Abstract
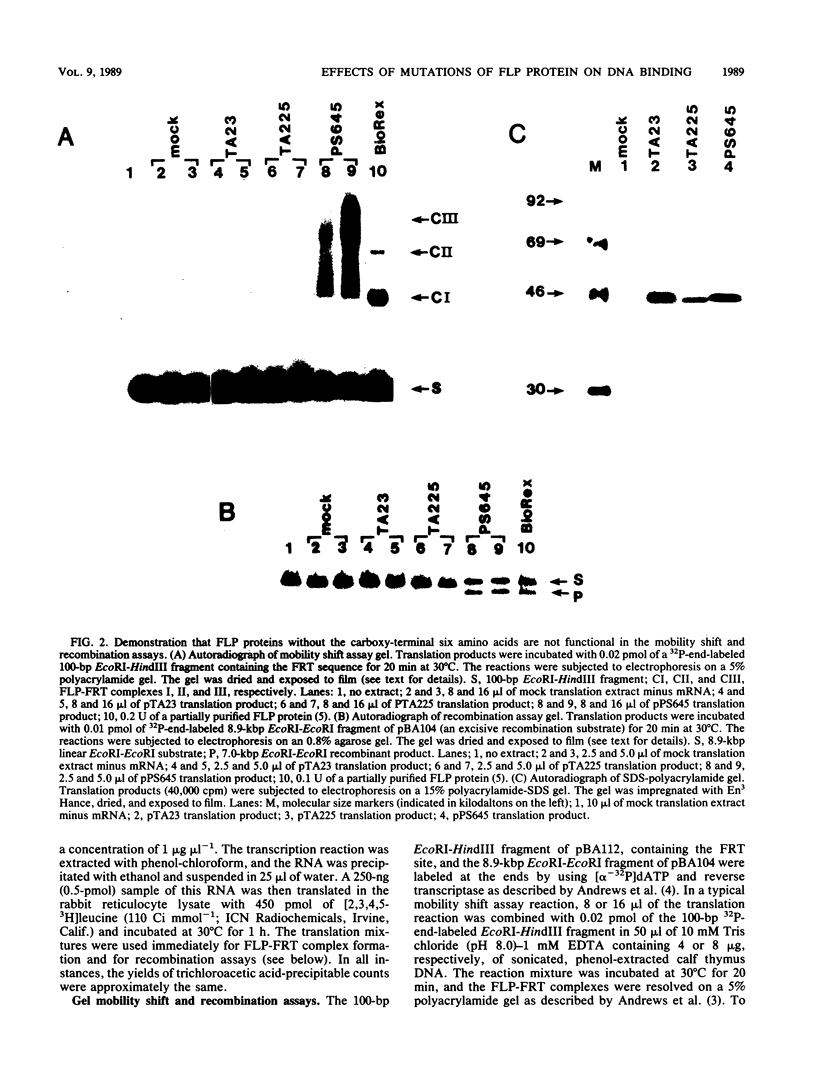
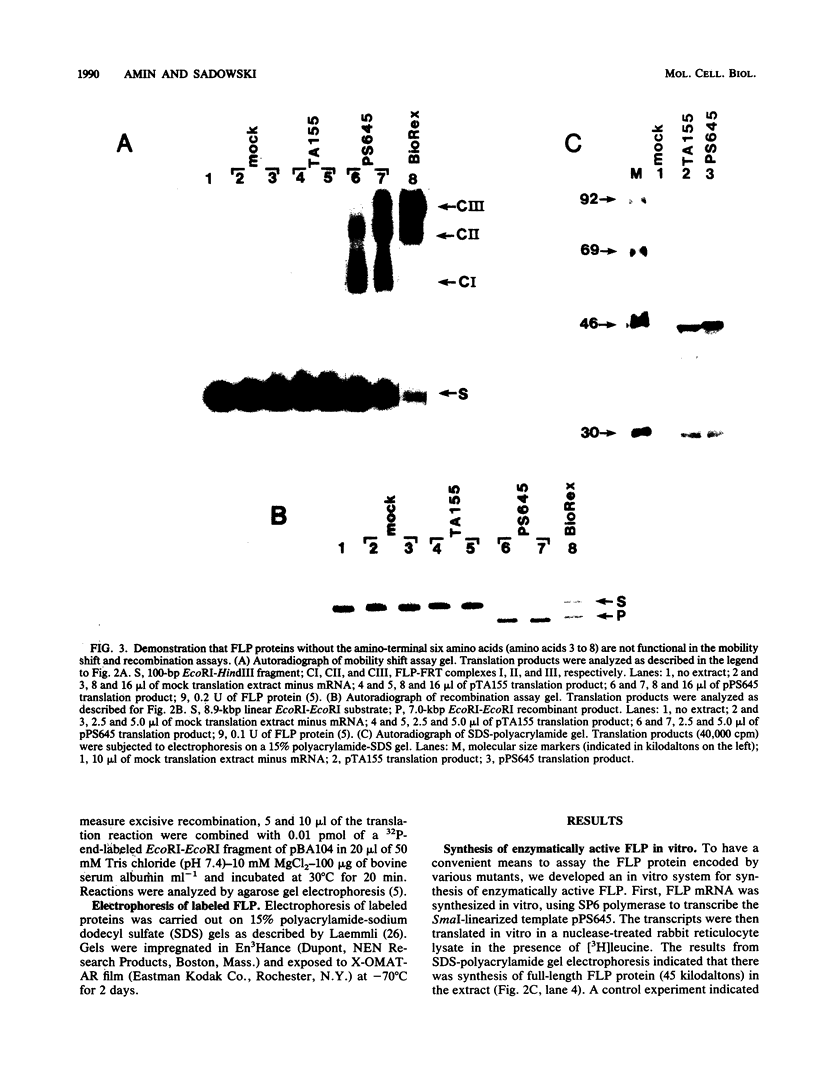
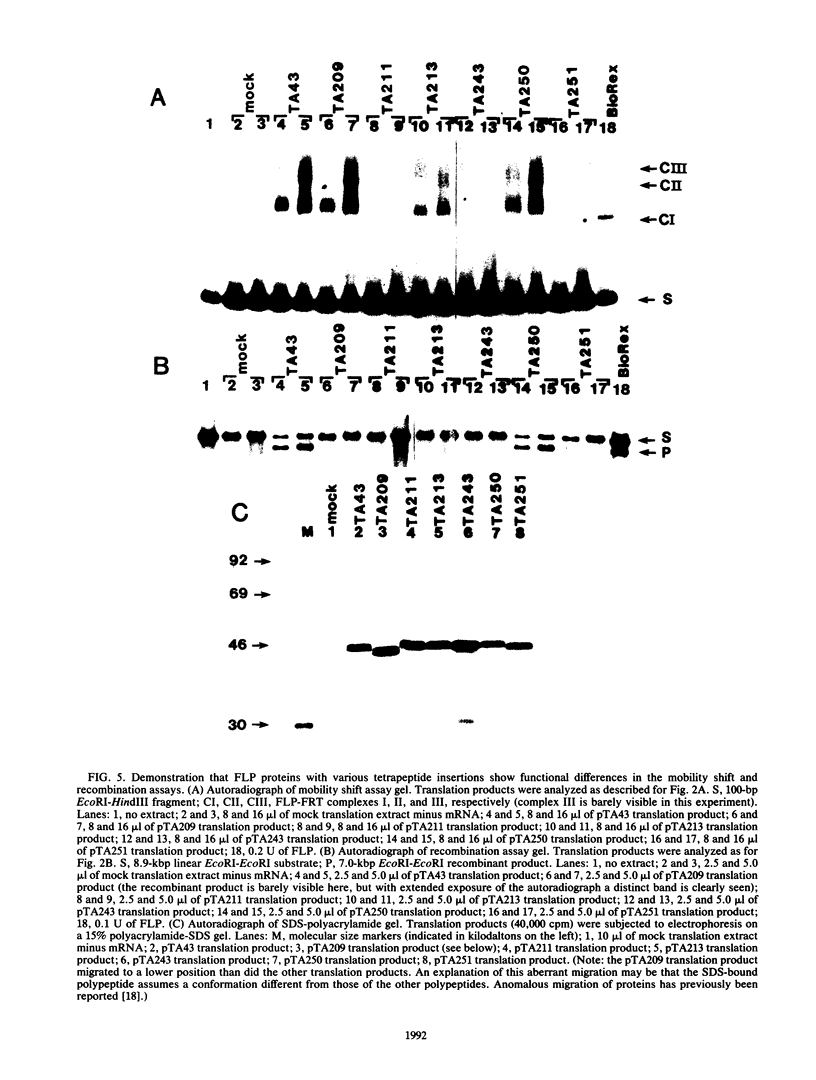
We have used an in vitro transcription and translation system to synthesize an enzymatically active FLP protein. The FLP mRNA synthesized in vitro by SP6 polymerase is translated efficiently in a rabbit reticulocyte lysate to produce enzymatically active FLP. Using this system, we assessed the effect of deletions and tetrapeptide insertions on the ability of the respective variant proteins synthesized in vitro to bind to the FLP recognition target site and to carry out excisive recombination. Deletions of as few as six amino acids from either the carboxy- or amino-terminal region of FLP resulted in loss of binding activity. Likewise, insertions at amino acid positions 79, 203, and 286 abolished DNA-binding activity. On the other hand, a protein with an insertion at amino acid 364 retained significant DNA-binding activity but had no detectable recombination activity. Also, an insertion at amino acid 115 had no measurable effect on DNA binding, but recombination was reduced by 95%. In addition, an insertion at amino acid 411 had no effect on DNA binding and recombination. On the basis of these results, we conclude that this approach fails to define a discrete DNA-binding domain. The possible reasons for this result are discussed.
