Abstract
Codon usage bias has been observed in almost all genomes and is thought to result from selection for efficient and accurate translation of highly expressed genes1–3. Codon usage is also implicated in the control of transcription, splicing and RNA structure4–6. Many genes exhibit little codon usage bias, which is thought to reflect a lack of selection for mRNA translation. Alternatively, however, non-optimal codon usage may have biological significance. The rhythmic expression and the proper function of the Neurospora FREQUENCY (FRQ) protein are essential for circadian clock function. Here we show that, unlike most genes in Neurospora, frq exhibits non-optimal codon usage across its entire open reading frame. Optimization of frq codon usage abolishes both overt and molecular circadian rhythms. Codon optimization not only increases FRQ level but surprisingly, also results in conformational changes in FRQ protein, altered FRQ phosphorylation profile and stability, and impaired functions in the circadian feedback loops. These results indicate that non-optimal codon usage of frq is essential for its circadian clock function. Our study provides an example of how non-optimal codon usage functions to regulate protein expression and to achieve optimal protein structure and function.
Eukaryotic circadian oscillators consist of autoregulatory circadian negative feedback loops. In the core circadian oscillator of Neurospora crassa, FRQ protein is a central component that functions as the circadian negative element with its partner FRH7–9. Two transcription factors, WHITE COLLAR (WC)-1 and WC-2, form a heterodimeric complex that activates frq transcription10. The FRQ-FRH complex inhibits WC complex activity by interacting with the WCs11,12. The level and stability of FRQ play a major role in setting period length, phase and clock-sensitivity to environmental signals7,8,13. In addition, FRQ promotes the expression of both WC proteins in an interlocked positive feedback loop10,14.
The protein-coding genes of Neurospora exhibit strong codon bias (Figure s1a). The third position of almost every codon family in this filamentous fungus has the preference C>G>T>A. Codon optimization enhances expression of a heterologous luciferase gene in Neurospora15,16. To establish that codon usage bias regulates protein expression, we identified the most abundant Neurospora proteins in whole cell extract by mass spectrometry analyses. The genes encoding the top100 most-abundant proteins (Table s1) exhibit much stronger codon bias than the rest of the protein coding genes (Figure s1b).
We classified all predicted Neurospora tRNA genes and predicted the relative translation elongation rate for each codon based on tRNA-gene copy numbers, which correlate with tRNA abundance, and the nature of anticodon-codon interactions4,17. The most preferred codon for each amino acid is always the codon with highest predicted translation elongation rate (Table s2). Therefore, to ensure efficient translation of abundant proteins, selection pressure favored a bias for codons that are translated by highly abundant tRNA species.
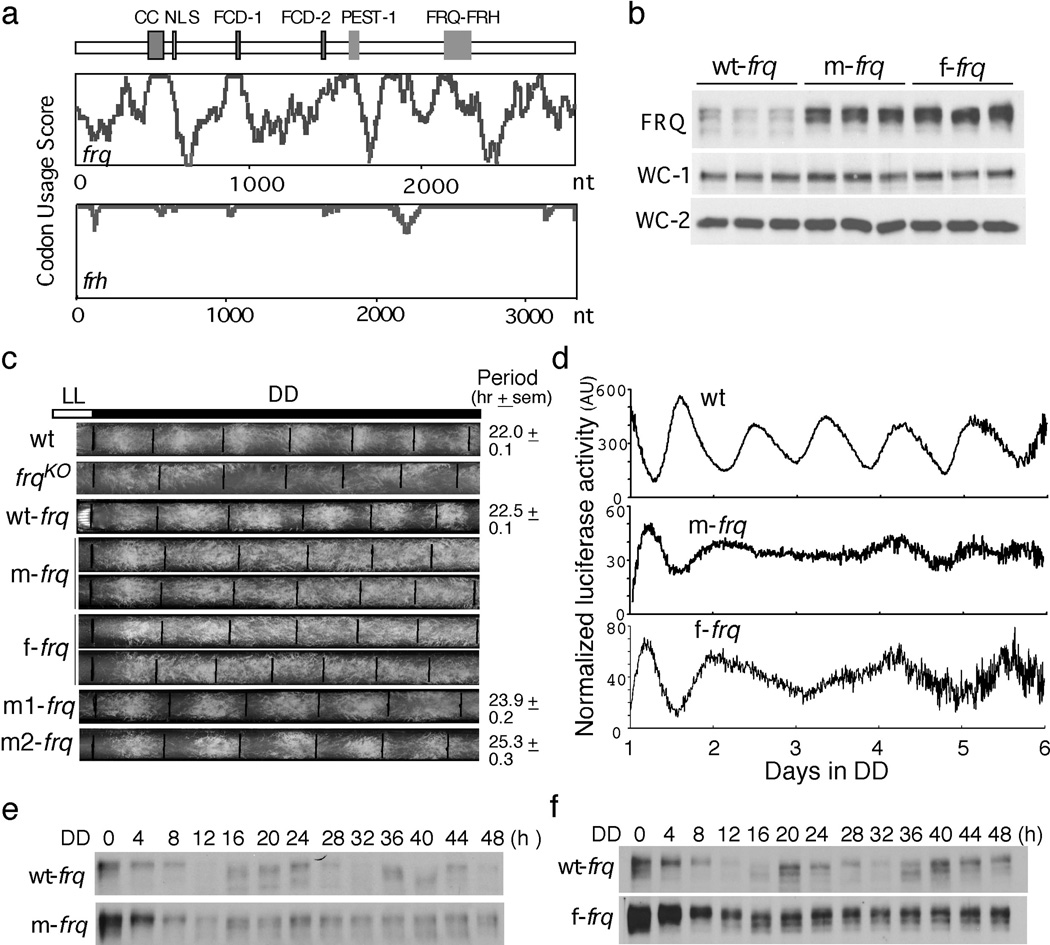
Many Neurospora genes exhibit little or no codon biases (Figure s1a). FRQ is a low abundance Neurospora protein. Its codon bias index18 (CBI; CBI = 0 indicates completely random codon usage) value of 0.08 indicates that frq has little codon bias (Figure s1b). A codon usage graph of the frq ORF shows that many regions have non-optimal usage (Figure 1a), whereas frh has good codon usage throughout its ORF.
