Abstract
We develop a method for pH-dependent fusion between liposomes and cellular membranes using pHLIP® (pH Low Insertion Peptide), which inserts into lipid bilayer of membrane only at low pH. Previously we establish the molecular mechanism of peptide action and show that pHLIP can target acidic diseased tissue. Here we investigate how coating of PEGylated liposomes with pHLIP might affect liposomal uptake by cells. The presence of pHLIP on the surface of PEGylated-liposomes enhanced membrane fusion and lipid exchange in a pH dependent fashion, leading to increase of cellular uptake and payload release, and inhibition of cell proliferation by liposomes containing ceramide. A novel type of pH-sensitive, “fusogenic” pHLIP-liposomes was developed, which could be used to selectively deliver various diagnostic and therapeutic agents to acidic diseased cells.
Keywords: acidosis, membrane-associated folding, peptide, fusion, endocytosis, delivery of payload, ceramide, pH-sensitive
INTRODUCTION
Liposomes are widely used nanocarries that allow encapsulation of various therapeutics and diagnostic agents [1]. Once internalized by cells, liposomes must disrupt or fuse with the endosomal membrane in order to release the payload into the cytoplasm. Endosomal trapping of liposomes (and their contents within) pose a significant challenge to cytoplasmic delivery of the drug payload. Thus, pH-sensitive functionalities have been incorporated into liposomes to promote the mixing of lipids (between the liposome and the endosomal membrane) under the acidic environment of the endosomes, which in turn facilitates payload release into the cytoplasm [2–8]. Meanwhile, to mask against opsonization and to increase circulation time in blood, liposomes are usually coated with poly(ethyleneglycol) (PEG) polymers [9–12]. Unfortunately, the presence of PEG on the surface of liposomes hinders drug release, even when liposomes have been internalized via receptor mediated endocytosis [13]. One approach to avoid this difficulty is to use pH-sensitive PEG polymers that disassemble in the endosomal low pH environment, thus exposing lipid bilayer membrane of the liposome for direct interaction with the membrane of the endosome [2, 6, 14].
Liposomes can be fashioned into targeted drug carriers. Current targeting strategies usually involve conjugation of liposomes with antibodies or other ligands. However, these methods have limitations due to the heterogeneous expression of the targeted receptors in diseased tissues. Alternatively, we have developed a nanotechnology platform to target the microenvironment of diseased tissues. One of the common characteristics of diseased tissues (cancerous tumors, ischemia, inflammation, arthritis, atherosclerosis) is an acidic extracellular environment. We designed peptides that insert into cellular membranes under slightly acidic pH but not at normal pH. These peptides, called pHLIP®s (pH Low Insertion Peptides), demonstrated a high ability of targeting acidic tissues in vivo, (including tumors, arthritic sites and ischemic myocardium)[15–19]. The targeting mechanism of pHLIP is based on its pH-triggered insertion into lipid bilayers that ultimately results in the formation of a transmembrane helix [20–23]. One promising application is to use pHLIP for targeting liposomes towards diseased tissues. Here, new formulations of pH-sensitive, “fusogenic”, PEGylated liposomes are developed by including pHLIP in the liposomal coating. We demonstrated that pHLIP-PEG-liposomes bind and fuse with cellular membranes at low pH, but not at normal pH, and promote cellular uptake.
MATERIALS AND METHODS
Materials
The N-capped pHLIP peptide, Acetyl-NH- ACEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT- CO2H, was prepared by standard solid phase synthesis and purified at the W.M. Keck Foundation Biotechnology Resources Laboratory at Yale. The lipids, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000](ammonium salt) (DSPE-PEG2000-Maleimide), 1,2-distearoyl-sn-glycero-3- phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG2000), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[lissamine rhodamine B sulfonyl] (Rhod-PE), N-octanoyl-sphingosine-1-{succinyl[methoxy(polyethylene glycol)750]} (C8-PEG750 ceramide) and N-hexanoyl-D-erythro-sphingosine (C6 Ceramide), were purchased from Avanti Polar Lipids, Inc (Alabaster, AL). The fluorescein labeled lipids, N-(fluorescein-5-thiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (FITC-DHPE), rhodamine labeled fatty acid, octadecyl rhodamine B chloride (R18), and fluorescent dyes: Wheat germ agglutinin Texas Red, ER-tracker green, mito-tracker deep red, 8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt (ANTS) and p-xylene-bis-pyridinium bromide (DPX) were from Life Technologies (Carlsbad, CA). The fatty acid conjugated with 1.4 nm gold nanoparticles, palmitoyl nanogold and HQ Silver™ Enhancement Kit were purchased from Nanoprobes, Inc (Yaphank, NY). DNA staining dye, propidium iodide (PI), was from Sigma-Aldrich Co, LLC (St. Louise, MO).