Full text
PDF
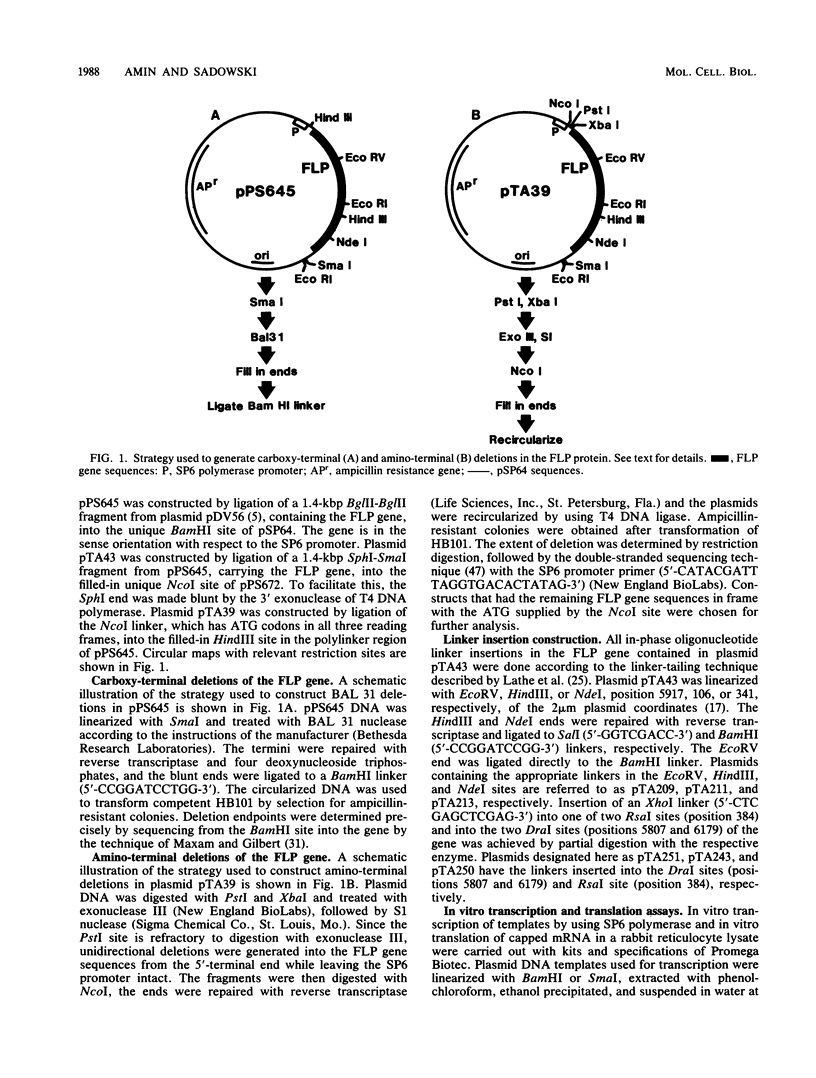




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Meguid S. S., Grindley N. D., Templeton N. S., Steitz T. A. Cleavage of the site-specific recombination protein gamma delta resolvase: the smaller of two fragments binds DNA specifically. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2001–2005. doi: 10.1073/pnas.81.7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. E., Ptashne M., Harrison S. C. A phage repressor-operator complex at 7 A resolution. Nature. 1985 Aug 15;316(6029):596–601. doi: 10.1038/316596a0. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., Beatty L. G., Sadowski P. D. Isolation of intermediates in the binding of the FLP recombinase of the yeast plasmid 2-micron circle to its target sequence. J Mol Biol. 1987 Jan 20;193(2):345–358. doi: 10.1016/0022-2836(87)90223-3. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Babineau D., Vetter D., Andrews B. J., Gronostajski R. M., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP protein of the 2-micron plasmid of yeast. Purification of the protein from Escherichia coli cells expressing the cloned FLP gene. J Biol Chem. 1985 Oct 5;260(22):12313–12319. [PubMed] [Google Scholar]
- Beatty L. G., Babineau-Clary D., Hogrefe C., Sadowski P. D. FLP site-specific recombinase of yeast 2-micron plasmid. Topological features of the reaction. J Mol Biol. 1986 Apr 20;188(4):529–544. doi: 10.1016/s0022-2836(86)80003-1. [DOI] [PubMed] [Google Scholar]
- Beatty L. G., Sadowski P. D. The mechanism of loading of the FLP recombinase onto its DNA target sequence. J Mol Biol. 1988 Nov 20;204(2):283–294. doi: 10.1016/0022-2836(88)90576-1. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Bruckner R. C., Cox M. M. Specific contacts between the FLP protein of the yeast 2-micron plasmid and its recombination site. J Biol Chem. 1986 Sep 5;261(25):11798–11807. [PubMed] [Google Scholar]
- Bruist M. F., Horvath S. J., Hood L. E., Steitz T. A., Simon M. I. Synthesis of a site-specific DNA-binding peptide. Science. 1987 Feb 13;235(4790):777–780. doi: 10.1126/science.3027895. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Berg J. M., Pabo C. O. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher A. B. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986 Mar 21;119(2):197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- Gronostajski R. M., Sadowski P. D. The FLP protein of the 2-micron plasmid of yeast. Inter- and intramolecular reactions. J Biol Chem. 1985 Oct 5;260(22):12328–12335. [PubMed] [Google Scholar]
- Gronostajski R. M., Sadowski P. D. The FLP recombinase of the Saccharomyces cerevisiae 2 microns plasmid attaches covalently to DNA via a phosphotyrosyl linkage. Mol Cell Biol. 1985 Nov;5(11):3274–3279. doi: 10.1128/mcb.5.11.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurstel S., Granger-Schnarr M., Daune M., Schnarr M. In vitro binding of LexA repressor to DNA: evidence for the involvement of the amino-terminal domain. EMBO J. 1986 Apr;5(4):793–798. doi: 10.1002/j.1460-2075.1986.tb04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen-Jacobson L., Lesser D., Kurpiewski M. The enfolding arms of EcoRI endonuclease: role in DNA binding and cleavage. Cell. 1986 May 23;45(4):619–629. doi: 10.1016/0092-8674(86)90294-1. [DOI] [PubMed] [Google Scholar]