Figure 1.
Codon optimization of frq results in high FRQ expression levels and loss of circadian rhythmicities. (a) Codon usage score plots of frq and frh obtained using Codon Usage 3.5. (b) Western blot showing the levels of FRQ and WCs in frq10, wt-frq (wt-frq); frq10, m-frq (m-frq) and frq10, f-frq (f-frq) strains. Three independent cultures in LL at room temperature were used. (c) Race tube analysis showing the conidiation phenotypes in DD. Black lines indicate the growth fronts every 24 h. (d) Luciferase reporter assay showing frq promoter activity of the indicated strains after one day in DD. The measurement of luciferase activity was normalized to subtract the baseline luciferase signal. (e and f) Western blots showing loss of FRQ expression rhythms in the codon-optimized strains. Densitometric analyses are shown below.
We created two constructs in which the N-terminal end (1–164 aa) of frq was codon optimized. In the m-frq construct, only the non-preferred codons were changed, while every codon was optimized in the f-frq construct. Predicted stability of RNA secondary structure was not significantly affected by the optimization (Table s3). These constructs and the wild-type frq construct (wt-frq) gene were transformed individually into a frq null strain (frq10). Both m-frq and f-frq strains have significantly higher levels of FRQ proteins in constant light (LL) than that of the wt-frq strain (Figures 1b & s2a). On the other hand, frq mRNA levels were comparable in these strains (Figure s2b). FRQ is known to up-regulate WC protein levels10,14. The WC-1 and WC-2 levels, however, were similar in these strains despite the much higher levels of FRQ in the optimized strains (Figures 1b, s2a, and s2c).
The wt-frq construct was able to fully rescue the arrhythmic conidiation rhythm of the frq10 strain in DD (Figure 1c), but both of the optimized frq strains exhibited arrhythmic conidiation phenotypes; these are not due to the modest changes in the ratios of two alternatively translated FRQ forms, since either form of FRQ alone can support robust rhythms19,20.
We created two additional constructs (m1-frq and m2-frq), in which only the N- or C-terminal segments of the optimized region of m-frq were optimized. The frq10 transformants carrying either construct exhibited long-period conidiation rhythms and had FRQ levels between those of wt-frq and m-frq strains (Figures 1c & s3a). These results suggest that the severe conidiation rhythm phenotypes of the m-frq and f-frq strains are due to the cumulative effect of codon optimization and are not likely due to mutation of a DNA/RNA element.
To examine circadian phenotypes at the molecular level, we introduced a luciferase reporter construct that is under the control of the frq promoter16 into wild-type, m-frq and f-frq strains. As shown in Figures 1d & s3b, the robust rhythmic luciferase activity was abolished in the optimized strains. FRQ protein levels also lost molecular rhythmicity in the optimized strains (Figures 1e, 1f, s3c & s3d): the overall levels of FRQ were high and circadian changes in FRQ abundance and phosphorylation profile were abolished. Together, these results indicate that the non-optimal codon usage of frq governs FRQ expression level and is essential for clock function.