Synthesis of DSPE-PEG2000-pHLIP
DSPE-PEG2000-pHLIP was synthesized by the covalent conjugation of DSPE-PEG2000-maleimide with the single cysteine residue at the N-terminus of pHLIP. In a typical coupling reaction, 600 nmol of peptide was mixed with 500 nmol of DSPE-PEG2000-Maleimide in 200 μL of methanol and left for overnight stirring at room temperature under argon. The existence of the product was ensured by SELDI-TOF mass-spectrometry analysis.
Liposome preparation
Liposomes were prepared by extrusion. A chloroform solution of the desired lipid mixture (1 μmol) including solution of DSPE-PEG2000-pHLIP in methanol was vortexed and evaporated using rotary evaporator, producing an even thin film. pHLIP is an unstructured peptide, which could be dissolved in organic solvents including methanol and chloroform. The lipid film was left under a vacuum for overnight to remove residuals of organic solvent. The hydration of the dry lipid film was accomplished by adding 1 mL of stock buffer solution (10 mM phosphate, 150 mM NaCl, pH 8) to the flask of lipids and agitating. Typically, a hydration time of 1 hr with vigorous vortexing was needed for pHLIP-containing liposome. After 10 freeze-thaw-vortex cycles, the resulting multilamellar liposome solution was extruded 15 or 31 times through 50, 100 or 200 nm polycarbonate filters and sterilized by filtering through 0.2 μm filter. The final lipid concentration was re-calculated by knowing the concentration of fluorescent lipids, which was obtained spectrophotometrically. Small amount of liposomes were diluted in methanol and absorbance of rhodamine (ε556=125,000 M−1cm−1) or fluorescein (ε495=88,000 M−1cm−1) was measured. When loading with propidium iodide (PI), ANTS or DPX was required, PI (4 mM), ANTS (25 mM) or DPX (90 mM) were added as a component of the rehydration solution. After liposome extrusion, free PI, ANTS or DPX were removed by dialysis using dialysis tube with MWCO of 8000 Da, followed by stirring in 1 L buffer solution for 24 hrs with 3 times buffer refreshing, or by fast spin semi-dry column using sephadex G25. In each case we ensure that ionic strength of solution within liposomes was the same as outside of liposomes. The ionic strength was regulated by addition of NaCl.
Cryo-electron microscopy
About 5 μL droplet of liposome solution was spread on an electron microscopy grid and preserved in a frozen-hydrated state by a rapid freezing in liquid ethane. The vitrification process was performed via FEI Vitrobot System (Hillsboro, OR), with the setting of a single blot of 3 sec, an offset of −1, and drain and wait time of 1 sec. Samples were imaged using JEOL 2100 TEM with an accelerating voltage of 200 kV at magnifications of 20,000x and 40,000x.
DLS measurements
The size of liposomes was measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS instrument with Malvern disposable cuvettes (Malvern USA, Southborough, MA) right after the extrusion of liposomes and in some cases, within a week after preparation. Samples were typically 200 nmol liposomes in 1 mL of aqueous buffer (10 mM phosphate, 150 mM NaCl, pH 8.0).
