Abstract
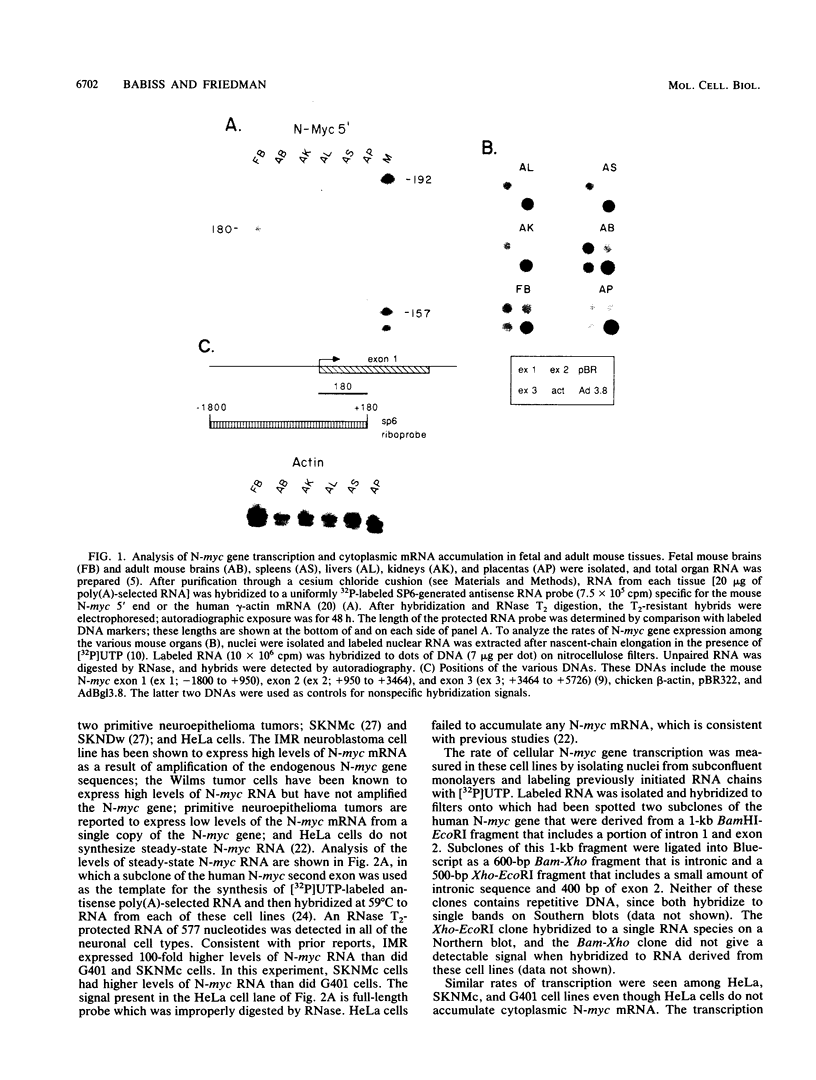
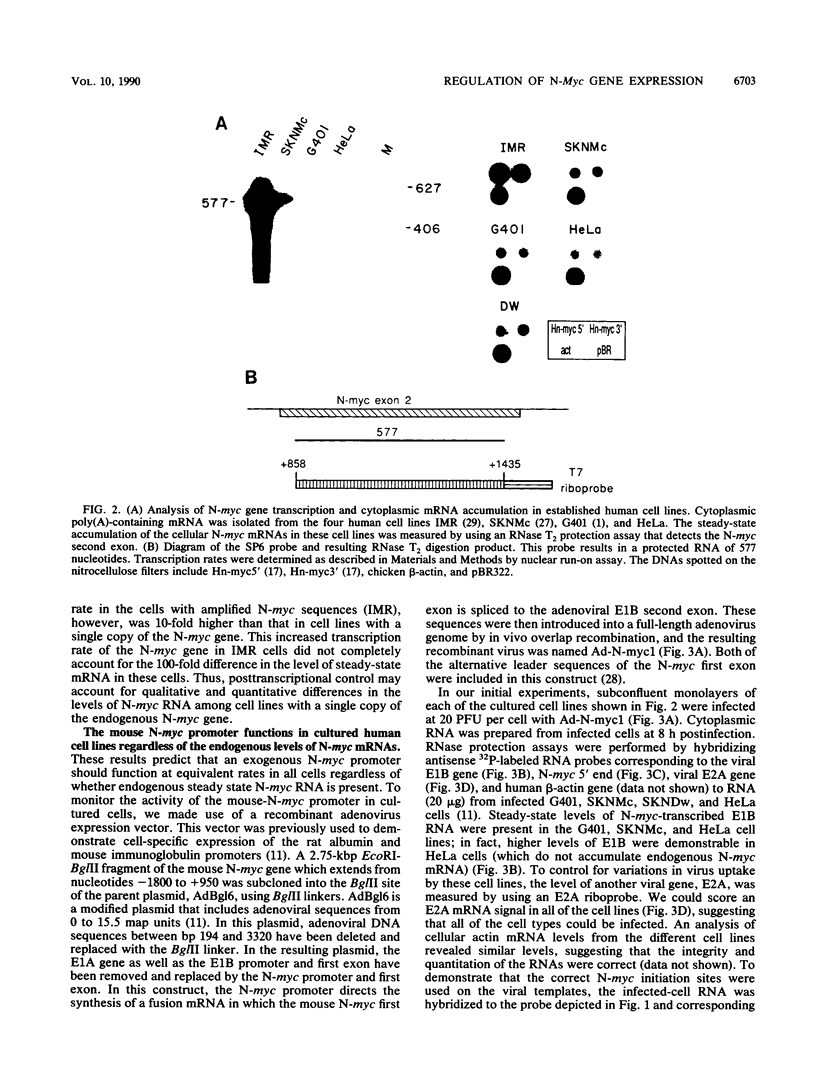
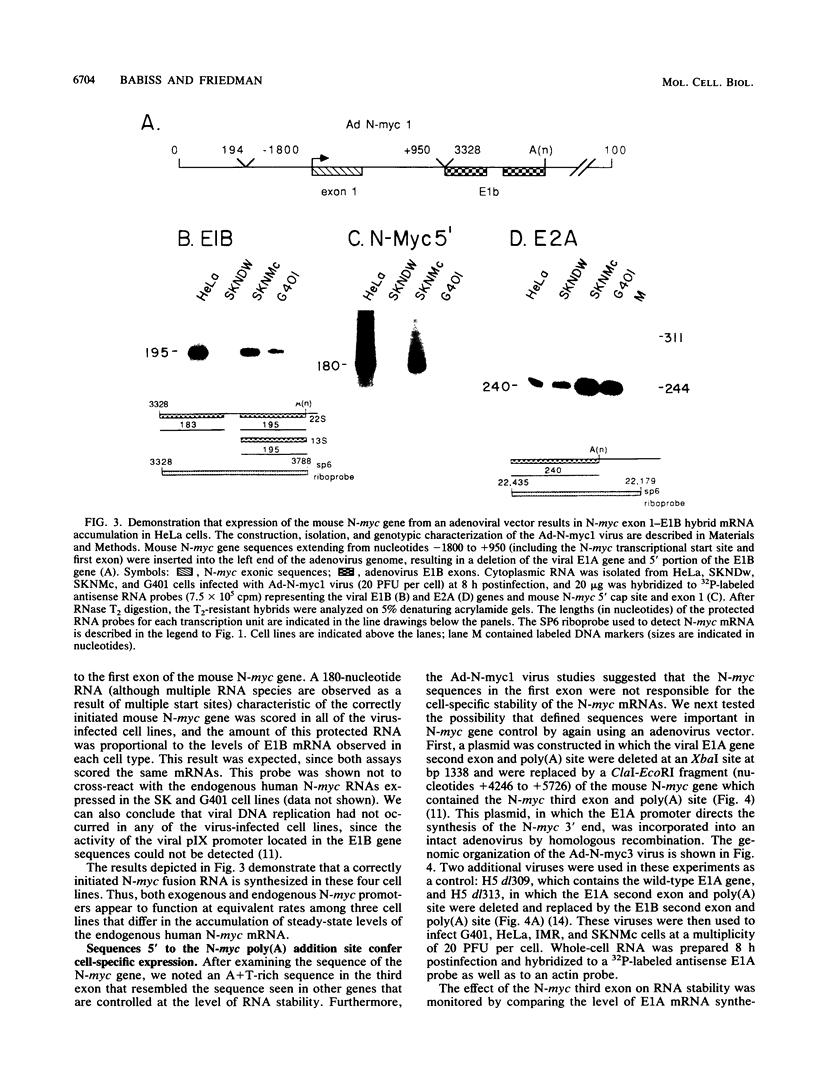
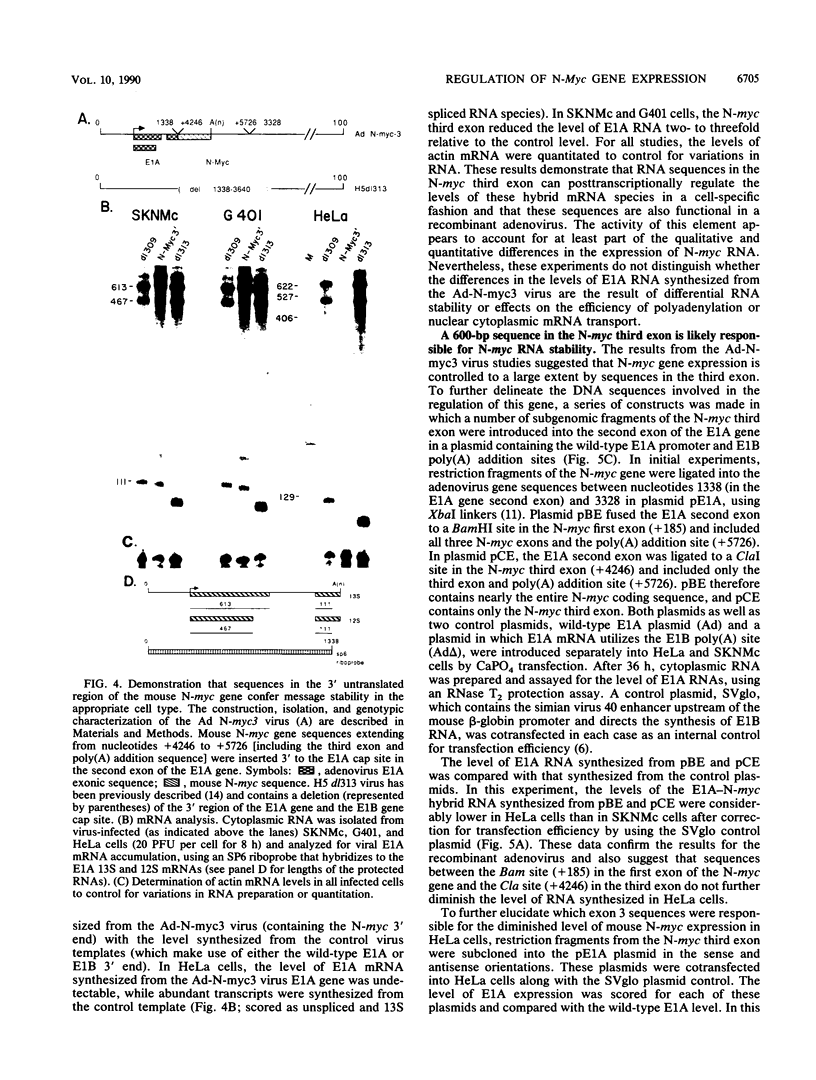
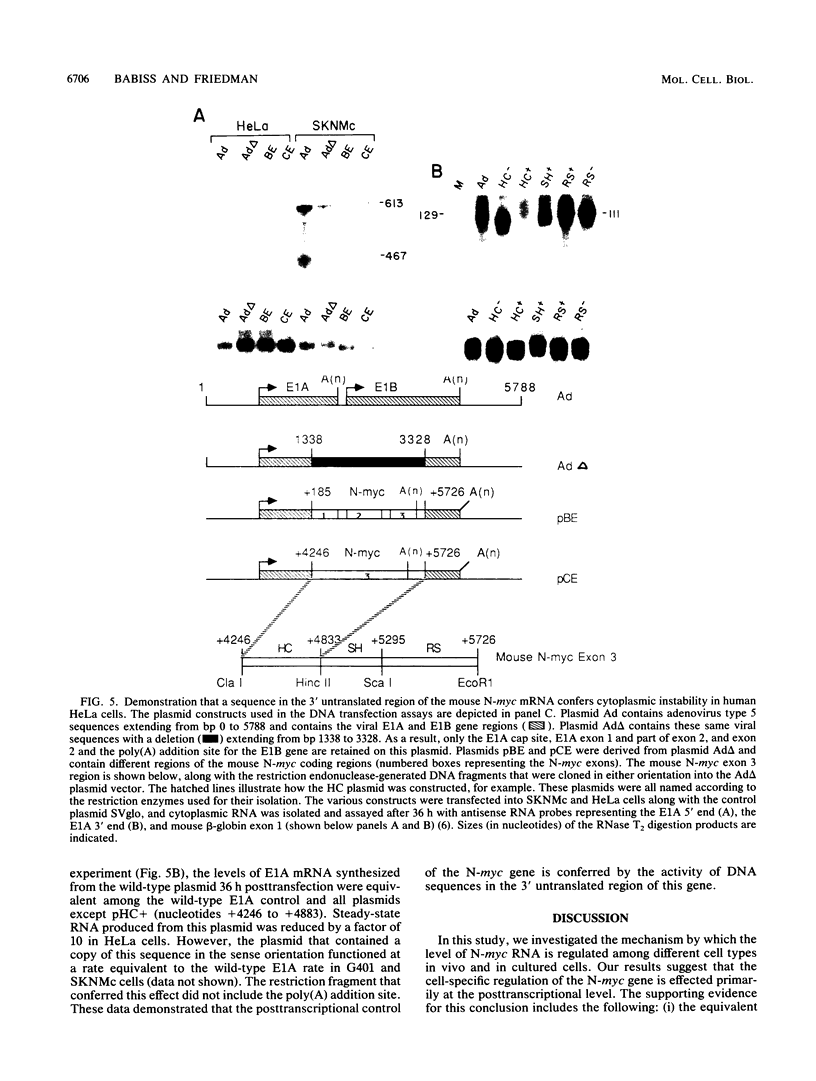
We present evidence that differences in the levels of N-myc mRNA among different cell types are the result of posttranscriptional control. First, we noted that while steady-state mouse N-myc mRNA could be detected only in fetal mouse brain, it was transcribed at an equivalent rate in adult brain, liver, spleen, and placenta and in fetal brain. Similarly, the human N-myc gene was transcribed at an equivalent rate in HeLa cells, which do not accumulate this RNA in the cytoplasm, and cell lines G401 (a Wilms tumor-derived cell line) and SKNMc (established from a primitive neuroepithelioma), which do express N-myc RNA. As expected, the N-myc promoter functioned at equivalent rates, as demonstrated by the level of a reporter gene, when introduced into these cell types by using a recombinant adenovirus vector. The suggestion that posttranscriptional mechanisms control the level of this RNA was supported by the observation that sequences in