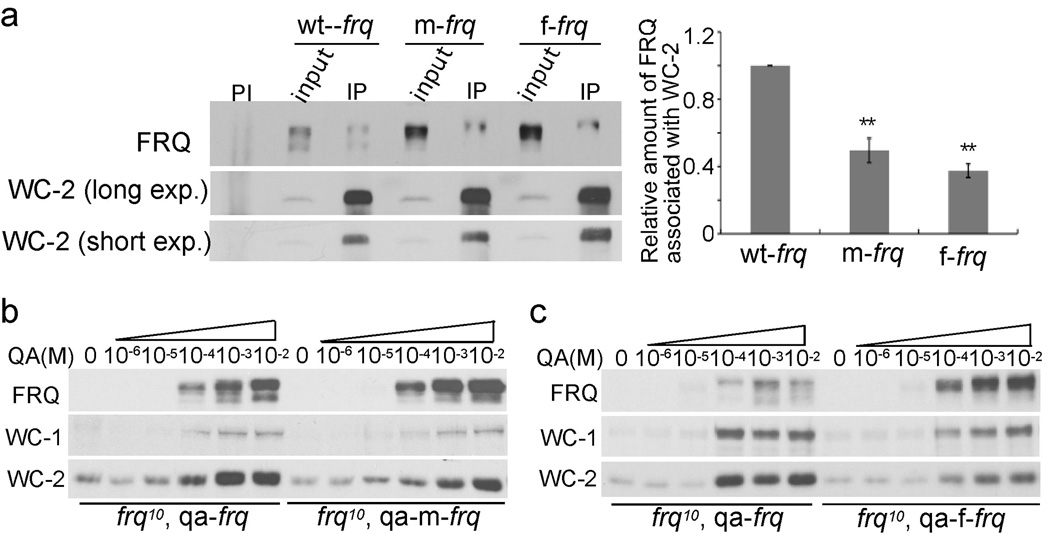
The loss of circadian rhythms in the optimized strains is surprising because we previously showed that high FRQ levels do not result in arrhythmicity10,21, suggesting that codon optimization causes defects in FRQ function. FRQ fulfills its function in the circadian negative feedback loop through its interaction with WCs11. Immunoprecipitation assays showed that the relative amounts of FRQ associated with WC-2 were significantly decreased in both optimized strains (Figure 2a), suggesting that the FRQ function in the negative feedback loop is impaired in the optimized strains.
Figure 2.
FRQ activities in circadian feedback loops are impaired in the frq codon-optimized strains. (a) Immunoprecipitation assay showing that FRQ has a decreased ability to interact with WC-2 in the codon-optimized strains. Densitometric analyses of results from four independent experiments are shown below. PI: pre-immune serum; IP: immunoprecipitation. Error bars indicate standard error (n=9). Asterisks indicate the p value <0.01. (b and c) Western blots showing the levels of WC and FRQ in the indicated strains at different concentrations of QA in LL.
FRQ also acts in a positive feedback loop by promoting WC expression10,14. Constructs qa-m-frq and qa-f-frq, in which either m-frq or f-frq is under the control of a quinic acid (QA)-inducible promoter, were introduced into the frq-null strain. As expected, FRQ levels were higher in the qa-m-frq and qa-f-frq strains than the control qa-frq strain at a given QA concentration (Figures 2b, 2c, s4 & s5). Induction of FRQ resulted in increased levels of WCs, demonstrating the role of FRQ in the positive feedback loop. At QA concentrations higher than 1×10−4 M, however, the qa-m-frq and qa-f-frq strains have lower levels of WC-1 and WC-2 than the qa-frq strain despite having higher FRQ levels, indicating that FRQ function in the positive feedback loop is also impaired in the codon-optimized strains.