Inter-liposomal fusion assay
R18-labeled liposomes (50 μL, 200 μM) were mixed with an equal volume of unlabeled POPC vesicles (lipid concentration was varied from 0 to 6 mM) in an aqueous buffer (10 mM phosphate, 150 mM NaCl, pH 8.0). The R18 fluorescence intensity was monitored on an ISS spectrofluorometer (Champaign, IL) at excitation and emission wavelengths set at 556 nm, and 590 nm, respectively, as the pH was lowered from 8 to 4 (via addition of small amounts of HCl). A circulating water bath was connected to cuvette holder to control sample temperature at 22°C. Complete fusion and mixing of lipids was achieved via 5 freeze-thaw cycles, the fluorescence intensity obtained afterwards was defined as ‘100% fusion’. The degree of liposome fusion in % was calculated according to the equation:
where F0 is the initial fluorescent intensity of liposome mixture at pH8 (no fusion), Fmax is the maximum fluorescent intensity of liposome mixture at pH 4 after 5 freeze-thaw cycles (100% fusion), and F is the fluorescent intensity of liposome mixture at pH 4 before freeze-thaw cycles.
In other fusion assay, 100 μM of 50 nm DOPC liposomes of ‘5:5’ formulation containing 5% R18 and encapsulated with 25 mM of fluorescent dye ANTS were mixed with equal volume of 2 mM of 200 nm POPC liposomes encapsulated with 90 mM of DPX, which is a quencher of ANTS fluorescence, in the 10 mM phosphate buffer of pH 8 containing of 40 mM NaCl. The pH was reduced by addition of aliquot of concentrated HCl. The increase of R18 fluorescence signal and decrease of ANTS signal due to the liposome fusion and mixing of ANTS with DPX were observed on the same liposomes. ANTS fluorescence was excited at wavelength of 360 nm and observed at 530 nm. In addition, changes of scattering signal monitored at 650 nm with excitation at 650 nm were recorded.
Cell lines
Lung carcinoma A549 and human cervix adenocarcinoma HeLa cell lines were obtained from American Type Culture Collection (Manassas, VA). Cells were authenticated, stored according to supplier’s instructions, and used within 3–4 months after frozen aliquots resuscitations. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 10 μg/mL of ciprofloxactin in a humidified atmosphere of 5% CO2 and 95% air at 37°C. By serial passages some cells were adapted for the growth in low pH medium (pH 6.5). The pH 6.5 media was prepared by mixing 13.5 g of dry DMEM powder with 0.2 g of sodium bicarbonate in 1 L of deionized water.
Cellular uptake of fluorescently labeled liposomes
Typically, a suspension of A549 cells was prepared by dissociating 4–6 days old monolayer grown in 75cm2 flask with trypin, EDTA (Gibco, Gaithersburg, MD). Trypsinized cells were counted using a hemacytometer and diluted to 1 × 106 cells/mL in serum-free media (for 1 hr incubation) or PBS (for 15 min incubation) as stock solutions. The liposomes (20 nmol), containing fluorescent probe lipids or fatty acid, were incubated with 2 × 105 cells in 1 mL of the desired solution for 1 hr or 15 min at 37°C or 4°C. In some studies, ATP-depleting media (DMEM + 20 mM NaN3/50 mM 2-deoxy-glucose) was added for 1 hr at 37°C prior to liposome addition and was kept throughout the liposome incubation period. After incubation, the cells were pelleted by centrifugation (2000 rpm, 4 min) at room temperature (or 4°C). Then the supernatant was removed and the pellet was resuspended in 1 mL of fresh serum-free media (or PBS) and centrifuged second time. The second pellet was resuspended in 100 μL of the same media. Sequentially, 20 μL of the cell suspension solution was loaded into a counting chamber and the fluorescent signal of individual cells was measured using Nexcelom Cellometer Vision (Lawrence, MA). For each measurement, fluorescent data of 2000–3000 cells were obtained and statistically analyzed. All samples were prepared at least in triplicate for each formulation of liposomes. The fluorescent signals of untreated cells were used as a common divisor for normalization of all the fluorescent data in each assay. Cell viability was confirmed by addition of trypan blue to cell sample. After cellometer counting, the residual cells were reseeded in collagen-coated cell dishes and viewed under the microscope after 24, 48 and 72 hrs.
