Abstract
Interferon-stimulated transcription is thought to occur mainly through the action of the JAK/STAT pathway. However, recent findings revealed an additional PI3K/Akt-dependent pathway, which contributes to the induction of a set of interferon-stimulated genes (ISGs) through the regulation of the transcriptional repressor EMSY.
Keywords: Akt, BRCA2, EMSY, IRF9, ISRE, JAK/STAT pathway, interferon-stimulated gene (ISG)
The interferon response is a paradigm of transcriptional regulation by extracellular stimuli. While the intracellular pathways that link interferon stimulation to its transcriptional output have been studied for many years, new research continues to add complexity to this picture. In this article, we will discuss studies that have refined our understanding of the interface between interferon signaling and transcription.
Interferons (IFNs) are secreted proteins that are induced by pathogen-associated molecular patterns (PAMPs) that bind to, and activate Toll-like receptors (TLRs) residing in the plasma membrane or in endosomal membranes.1 Alternatively, they are induced by signals initiated by the binding of PAMPs to the cytoplasmic RNA helicases, retinoic acid-inducible gene I (RIG-I) and melanoma differentiation associated protein 5 (MDA5). Interferons fall mainly into three classes: Type I interferons belong to the helical cytokine family and they are characterized by a five α-helix bundle that is held together by two disulfide bonds. They are small (165–200 amino acid) non-glycosylated proteins and they include IFNα (leucocyte IFN), which consists of 13 subtypes and IFNβ (fibroblast IFN), which consists of only one subtype. The only type II IFN is IFNγ, a 140 amino acid glycosylated protein that is produced by natural killer cells (NK cells) and activated T cells. The type III IFNs include IFNλ, which consists of three subtypes that are co-produced with IFNβ but signal through a different receptor system (Table 1). IFNs bind to the extracellular domains of heterodimeric interferon receptor complexes to activate intracellular signaling pathways. Type I IFNs, which we will focus on here, bind to IFNAR1/IFNAR2 heterodimers. Their affinity to IFNAR2 is significantly higher and their interaction with the receptor tends to start by binding to this receptor subunit. Binding of Type I IFNs to their receptors results in the activation of the tyrosine kinases JAK1 and TYK2.1,2
Table 1. There are three types of interferons. Different IFN types signal through distinct receptors and signaling pathways.
| Type | Produced by | Receptor | Signal Transducers |
|---|---|---|---|
| Type I: IFN-α IFN-β Other* |
Broadly expressed |
IFNAR ½ |
STAT1 STAT2 lRF9 |
| Type II: IFN-γ |
NK cells Activated T cells |
IFNGR1/2 |
STAT1 |
| Type III: IFN-λ | Broadly expressed | IL10R2/ IFNLR1 |
STAT1 STAT2 IRF9 |
Several additional type I IFN sub-types are produced in a species-specific manner
The JAK/STAT pathway has long been considered the primary output of Type I IFN stimulation. Tyrosine phosphorylation of STAT1 and STAT2 by JAK1 and TYK2, results in the formation of a trimeric complex of STAT1, STAT2 and IRF9 (also known as ISGF3), which then translocates to the nucleus and binds to DNA. ISGF3 targets promoters of interferon-stimulated genes (ISGs) by binding to a DNA sequence known as the interferon stimulated response element (ISRE). Binding of this complex to ISG promoters results in the induction of ISGs.3
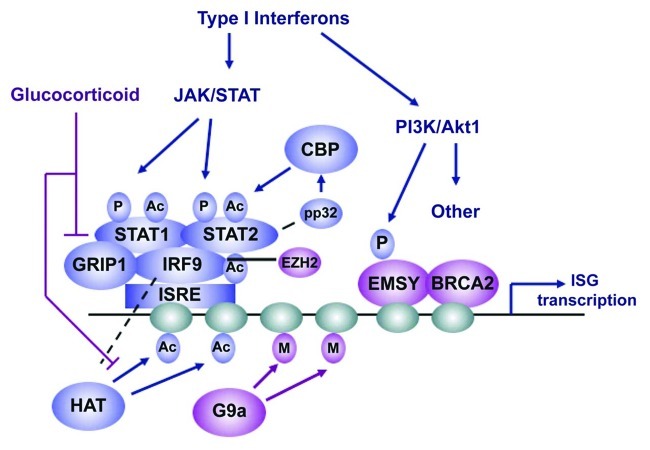
In addition to STAT1/2 phosphorylation, other post-translational modifications are necessary for ISG induction (Fig. 1). All three components of the ISGF3 complex are acetylated by CBP, which enhances the ISGF3 DNA binding activity.4 CBP has also been shown to interact with STAT2, and to play an obligatory role in the induction of ISGs.5 Inhibition of CBP by the adenovirus E1A protein is in fact sufficient to inhibit the IFN response in the course of adenovirus infection. In addition, the nuclear scaffolding protein pp32 interacts with STAT1/2 in response to interferon treatment and induces ISG expression via enhancement of histone acetylation within ISG promoters.6 It is