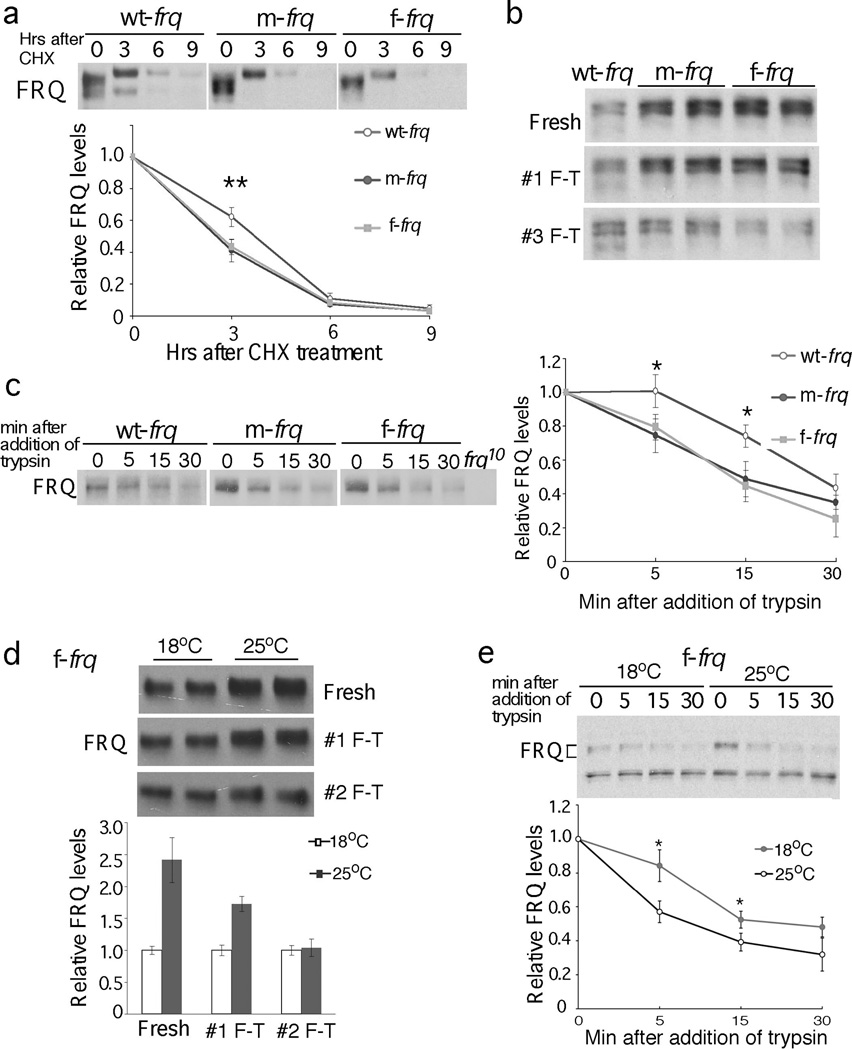
The impaired FRQ functions despite higher FRQ levels in codon-optimized strains suggest that FRQ’s structural conformation is altered. FRQ becomes progressively phosphorylated before its degradation7. Figure 3a shows that, in both m-frq and f-frq strains, FRQ was less stable than in the wild-type strain after the addition of cycloheximide (CHX). The difference in FRQ stability was most pronounced after 3 hr of CHX treatment, a time when FRQ became hyperphosphorylated.
Figure 3.
FRQ protein in the codon-optimized strains is less stable and more sensitive to trypsin digestion. (a) Western blots showing FRQ degradation after CHX treatment (10 µg/ml). A longer exposure for the wt-frq strain was used so that the FRQ signals at time 0 are comparable in three strains. Densitometric analyses of results of four independent experiments are shown. Error bars, standard deviation. (b & c) Western blots showing sensitivity of FRQ from codon-optimized strains to freeze-thaw cycles (b) and trypsin (1 µg/ml) digestion (c). A longer exposure for the wt-frq strain was used in (c). Densitometric analyses of FRQ levels of three independent experiments in are shown. (d & e) Western blots showing that FRQ from the f-frq strain grown at 18°C is more resistant to freeze and thaw cycles (d, n=2) and to trypsin digestion (e, n=4) than that from 25°C. Two asterisks indicate p value <0.01, and one asterisk indicates p value <0.05.
FRQ from the optimized strains was also less stable after protein extraction in vitro after freeze-thaw cycles. Although FRQ levels in freshly isolated samples were significantly higher in optimized strains, they decreased rapidly after freeze-thaw cycles (Figure 3b). In contrast, expression of wt-frq in a wc-2KO strain to a level that is comparable to that of the optimized frq strains did not affect the freeze-thaw sensitivity of FRQ (Figure s6), indicating that the change in FRQ stability in the optimized strains is not due to high FRQ level or its reduced ability to interact with WCs. Furthermore, limited trypsin digestion showed that full-length FRQ in either optimized strain was more sensitive to trypsin digestion than in the control strain (Figure 3c).
We reasoned that the changes in FRQ conformation in the optimized strains are due to an increase of translation rate as a result of codon optimization. Thus, we examined whether FRQ in the f-frq strain can be rescued by decreasing protein translation rate at 18°C because low temperature was known to reduce FRQ expression21 (Figure 4d). The low temperature treatment was not successful in restoring the circadian conidiation rhythm of the f-frq strain (data not shown), which was not surprising because 18°C is near the low limit of temperature range permissive for rhythmicity22. FRQ in the f-frq strain at 18°C, however, is much less sensitive to freeze thaw cycles and to trypsin digestion than at 25°C (Figure 3d & 3e). These results suggest that frq codon optimization changes FRQ structure as a consequence of increasing translation rate.
Figure 4.