Fluorescence microscopy
HeLa and A549 cells adapted for low pH growth (pH 6.5 and pH 6.2, respectively) were seeded in collagen-coated cell dishes (5,000 and 15,000 cells/dish, respectively). After 24 hr the culture medium was removed and cells were incubated with 6 nmol of fluorescein-labeled liposome, DOPC/DSPE- PEG2000-pHLIP/DSPE-PEG2000/fluorescein-DHPE (85:5:5:5) in 240 μL of PBS (pH 6.3) for 1 hr at 37°C under 5% CO2, followed by 3 times washing with sterile PBS (pH 7.4). Sequentially, wheat germ agglutinin Texas Red (10 μg/mL) was used to stain the plasma membranes of live cells for co-localization studies. For the cells treated with R18 labeled liposome, ER-tracker green and Mito-tracker Deep Red FM were used to stain endoplasmic reticulum (ER) and mitochondria of live cells. Fluorescent images were acquired with a Retiga CCD camera (Qimaging, Burnaby, BC, Canada) mounted to an inverted Olympus IX71 microscope (Olympus America, Inc., Center Valley, PA). Fluorescent images from different stains were obtained in the same field of view with corresponding filters.
Cellular uptake of nanogold labeled liposomes
A549 cells adapted for low pH media, grown to 80–90% confluence on 8-well chamber slide were incubated in 200 μL serum-free DMEM (pH 6.5) with 12 nmol of nanogold labeled liposome for 15 min at 37°C under 5% CO2. After incubation, the medium was removed and cells were washed three times with sterile PBS (pH7.4) and fixed in cold methanol for 10 min, followed by washing twice with sterile PBS (pH7.4) and once with distilled H2O. After air-drying, cells were treated with silver enhancement solution (following manufacture’s instruction) and viewed under the light microscope.
Delivery of PI
A549 cells adapted for low pH media were seeded in collagen-coated cell dishes (5,000 cells/dish). After 24 hrs, the culture medium was removed and cells were treated with 10 nmol of PI loaded liposome DOPE/pHLIP-PEG-DSPE/DSPE-PEG2000/fluorescein-DHPE (85:5:5:5 or 85:0:10:5) in 100 μL of serum-free DMEM (pH 6.5) for 1 hr at 37°C under 5% CO2. After incubation, the medium was removed and cells were washed three times with sterile PBS (pH 7.4) and viewed under fluorescence microscope.
Cytotoxicity Assay
A549 cells grown in standard growth medium of pH 7.4 and A549 cells adapted for low pH (pH 6.2) growth medium were loaded in the wells of 96-well plates (~5,000 and 7,000 cells per well, respectively) and incubated overnight. Next day cells were treated with increasing concentrations (0, 10, 50, 100, 200, 400 μM) of DOPE and DOPC/pHLIP-PEG-DSPE/DSPE-PEG2000 (85:5:5:5 or 85:0:10:5) for 2 hrs followed by the addition of equal volume of growth medium with 20% of serum of the same pH (to have 10% FBS of final concentration, since constructs were not removed). Cell viability was assessed after 1 and 2 days of incubation with constructs using the colorimetric reagent (CellTiter 96 AQueous One Solution Assay, Promega), which was added for 1 hr to cells followed by measuring absorbance at 490 nm. All samples were prepared in triplicate.
Inhibition of cell proliferation by liposomal ceramide
A549 cell suspension (160,000 cells) were incubated with 160 nmol of liposomal ceramide in 1 mL of 10 mM phosphate buffer (150 mM NaCl, 1 mM MgCl2 and 1 mM CaCl2) at pH 6.1 or pH 8 for 1 hr at 37°C under 5% CO2. After incubation, cells were pelleted by centrifugation (2000 rpm, 4 min) at room temperature followed by the removal of supernatant. The pellet was resuspended in 1 mL of fresh serum-free media and centrifuged second time. The second pellet was reseeded into 96-well tissue culture plates, 5,000 cells per well with 250 μL medium. After 72 hrs, the medium was removed and Cell Titer 96 Aqueous One solution (Promega Co., Madison, WI) was added to each well. The viability of cells was measured by using a SpectraMax plate reader (Molecular Devices, Inc. Sunnyvale, CA) at 490 nm. Relative viability was calculated with cells treated only with medium alone as a control.