- Jobling S. A., Gehrke L. Enhanced translation of chimaeric messenger RNAs containing a plant viral untranslated leader sequence. Nature. 1987 Feb 12;325(6105):622–625. doi: 10.1038/325622a0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lathe R., Kieny M. P., Skory S., Lecocq J. P. Linker tailing: unphosphorylated linker oligonucleotides for joining DNA termini. DNA. 1984;3(2):173–182. doi: 10.1089/dna.1984.3.173. [DOI] [PubMed] [Google Scholar]
- Lebreton B., Prasad P. V., Jayaram M., Youderian P. Mutations that improve the binding of yeast FLP recombinase to its substrate. Genetics. 1988 Mar;118(3):393–400. doi: 10.1093/genetics/118.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh E., Reiss N., Berent E., Ullrich A., Schlessinger J. An insertional mutant of epidermal growth factor receptor allows dissection of diverse receptor functions. EMBO J. 1987 Sep;6(9):2669–2676. doi: 10.1002/j.1460-2075.1987.tb02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Becktel W. J., Matthews B. W. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature. 1988 Aug 4;334(6181):406–410. doi: 10.1038/334406a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Pargellis C. A., Hasan N. M., Bushman E. W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988 Sep 23;54(7):923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- Moskaluk C., Bastia D. DNA bending is induced in an enhancer by the DNA-binding domain of the bovine papillomavirus E2 protein. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1826–1830. doi: 10.1073/pnas.85.6.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. A., Scarpa M., Rossi N., Cesareni G. Antagonistic controls regulate copy number of the yeast 2 mu plasmid. EMBO J. 1987 Dec 20;6(13):4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Parsons R. L., Prasad P. V., Harshey R. M., Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol Cell Biol. 1988 Aug;8(8):3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Horensky D., Young L. J., Jayaram M. Substrate recognition by the 2 micron circle site-specific recombinase: effect of mutations within the symmetry elements of the minimal substrate. Mol Cell Biol. 1986 Dec;6(12):4329–4334. doi: 10.1128/mcb.6.12.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Murray A. W., Szostak J. W. Roles of the 2 microns gene products in stable maintenance of the 2 microns plasmid of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi S., Yamamoto K. R. Functional dissection of the hormone and DNA binding activities of the glucocorticoid receptor. EMBO J. 1987 May;6(5):1309–1315. doi: 10.1002/j.1460-2075.1987.tb02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Stone J. C., Atkinson T., Smith M., Pawson T. Identification of functional regions in the transforming protein of Fujinami sarcoma virus by in-phase insertion mutagenesis. Cell. 1984 Jun;37(2):549–558. doi: 10.1016/0092-8674(84)90385-4. [DOI] [PubMed] [Google Scholar]
- Vetter D., Andrews B. J., Roberts-Beatty L., Sadowski P. D. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7284–7288. doi: 10.1073/pnas.80.23.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert F. C., Broach J. R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986 Aug 15;46(4):541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987 Jun 18;327(6123):591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]