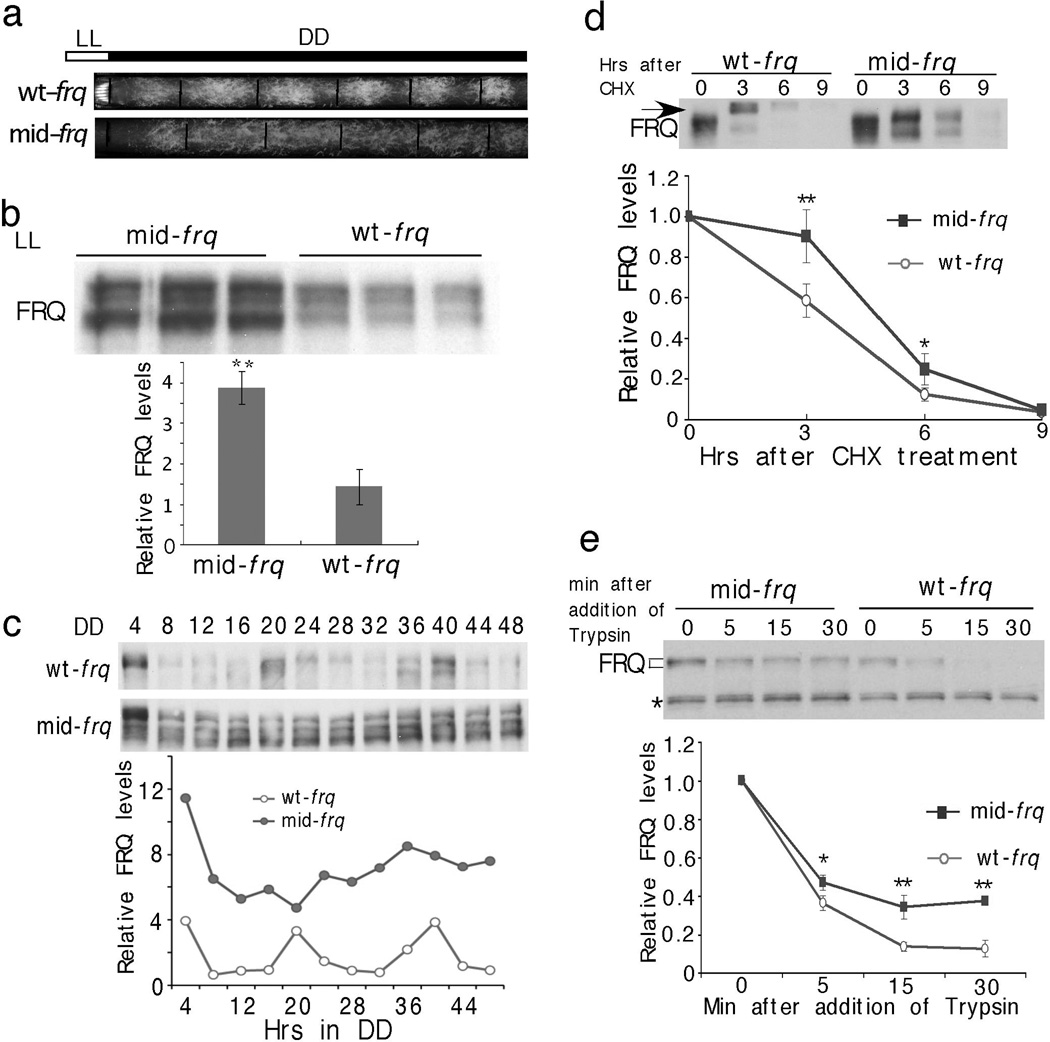
Codon optimization of the middle region of FRQ impairs FRQ phosphorylation and stabilizes FRQ. (a) Race tube analysis showing the conidiation phenoypes of indicated strains in DD. (b) Western blot showing FRQ expression profile in LL. Three independent samples were shown. (c) Western blots showing FRQ expression profile in DD. Densitometric analysis of the result is shown below. (d) Western blot showing the degradation of FRQ after CHX treatment. The arrow indicates the hyperphosphorylated forms of FRQ after the addition of CHX in the wt-frq strain. Densitometric analyses from four independent experiments are shown below. (e) Western blot showing reduced sensitivity of FRQ from mid-frq to trypsin digestion (2 µg/ml). Densitometric analyses of three independent experiments are shown. Error bars, Standard deviations.