RESULTS
Liposome composition and characterization
The main goal of our investigation is to demonstrate that pHLIP can promote cellular uptake of PEGylated liposomes and enhance lipid mixing and fusion between liposomal and cellular membranes. It is well established that PE lipids (such as DOPE), which are minimally hydrated and have small headgroups, possess high fusogenic propensity [24]. In our study, we tested both fusogenic (DOPE-based) and non-fusogenic (DOPC-based) liposomes.
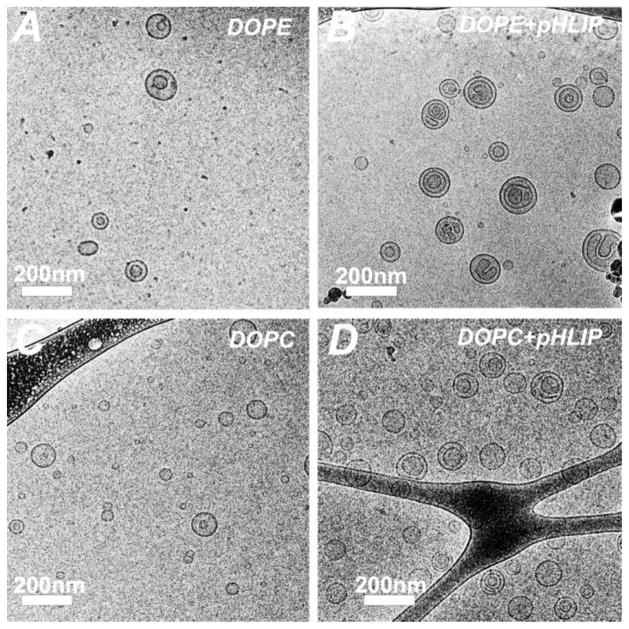
To introduce pHLIP in a liposome coat, we conjugated DSPE-PEG2000maleimide lipid with Cys residue in the N-terminal region of pHLIP. Due to the fact that pHLIP is an unstructured polypeptide, it could be dissolved in organic solvent with no harm to its structure. The conjugation product was characterized by SELDI-TOF mass spectrometry (Figure 1). The narrow peak at 4154 Da corresponds to pHLIP, and the broad peak centered at ~7000 Da corresponds to the conjugation product DSPE-PEG2000-pHLIP. DSPE-PEG2000-pHLIP was used for preparation of liposomes containing pHLIP. The detailed composition of liposomes used in the study is given in Table 1. The first number in the formulation code refers to the mole percentage of pHLIP-PEG lipids (DSPE-PEG2000-pHLIP), whereas the second number refers to the amount of PEG lipids (DSPE-PEG2000). The different formulations of liposomes were characterized by cryo-TEM and DLS. Special attention was given to the stability of DOPC/DOPE ‘5:5’ and ‘10:0’ formulations containing 5% and 10% DSPE-PEG2000-pHLIP, respectively. The average size of DOPC and DOPE liposomes of ‘5:5’ formulation was about 125 nm in diameter with PDI (polydispersity index) of about 0.06 and Zeta potential of about -16 mV. The measurements were performed within several days after liposomes preparations and numbers represent averaged values. The average size of DOPE and DOPC liposomes containing 10% DSPE-PEG2000-pHLIP was larger in size. The DOPE liposomes, were stable for several days with average size of 150 nm, PDI of 0.06 and Zeta-potential of -19.5 mV. However, the DOPC‘10:0’ liposome formulation was very unstable with increased size of liposomes of 200 nm. Figure 2 shows cryo-TEM images of DOPC/DOPE liposomes with and without pHLIP in liposome coat. These liposomes have formulations “0:10” (an example of a control liposome without pHLIP) and “5:5” (an example of a pHLIP-containing liposome) (Table 1). The TEM images illustrate that our liposome samples consist of a mixture of small unilamellar (<100 nm) and multilamellar vesicles, mostly with two bilayers (~100 nm). pHLIP-containing liposomes show more complex multilamellar structures, especially for liposomes consisting of fusogenic DOPE lipids (Figure 2B), which is most probably due to the pHLIP-induced membrane fusion. Since pHLIP has high affinity to the lipid bilayer of membrane, fusion promoted by pHLIP results in formation of multilamelar structures rather than bigger liposomes. Such multilamelar structures allow interaction of pHLIP peptides with lipid bilayer of the same liposome.