interesting that in addition to histone acetyltransferases, histone deacetylases HDAC1/2 are also required for the function of the ISGF3 complex.7,8 Indeed, trichostatin A, an HDAC inhibitor, blocks ISG induction by preventing the formation of the ISGF3 complex in response to IFN. However, the specific targets of deacetylation have not been identified.9 STAT1 has also been reported to undergo arginine methylation on R31, by the arginine methyltransferase PRMT1. Methylation of STAT1 at this site prevents the binding of PIAS1, an inhibitor of STAT-dependent transcription.10 It also inhibits the binding of the tyrosine phosphatase TcPTP to STAT1 and blocks STAT1 dephosphorylation.11 However, the arginine methylation of STAT1 has been questioned and remains to be confirmed.12,13
Figure 1. Type I interferons activate both the JAK/STAT and PI3K/Akt pathways to activate ISG transcription. STAT1/2 and EMSY are phosphorylated in response to IFN (P). Both acetylation (Ac) and methylation (M) of histones controls ISG expression. Proteins that activate ISG transcription are shown in blue. Proteins that repress transcription are shown in purple.
H3K9 dimethylation within ISG promoters has been shown to inhibit promoter activation in response to both IFN stimulation and viral infection.14 The histone methyltransferase G9a, which catalyzes this modification, appears to be at least partially responsible for the H3K9me2-mediated ISG repression.14 Loss of G9a increases ISG expression and causes resistance to viral infection. Other studies revealed a functional interaction between ISGF3 and the H3K27 methyltransferase EZH2,15 and they showed that this interaction contributes to the downregulation of the tumor suppressor p73 in hepatocytes treated with IFNα. STAT2 colocalizes with EZH2 on the p73 promoter after IFNα treatment and this results in the increased abundance of H3K27me3 and in transcriptional repression. The role of DNA methylation in IFN-regulated transcription has not been extensively investigated. The promoter of one ISG, the antiviral gene IFITM3, has been reported to undergo DNA demethylation in response to IFNα stimulation, followed by re-methylation after the termination of the activation signal.16 Studies on the role of chromatin and DNA modifications in the IFN response are still at an early stage, and additional mechanisms contributing to the induction of ISGs should be expected in the future.
The crosstalk of the canonical IFN pathway with other signaling pathways has not been extensively studied. One pathway that interacts with the IFN response pathway is activated by glucocortocoid stimulation and blocks IFN-induced transcription without targeting the JAK/STAT pathway directly.17 Instead, it acts through the glucocortocoid receptor (GR) and the GR binding protein GRIP1, which potentiates ISG expression by acting as a cofactor of the ISGF3 complex. Glucocorticoid treatment also blocks IFN-induced histone acetylation within ISG promoters.17
The Akt protein kinase family consists of three members, Akt1, Akt2, and Akt3, which are activated by a variety of extracellular and intracellular PI-3K-dependent signals. Following activation, they phosphorylate numerous substrates to regulate a variety of cellular processes, including cell proliferation, survival, metabolism, and migration.18 IFN activates PI-3K and Akt via a pathway that starts with the binding of p85, the PI-3K regulatory subunit, to the IFN receptor. Studies published several years ago had shown that PI3K and Akt are required for the full activation of the interferon response by IFNα or IFNγ19. Knocking out p85 inhibited the transcriptional induction of ISG15 in response to interferon.19 Moreover, knocking out Akt1 and Akt2 in murine fibroblasts inhibited mTOR signaling and reduced the translation of ISG15, resulting in an impaired antiviral response.20 Finally, expression of a dominant negative form of Akt also inhibited the transcriptional response to IFNβ. These studies suggested that the PI3K/Akt pathway compliments the JAK/STAT pathway in interferon signaling. However, the mechanism through which PI3K regulates ISG transcription was not determined.