To determine whether the codon-usage effect on FRQ is limited to the N-terminus of FRQ, we created a mid-frq strain, in which the middle region (aa 185–530) of frq ORF is optimized. This region contains two CK1 interaction domains and most of the phosphorylation sites that are important for FRQ stability and period determination23,24. As with m-frq and f-frq strains, the mid-frq strain exhibited arrhythmic conidiation (Figure 4a). In the mid-frq strain, FRQ levels were high, and FRQ levels and phosphorylation profile were not rhythmic (Figure 4b & 4c). No significant difference in frq mRNA was observed (Figure s7). Importantly, in the mid-frq strain, FRQ accumulated in hypophosphorylated forms, was more stable after CHX treatment and was more resistant to trypsin digestion (Figure 4b–e). In addition, the CHX-triggered rapid hyperphosphorylation of FRQ in the wt-frq strain was abolished in the mid-frq strain22. These results strongly indicate that the non-optimal codon usage of frq is important for FRQ structural conformation required for proper phosphorylation and stability. The opposite molecular phenotypes of mid-frq and the N-terminal optimized strains indicate that changes in FRQ structural conformation in these strains are due to location-specific codon optimization.
In a companion study, codon optimization of kaiBC clock genes in cyanobacteria results in high Kai protein levels and impaired cell growth at cool temperatures (Xu et al, the companion paper; ref 25), suggesting that non-optimal codon usage is a shared adaptive mechanism in both prokaryotes and eukaryotes.
Our study suggests that codon usage regulates not only protein expression level but also its structure and function. Protein folding, which occurs cotranslationally, requires protein chaperones and sufficient amount of time17. Codon optimization results in increased translation rates and thus reduces the time available for folding. Bioinformatics analyses and heterologous protein expression studies previously implicated codon usage as a factor that regulates protein folding26–28. In addition, a single rare codon in the human MDR1 gene results in altered drug and inhibitor interaction29.
In sharp contrast with the cyanobacterial Kai proteins30, most of the FRQ protein is predicted to be disordered (Figure s8). Interestingly, the known domains of FRQ all have relatively low disorder tendencies and fall in regions where codon usage scores are relatively high, suggesting that a fast translation rate in these well-structured regions does not interfere with protein folding. On the other hand, although the disordered regions may not form stable structures by themselves, they are critical for proper FRQ phosphorylation and stability23,24. They may serve as platforms for inter- or intramolecular protein-protein interactions, which may require more time for protein folding than well-structured domains. Therefore, frq non-optimal codon usage should be a mechanism that allows the proper folding of FRQ by reducing translation rate in these predicted disordered regions.
Methods
Strains and Growth Conditions
Neurospora strains used in this study were 87-3 (bd a; clock wild-type), 303-3 (bd, frq10, his-3)30 and different frq N-terminal codon optimized strains created in this study. Strain 303-3 (bd, frq10, his-3) was used as the host strain for various his-3 targeting constructs. The frq10, bd, wt-frq (frq10 containing a wild-type frq gene at the his-3 locus) strain was used as the control in this study.
Growth conditions have been described previously 31. Liquid cultures were grown in minimal medium (1×Vogel’s, 2% glucose). When QA was used, liquid cultures were grown in (10−6M to 10−2M) QA (pH 5.8), 1×Vogel’s, 0.1% glucose and 0.17% arginine. Race tube media contained 1×Vogel’s, 0.1% glucose, 0.17% arginine, 50 ng/ml biotin, and 1.5% agar. For rhythmic experiments, the Neurospora cultures were transferred from LL to DD at time 0 and were harvested in constant darkness at the indicated time (hours). Calculations of period length were performed as described 22.
frq codon optimization, codon usage score plot and indices calculation
frq codon optimization was performed for the N-terminal part (1–498 nt) or the middle region (553–1590 nt) of the ORF. The nucleotide sequences of the optimized frq are shown in Figures s9 & s10. Codons were optimized based on Neurospora crassa codon usage frequency and the predicted most efficient codon based on tRNA copy numbers. 65 codons were optimized in the m-frq construct while 94 codons were optimized in the f-frq construct. Sequences surrounding an alternative frq 3’ splice site in this region were not mutated.
Codon usage score plot was obtained using Codon Usage 3.5 (developed by Conrad Halling) using window size of 35 and logarithmic range of 3. Neurospora crassa codon usage frequency table was obtained from http://www.kazusa.or.jp/codon/. To calculate codon bias index (CBI), Frequency of Optimal Codons (Fop), ENC and GC content, codonw in the Mobyle Portal website (http://mobyle.pasteur.fr/cgi-bin/portal.py#forms::codonw) was used 18,32. CBI will equal 1.0 if a gene has extreme codon bias and will equal 0 if codon usage is completely random. If the number of optimal codons are less than expected by random change, the CBI value will be a negative value. Neurospora genome sequences were downloaded from the Broad Institute Neurospora crassa database (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html). The top 100 abundant proteins were identified by mass spectrometry analyses and ranked by their emPAI values 33 (Table s1).