Figure 1. SELDI-TOF mass spectrum of DSPE-PEG2000-pHLIP.
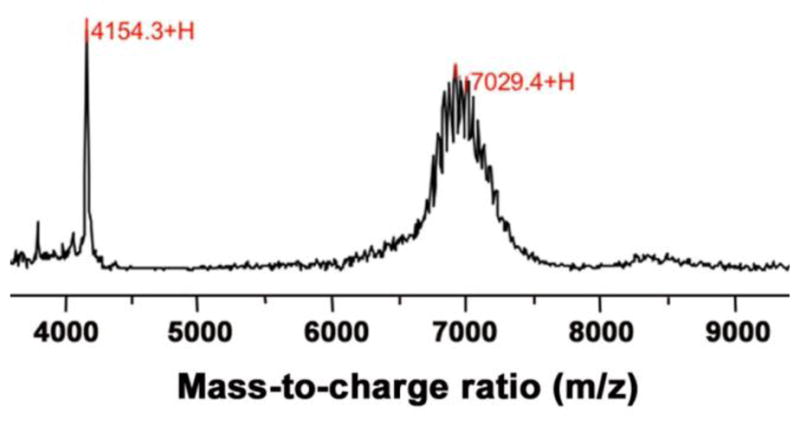
The narrow peak at 4154 corresponds to the pHLIP peptide. The broad peak centered at ~7000 Da corresponds to the conjugated product DSPE-PEG2000-pHLIP. The individual spikes on the broad peak differ from each other by the repeating unit(s) of PEG (mass: 44 Da). The marked mass at 7029 corresponds to the expected mass of the product minus one unit of PEG (7074 – 44 = 7030 Da).
Table 1.
Formulation of liposomes used in various experiments
| Formulations of fluorescently labeled liposomes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Liposome codea | ‘0:10’ | ‘1:9’ | ‘5:5’ | ‘10:0’ | ‘0:5’ | ‘1:4’ | ‘5:0’e; | ‘0:1’ | ‘1:0’ |
| DOPC (or DOPE) | 83%b | 83% | 83% | 83% | 88% | 88% | 88% | 92% | 92% |
| DSPE-PEG2000-pHLIP | 0% | 1% | 5% | 10% | 0% | 1% | 5% | 0% | 1% |
| DSPE-PEG2000 | 10% | 9% | 5% | 0% | 5% | 4% | 0% | 1% | 0% |
| Fluorescent lipidsc | 7% | 7% | 7% | 7% | 7% | 7% | 7% | 7% | 7% |
| Formulations of nanogold labeled liposomes | ||
|---|---|---|
| Control-liposome | pHLIP-liposome | |
| DOPE | 86% | 86% |
| DSPE-PEG2000 | 10% | 0% |
| DSPE-PEG2000-pHLIP | 0% | 10% |
| Palmitoyl Nanogold | 2.5% | 2.5% |
| R18 | 1.5% | 1.5% |
| Formulations of ceramide liposome | ||
|---|---|---|
| Control-liposome | pHLIP-liposome | |
| DOPC | 37% | 37% |
| DOPE | 17.5% | 17.5% |
| DSPE-PEG2000 | 7.5% | 2.5% |
| DSPE-PEG2000-pHLIP | 0% | 5% |
| C8-PEG750 ceramide | 7.5% | 7.5% |
| C6 ceramide | 30% | 30% |
| R18 | 0.5% | 0.5% |
Notes:
Ratio in liposome code refers to molar ratio between pHLIP-PEG-lipids and PEG-lipids.
Percentages indicate mole fractions.
Fluorescent lipids are R18, Rho-PE, or fluorescein-DHPE.
Figure 2. Cryo-TEM images of R18- labeled liposomes.
