Google Scholar]
- 43.Grey A. Skeletal consequences of thiazolidinedione therapy. Osteoporos Int. 2008;19:129–137. doi: 10.1007/s00198-007-0477-y. [DOI] [PubMed] [Google Scholar]
- 44.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]