Plasmid Constructs and Neurospora Transformation
pKAJ120 (containing the entire wild-type frq gene including its promoter and a his-3 targeting sequence) and pBA50 (containing the wild-type frq gene under the control of the qa-2 promoter and a his-3 targeting sequence) were used as the parental plasmids to create the optimized frq constructs 31. Optimized frq sequences (synthesized by Genscript) were subcloned into parental plasmids to replace the wild-type frq gene to generate m-frq, f-frq and mid-frq constructs. In the m1-frq construct, only the codons upstream of the predicted intron branch point were optimized as m-frq. For the m2-frq construct, only the codons downstream of the intron 3’ end were optimized as m-frq. The resulting constructs were transformed into strain 303-3 (bd, frq10, his-3) by electroporation and targeted to the his-3 locus 34. Homokaryon strains were obtained by microconidia purification.
Protein and RNA Analyses
Protein extraction, Western blot analysis, and immunoprecipitation assays were performed as previously described 35–37. Equal amounts of total protein (50µg) were loaded in each lane of SDS-PAGE gels (7.5% SDS-PAGE gels containing a ratio of 37.5:1 acrylamide/bisacrylamide). Densitometry of the signal was performed by using Image J.
RNA extraction and qRT-PCR were performed as previously described 38,39. For qRT-PCR, the primer sequences used for frq were 5’-GGAGGAGTCGATGTCACAAGG-3’ (forward) and 5’-CACTTCGAGTTACCCATGTTGC-3’ (reverse). The Neurospora β-tubulin gene was used as an internal control. The primer sequences specific for tubulin were 5’-GCGTATCGGCGAGCAGTT-3’(forward) and 5’-CCTCACCAGTGTACCAATGCA-3’ (reverse).
frq mRNA secondary structure and folding energy was predicted by mfold program (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
Mass spectrometric analyses and database search
The Neurospora proteins were separated on a 4–20% SDS-PAGE gradient gel. The whole protein gel lane was sliced equally into 14 gel blocks from top to bottom. Each gel block was de-stained and then digested in-gel with sequencing grade trypsin (10 ng/µL trypsin in 50 mM ammonium bicarbonate, pH 8.0) at 37 °C overnight. The resulting tryptic peptides from each gel block were extracted with 5% formic acid/50% acetonitrile and 0.1% formic acid/75% acetonitrile sequentially and then concentrated to ~ 20 µl in a CentriVap system (Labconco, Missouri, USA). The extracted peptides from each sample were separated by a home-made analytical capillary column (50 µm × 10 cm) packed with 5 µm spherical C18 reversed phase material (YMC, Kyoyo, Japan). An Agilent 1100 binary pump was used to generate HPLC gradient as follows: 0–5% B in 5 min, 5–40% B in 55 min, 40–100% B in 15 min (A = 0.2 M acetic acid in water; B = 0.2 M acetic acid/70% acetonitrile). The eluted peptides were sprayed directly into a LTQ mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a nano-ESI ion source. The mass spectrometer was operated in data-dependent mode (MS scan mass range was from 350 to 2000 Da, the top 5 most abundant precursor ions from each MS scan were selected for MS/MS scans, and dynamic exclusion time was 30 seconds). The mass spectrometric data from all 14 samples were combined and searched against Neurospora protein database on an in-house Mascot server (Matrix Science Ltd., London, UK). The major search parameters were as below: protein N-terminal acetylation and methionine oxidation were included as variable modifications; two missed cleavage sites were allowed; precursor ion mass tolerance was set as 3 Da; fragment ion mass tolerance was 0.8 Da. Only peptides with E-value above 0.01 were retained. The emPAI (Exponentially Modified Protein Abundance Index) 33 was calculated for each protein by the Mascot software.
Luciferase Reporter Assay
The luciferase reporter construct (frq-luc-bar) was generated by insertion of the 4.7 kb BamHI-NotI fragment of pfrq-luc-I (a generous gift from Dr. Jay Dunlap) 16 into the corresponding sites of pBARKS1. This construct, which contains the luciferase gene under the control of the frq promoter and the bar gene, was transformed into 87-3 (bd, a), wt-frq (frq10, bd, wt-frq), m-frq (frq10, bd, m-frq) and f-frq (frq10, bd, f-frq) strains. Transformants were selected using the basta/ignite (200 µg/mL) resistance conferred by the bar gene.