Recent data from our lab provided evidence for a novel Akt-dependent mechanism by which IFN and other growth factors regulate ISG expression.21 The central component of this pathway is EMSY, a BRCA2-interacting, chromatin associated protein that is amplified in human mammary adenocarcinomas.22 EMSY is one of the Akt1 phosphorylation targets identified in a recent phosphoproteomics screen designed to identify novel Akt isoform-specific phosphorylation substrates (manuscript in preparation). Because of earlier studies showing that BRCA2 regulates the expression of a set of ISGs (IFITM1, ISG15 and Viperin),23 we overexpressed or knocked down EMSY in human epithelial cells in culture and we observed that it functions as an ISG repressor. Overexpression of the phosphorylation site mutant (EMSYS209A) in the same cells revealed that EMSYS209A is a stronger ISG repressor and suggested that the phosphorylation of EMSY by Akt1 may relieve the repression. This was confirmed by overexpression of constitutively active Akt1, which was shown to drive expression of the same subset of ISGs21 in human epithelial cells that were either unmodified or they were engineered to express wild type EMSY, but not in human epithelial cells engineered to express EMSYS209A. In agreement with this conclusion, constitutively active Akt2 only minimally upregulated ISG expression. Moreover, the knockdown of Akt1 impaired ISG expression, while the knockdown of Akt2 did not.
ISGs are expressed at low levels in cells growing normally in complete media, even in the absence of IFN stimulation. The low level expression of ISGs in these cells is due to IGF and other growth factors normally present in the serum and depends on EMSY phosphorylation by Akt1. Type I IFN stimulation and viral infection further induce ISG expression. Moreover, full induction of ISGs by IFN also depends on EMSY phosphorylation by Akt1. More important, EMSY phosphorylation by constitutively active Akt1, but not Akt2, blocked the replication of two unrelated viruses, Herpes simplex virus-1 (HSV-1), a DNA herpesvirus and Vesicular stomatitis virus (VSV) (a ds RNA rhabdovirus) in human epithelial cells. These findings suggest that Akt1 inhibits viral replication by regulating the IFN response and that the role of Akt in the regulation of this pathway is isoform-specific. These findings are also reminiscent of the results of other studies showing that Akt1 and Akt2 have very different, and even opposing, roles in the regulation of pro-inflammatory Toll-like receptor signaling and macrophage function.24,25
Although it is clear that phosphorylation of EMSY by Akt1 relieves ISG transcriptional repression, the precise mechanism through which EMSY phosphorylation affects transcription is unclear; nevertheless, EMSY-mediated repression of ISGs requires its interacting partner BRCA2. Based on this data, we conclude that EMSY phosphorylation by Akt1 is required for full activation of a subset of ISGs by interferon or viral infection. The full induction of this subset of ISGs therefore in response to IFN stimulation occurs only after de-repression, mediated by Ser209 phosphorylation. This additional step may be necessary to prevent inappropriate expression of ISGs or to limit basal ISG expression, which may be damaging to tissues in the absence of an IFN-inducing signal.
EMSY has been reported to bind to a number of proteins involved in chromatin remodeling, modification, and transcriptional regulation. These include the heterochromatin-associated protein CBX1 and the NCoR-associated repressor ZMYND11.22 It is also found in complex with the zinc finger protein NIF-1 and is required for ligand-dependent upregulation of RARα target genes.26 The NIF-1, EMSY-containing complex also includes DBC1 (Deleted in breast cancer-1), which is known to regulate several histone-modifying enzymes.27,28 Finally, EMSY is found in complex with the histone chaperones ASF1 and NAP1, the co-repressor SIN3A, the histone deacetylases RPD3, the H3K36me3-binding protein MRG15, and the H3K4 demethylase KDM5.29 The effects of EMSY phosphorylation on chromatin modification and remodeling and the potential role of these functions on EMSY-mediated ISG regulation, remain to be determined.
Our increasing understanding of the mechanisms that control IFN-stimulated gene expression can lead to a more nuanced understanding the physiological significance of the pathway. For example, our data indicate that ISGs can respond directly to serum stimulation through PI3K/Akt1. This may indicate a greater role for ISGs in non-immune functions than had been previously appreciated. The regulation of a subset of ISGs by PI3K/Akt may contribute to the induction of distinct IFN responses, depending on the signaling landscape of the cell. It will be interesting to fully survey ISGs to identify those that respond to PI3K/Akt and to determine what distinguishes these genes from the ones that may not require Akt to respond to IFN, or to growth factors.
Interferons are important contributors to the biology of cancer. They stimulate the infiltration of tumors with NK cells and they promote the expression of MHC molecules and antigen presentation by dendritic cells.30 They also inhibit angiogenesis31 and promote tumor cell apoptosis.32 Although the antitumor effects of IFNs are well documented, the role of ISGs whose full activation depends on the IFN/Akt1/EMSY pathway in mediating these effects is not clear. IFITM1 has been shown to promote both proliferation and invasiveness.33-35 Also, in some settings IFITM1 and ISG15 enhance, while in other settings they decrease, the sensitivity of tumor cells to radiotherapy and chemotherapy.36-38 Future studies should address the spectrum of Akt1-sensitive ISGs in IFN-responsive malignancies and the effects of these ISGs on the therapeutic effectiveness of IFNs in these tumors.