the N-myc third exon specifically decreased the level of E1A mRNA when these sequences were placed downstream of the E1A promoter in a recombinant adenovirus. Finally, we further localized these sequences to a 600-bp fragment of the third exon by introducing various subclones of this sequence downstream of the E1A promoter in both viral and plasmid vectors.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babich A., Nevins J. R. The stability of early adenovirus mRNA is controlled by the viral 72 kd DNA-binding protein. Cell. 1981 Nov;26(3 Pt 1):371–379. doi: 10.1016/0092-8674(81)90206-3. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Grayson D. R., Darnell J. E., Jr The cell-specific enhancer of the mouse transthyretin (prealbumin) gene binds a common factor at one site and a liver-specific factor(s) at two other sites. Mol Cell Biol. 1988 Jan;8(1):81–90. doi: 10.1128/mcb.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- DePinho R. A., Legouy E., Feldman L. B., Kohl N. E., Yancopoulos G. D., Alt F. W. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1827–1831. doi: 10.1073/pnas.83.6.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman E., Krauter K., Walling L., Weinberger C., Ray M., Darnell J. E., Jr Transcriptional control in the production of liver-specific mRNAs. Cell. 1981 Mar;23(3):731–739. doi: 10.1016/0092-8674(81)90436-0. [DOI] [PubMed] [Google Scholar]
- Friedman J. M., Chung E. Y., Darnell J. E., Jr Gene expression during liver regeneration. J Mol Biol. 1984 Oct 15;179(1):37–53. doi: 10.1016/0022-2836(84)90305-x. [DOI] [PubMed] [Google Scholar]
- Jakobovits A., Schwab M., Bishop J. M., Martin G. R. Expression of N-myc in teratocarcinoma stem cells and mouse embryos. Nature. 1985 Nov 14;318(6042):188–191. doi: 10.1038/318188a0. [DOI] [PubMed] [Google Scholar]
- Jones N., Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979 Jul;17(3):683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl N. E., Gee C. E., Alt F. W. Activated expression of the N-myc gene in human neuroblastomas and related tumors. Science. 1984 Dec 14;226(4680):1335–1337. doi: 10.1126/science.6505694. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Kanda N., Schreck R. R., Bruns G., Latt S. A., Gilbert F., Alt F. W. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983 Dec;35(2 Pt 1):359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Legouy E., DePinho R. A., Nisen P. D., Smith R. K., Gee C. E., Alt F. W. Human N-myc is closely related in organization and nucleotide sequence to c-myc. Nature. 1986 Jan 2;319(6048):73–77. doi: 10.1038/319073a0. [DOI] [PubMed] [Google Scholar]
- Krystal G. W., Armstrong B. C., Battey J. F. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990 Aug;10(8):4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau M. M., Brooks B. J., Battey J., Sausville E., Gazdar A. F., Kirsch I. R., McBride O. W., Bertness V., Hollis G. F., Minna J. D. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985 Nov 7;318(6041):69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- Nisen P. D., Zimmerman K. A., Cotter S. V., Gilbert F., Alt F. W. Enhanced expression of the N-myc gene in Wilms' tumors. Cancer Res. 1986 Dec;46(12 Pt 1):6217–6222. [PubMed] [Google Scholar]
- Powell D. J., Friedman J. M., Oulette A. J., Krauter K. S., Darnell J. E., Jr Transcriptional and post-transcriptional control of specific messenger RNAs in adult and embryonic liver. J Mol Biol. 1984 Oct 15;179(1):21–35. doi: 10.1016/0022-2836(84)90304-8. [DOI] [PubMed] [Google Scholar]
- Schwab M., Alitalo K., Klempnauer K. H., Varmus H. E., Bishop J. M., Gilbert F., Brodeur G., Goldstein M., Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983 Sep 15;305(5931):245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- Schwab M., Varmus H. E., Bishop J. M. Human N-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature. 1985 Jul 11;316(6024):160–162. doi: 10.1038/316160a0. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Bishop J. M. Alternative processing of RNA transcribed from NMYC. Mol Cell Biol. 1987 Dec;7(12):4266–4272. doi: 10.1128/mcb.7.12.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumilowicz J. J., Nichols W. W., Cholon J. J., Greene A. E. Definition of a continuous human cell line derived from neuroblastoma. Cancer Res. 1970 Aug;30(8):2110–2118. [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman K., Legouy E., Stewart V., Depinho R., Alt F. W. Differential regulation of the N-myc gene in transfected cells and transgenic mice. Mol Cell Biol. 1990 May;10(5):2096–2103. doi: 10.1128/mcb.10.5.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]