LumiCycle (ACTIMETRICS) was used for the luciferase assay using a protocol similar to previously described 16. The AFV (autoclaved FGS-Vogel’s) medium contained 1×FGS (0.05% fructose, 0.05% glucose, 2% sorbose), 1×Vogel’s medium, 50 µg/L biotin, and 1.8% agar. Firefly luciferin (BioSynt L-8200 D-luciferin firefly [synthetic] potassium salt) was added to the medium after autoclaving (final concentration of 50 µM). One drop of conidia suspensions in water were placed on AFV medium and grown in constant light (LL) overnight. The cultures were then transferred to darkness, and luminescence was recorded in real time (DD) using a LumiCycle after one day in DD. The data was then normalized with LumiCycle Analysis software by subtracting the baseline luciferase signal which increases as cell grows. Therefore, the normalized luciferase signals reflect the amplitude of the rhythm and does not reflect the absolute luciferase signal. Under our experimental condition, luciferasae signals are highly variable during the first day in the LumiCycle and become stabilized afterwards, which is likely due to an artifact caused by the light-dark transfer of the cultures. Thus, the results presented were recorded after one day in DD.
Protein Stability Assay
The liquid cultures of Neurospora strains were grown in LL for 1 day prior to the addition of cycloheximide (CHX, final concentration 10µg/mL). Cells were harvested at the indicated time points.
For the trypsin sensitivity assay, protein extracts were diluted to a protein concentration of 2.5µg/µl. 100µl extracts were treated with trypsin (final concentration 1µg/ml) at 25°. A 20µl sample was taken from the reaction at each time point (0, 5min, 15min and 30min) after addition of trypsin. Protein samples were mixed with protein loading buffer and resolved by SDS-PAGE. To compare trypsin sensitivity of FRQ from different strains, experiments were performed side-by-side and the protein samples were transferred to the same membrane for western blot analysis.
Supplementary Material
Acknowledgements
We thank Haiyan Yuan and Qianhong Ye for technical assistance and Dr. Jay Dunlap for providing the pfrq-luc-I construct. We apologize to those colleagues whose studies we could not cite due to space limitation. This work was supported by grants from the National Institutes of Health to Y. L. (GM068496 & GM062591) and M.S.S. (GM47498) and from the Welch Foundation (I-1560) to Y.L.
Footnotes
Contributions
Y. L., M.Z., and G.J. designed the research. M.Z., G.J., J.C., M.C., S. C., J. M. B. performed experiments. M.Z., G.J., J. M. B., M. S. S., and Y. L. analyzed the results. Y. L. and M.Z. wrote the paper; G. J., J. M. B. and M. S. S. edited and commented on the manuscript.
References
- 1.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 2.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 5.Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannarozzi G, et al. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 7.Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- 8.Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. 2012;36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng P, He Q, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng P, Yang Y, Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. USA. 2001;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Q, et al. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes & Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafmeier T, et al. Transcriptional feedback of neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Huang G, Wang L, Liu Y. Molecular mechanism of suppression of circadian rhythms by a critical stimulus. Embo J. 2006;25:5349–5357. doi: 10.1038/sj.emboj.7601397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- 15.Morgan LW, Greene AV, Bell-Pedersen D. Circadian and light-induced expression of luciferase in Neurospora crassa. Fungal Genet Biol. 2003;38:327–332. doi: 10.1016/s1087-1845(02)00562-5. [DOI] [PubMed] [Google Scholar]
- 16.Gooch VD, et al. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell. 2008;7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer PS, Siller E, Anderson JF, Barral JM. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J Mol Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 19.Colot HV, Loros JJ, Dunlap JC. Temperature-modulated Alternative Splicing and Promoter Use in the Circadian Clock Gene frequency. Mol. Biol. Cell. 2005;22:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diernfellner A, et al. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 2007;581:5759–5764. doi: 10.1016/j.febslet.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Merrow MM, Loros JJ, Dunlap JC. How temperature changes reset a circadian oscillator. Science. 1998;281:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Garceau N, Loros JJ, Dunlap JC. Thermally regulated translational control mediates an aspect of temperature compensation in the Neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 23.Tang CT, et al. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc Natl Acad Sci U S A. 2009;106:10722–10727. doi: 10.1073/pnas.0904898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol. 2009;26:1571–1580. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol. 2010;396:1310–1318. doi: 10.1016/j.jmb.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 28.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 29.Kimchi-Sarfaty C, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 30.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cha J, Yuan H, Liu Y. Regulation of the activity and cellular localization of the circadian clock protein FRQ. J Biol Chem. 2011;286:11469–11478. doi: 10.1074/jbc.M111.219782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aronson B, Johnson K, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation in the clock gene frequency . Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 32.Wright F. The 'effective number of codons' used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 33.Ishihama Y, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Bell-Pedersen D, Dunlap JC, Loros JJ. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol. Cell. Biol. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora . EMBO J. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garceau N, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 37.Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crosthwaite SK, Loros JJ, Dunlap JC. Light-Induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 39.Choudhary S, et al. A double-stranded-RNA response program important for RNA interference efficiency. Mol Cell Biol. 2007;27:3995–4005. doi: 10.1128/MCB.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.