The preceding data may have implications in the design of therapeutic interventions for cancer and other human diseases. One type of intervention that is being explored for the treatment of human cancer is the infection of the tumor cells with oncolytic viruses. Among such viruses are variants of HSV1 and VSV,39,40 whose replication is strongly inhibited by Akt1 via the Akt1/EMSY arm of the IFN response. Based on this information, it is expected that oncolytic anti-cancer viruses will synergize with Akt1 inhibitors for the targeting of cancer cells. Other studies are exploring whether ISGs, such as ISG15, whose induction by IFN depends on Akt1, can be targeted by immunotherapy and whether their targeting can be applied therapeutically in cancer.41 Finally, IFNs themselves have been shown to possess strong anticancer activities. The antitumor activities of IFNs have been exploited therapeutically for the treatment of human cancer, primarily for the treatment of melanomas, hairy cell leukemia and non-Hodgkin’s lymphomas.42 Given that Akt1 is required for the IFN response, the frequent activation of Akt1 in tumor cells may render them more sensitive than normal cells to the IFN treatment. By altering the response to IFN, the constitutive activation of Akt1 in cancer cells may also modulate the antitumor effects of the immune system responding to the tumor challenge naturally or after activation by immunotherapy. Other human diseases responding to IFN treatment include diseases caused by viruses, such as hepatitis B and C and autoimmune diseases, such as multiple sclerosis.42 Our data suggest that the therapeutic efficacy of IFN in these diseases may be enhanced via the concerted stimulation of the activity of Akt1 in the target cells.
In summary, our recent findings provide evidence for an additional Akt1 and EMSY dependent pathway that is required for the full activation of the IFN response. Akt1 activation plays a critical role in IFN-directed transcriptional regulation, and secondarily, in the regulation of the antiviral and other biological activities of IFNs. The identification of EMSY as an effector of the PI3K/Akt pathway and as a regulator of ISG expression indicates a need for a more detailed analysis ISG transcriptional regulation that goes beyond the JAK/STAT pathway.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21904
References
- 1.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Weerd NA, Nguyen T. The interferons and their receptors--distribution and regulation. Immunol Cell Biol. 2012;90:483–91. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath CM, Stark GR, Kerr IM, Darnell JE., Jr. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–64. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, et al. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–7. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 6.Kadota S, Nagata K. pp32, an INHAT component, is a transcription machinery recruiter for maximal induction of IFN-stimulated genes. J Cell Sci. 2011;124:892–9. doi: 10.1242/jcs.078253. [DOI] [PubMed] [Google Scholar]
- 7.Icardi L, Lievens S, Mori R, Piessevaux J, De Cauwer L, De Bosscher K, et al. Opposed regulation of type I IFN-induced STAT3 and ISGF3 transcriptional activities by histone deacetylases (HDACS) 1 and 2. FASEB J. 2012;26:240–9. doi: 10.1096/fj.11-191122. [DOI] [PubMed] [Google Scholar]
- 8.Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marié I, et al. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:9578–83. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Génin P, Morin P, Civas A. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J Virol. 2003;77:7113–9. doi: 10.1128/JVI.77.12.7113-7119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–41. doi: 10.1016/S0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu W, Mustelin T, David M. Arginine methylation of STAT1 regulates its dephosphorylation by T cell protein tyrosine phosphatase. J Biol Chem. 2002;277:35787–90. doi: 10.1074/jbc.C200346200. [DOI] [PubMed] [Google Scholar]
- 12.Komyod W, Bauer UM, Heinrich PC, Haan S, Behrmann I. Are STATS arginine-methylated? J Biol Chem. 2005;280:21700–5. doi: 10.1074/jbc.C400606200. [DOI] [PubMed] [Google Scholar]
- 13.Meissner T, Krause E, Lödige I, Vinkemeier U. Arginine methylation of STAT1: a reassessment. Cell. 2004;119:587–9, discussion 589-90. doi: 10.1016/S0092-8674(04)01090-6. [DOI] [PubMed] [Google Scholar]
- 14.Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, Chen MS, et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med. 2012;209:661–9. doi: 10.1084/jem.20112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Testoni B, Schinzari V, Guerrieri F, Gerbal-Chaloin S, Blandino G, Levrero M. p53-paralog DNp73 oncogene is repressed by IFNα/STAT2 through the recruitment of the Ezh2 polycomb group transcriptional repressor. Oncogene. 2011;30:2670–8. doi: 10.1038/onc.2010.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott R, Siegrist F, Foser S, Certa U. Interferon-alpha induces reversible DNA demethylation of the interferon-induced transmembrane protein-3 core promoter in human melanoma cells. J Interferon Cytokine Res. 2011;31:601–8. doi: 10.1089/jir.2010.0134. [DOI] [PubMed] [Google Scholar]
- 17.Flammer JR, Dobrovolna J, Kennedy MA, Chinenov Y, Glass CK, Ivashkiv LB, et al. The type I interferon signaling pathway is a target for glucocorticoid inhibition. Mol Cell Biol. 2010;30:4564–74. doi: 10.1128/MCB.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A, et al. Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling. J Immunol. 2008;181:7316–23. doi: 10.4049/jimmunol.181.10.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, et al. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci U S A. 2008;105:4808–13. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ezell SA, Polytarchou C, Hatziapostolou M, Guo A, Sanidas I, Bihani T, et al. The protein kinase Akt1 regulates the interferon response through phosphorylation of the transcriptional repressor EMSY. Proc Natl Acad Sci U S A. 2012;109:E613–21. doi: 10.1073/pnas.1115029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–35. doi: 10.1016/S0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 23.Tripathi MK, Chaudhuri G. Down-regulation of UCRP and UBE2L6 in BRCA2 knocked-down human breast cells. Biochem Biophys Res Commun. 2005;328:43–8. doi: 10.1016/j.bbrc.2004.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–31. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–22. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garapaty S, Xu CF, Trojer P, Mahajan MA, Neubert TA, Samuels HH. Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation. J Biol Chem. 2009;284:7542–52. doi: 10.1074/jbc.M805872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem. 2009;284:10361–6. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, et al. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–93. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S. Neutrophils responsive to endogenous IFN-beta regulate tumor angiogenesis and growth in a mouse tumor model. J Clin Invest. 2010;120:1151–64. doi: 10.1172/JCI37223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokrovskaja K, Panaretakis T, Grandér D. Alternative signaling pathways regulating type I interferon-induced apoptosis. J Interferon Cytokine Res. 2005;25:799–810. doi: 10.1089/jir.2005.25.799. [DOI] [PubMed] [Google Scholar]
- 33.Yu F, Ng SS, Chow BK, Sze J, Lu G, Poon WS, et al. Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J Neurooncol. 2011;103:187–95. doi: 10.1007/s11060-010-0377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatano H, Kudo Y, Ogawa I, Tsunematsu T, Kikuchi A, Abiko Y, et al. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin Cancer Res. 2008;14:6097–105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Goh SH, Song N, Hwang JA, Nam S, Choi IJ, et al. Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am J Pathol. 2012;181:43–52. doi: 10.1016/j.ajpath.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 36.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490–5. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai SD, Wood LM, Tsai YC, Hsieh TS, Marks JR, Scott GL, et al. ISG15 as a novel tumor biomarker for drug sensitivity. Mol Cancer Ther. 2008;7:1430–9. doi: 10.1158/1535-7163.MCT-07-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fumoto S, Shimokuni T, Tanimoto K, Hiyama K, Otani K, Ohtaki M, et al. Selection of a novel drug-response predictor in esophageal cancer: a novel screening method using microarray and identification of IFITM1 as a potent marker gene of CDDP response. Int J Oncol. 2008;32:413–23. [PubMed] [Google Scholar]
- 39.Longo SL, Griffith C, Glass A, Shillitoe EJ, Post DE. Development of an oncolytic herpes simplex virus using a tumor-specific HIF-responsive promoter. Cancer Gene Ther. 2011;18:123–34. doi: 10.1038/cgt.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moussavi M, Fazli L, Tearle H, Guo Y, Cox M, Bell J, et al. Oncolysis of prostate cancers induced by vesicular stomatitis virus in PTEN knockout mice. Cancer Res. 2010;70:1367–76. doi: 10.1158/0008-5472.CAN-09-2377. [DOI] [PubMed] [Google Scholar]
- 41.Wood LM, Pan ZK, Seavey MM, Muthukumaran G, Paterson Y. The ubiquitin-like protein, ISG15, is a novel tumor-associated antigen for cancer immunotherapy. Cancer Immunol Immunother. 2012;61:689–700. doi: 10.1007/s00262-011-1129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Curr Med Chem. 2007;14:1279–89. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]