The control liposomes without pHLIP are shown on panels (A) (DOPE/DSPE- PEG2000/R18, 83:10:7) and (C) (DOPC/DSPE-PEG2000/R18, 83:10:7); and the pHLIP coated liposomes are shown on panels (B) (DOPE/DSPE- PEG2000/DSPE-PEG2000-pHLIP/R18, 83:5:5:7) and (D) (DOPC/DSPE- PEG2000/DSPE-PEG2000-pHLIP/R18, 83:5:5:7). The images were obtained using JEOL 2100 TEM with an accelerating voltage of 200 kV. The total lipid concentration of samples shown is 1 mM.
Inter-liposomal fusion
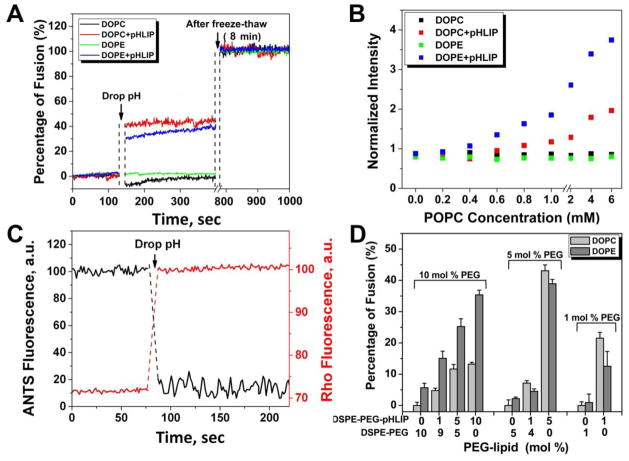
First, we investigated pHLIP-induced membrane fusion on the model system of phospholipid vesicles. Inter-liposomal fusion was monitored by fluorescence increase of octadecyl rhodamine B (R18), which was part of liposome composition. When rhodamine dye molecules are in close proximity to each other, such as in the case of high concentration (7 mol % of R18) in two-dimensional membrane space, the self-quenching occurs [25]. The fluorescence increase is observed when liposomes containing R18 fuse with unlabeled POPC liposomes, which leads to the decrease of effective concentration of R18, that is, an increase of averaged distance between rhodamine dyes. Four types of R18-labeled liposomes were studied, i.e. DOPC- or DOPE-based PEG-liposomes with or without pHLIP (Figure 3).
Figure 3. Inter-liposomal fusion assays.
(A) Increase of rhodamine fluorescence reflects fusion of membranes. The fluorescence changes were monitored in the solution of R18-labled DOPC or DOPE liposomes with or without pHLIP in liposome coat (200 μM total lipid concentration) mixed with equal volume of 6 mM POPC at pH 8. The addition of POPC liposomes to the solution of R18-labeled DOPC or DOPE liposomes at pH 8 leads to the fluorescence decrease due to the dilution (data not shown). Spectral signal at pH 8 was taken as ‘0% fusion’, the fluorescence was monitored several minutes after pH drop to pH 4, followed by 5 freeze-thaw cycles performed to induce complete fusion (‘100% fusion’). Liposome formulations are ‘5:0’ (with pHLIP) and ‘0:5’ (without pHLIP). (B) Dependence of fluorescence intensity changes of rhodamine on concentration of POPC liposomes when mixed with 200 μM (total lipid concentration) of R18-labled DOPC or DOPE liposomes with or without pHLIP in liposome coat. Liposome formulations are ‘10:0’ (with pHLIP) and ‘0:10’ (without pHLIP). (C) To confirm that increase of rhodamine fluorescence occurs due to fusion of liposomes, another assay was carried. 50 nm DOPC liposomes of ‘5:5’ formulation with entrapped ANTS at pH 8 were mixed with 200 nm POPC with entrapped DPX, a quencher of ANTS fluorescence. The DOPC liposomes also contained 5% of R18. The increase of rhodamine fluorescence in a result of pH drop from pH 8 to pH 4 (red line) was accompanied by the decrease of ANTS fluorescence (black line) in a result of liposomes fusion and mixing of ANTS and DPX. (D) The efficiency of pHLIP-mediated inter-liposomal fusion depends on the amount of PEGylated lipids (with 5 mol % of PEG-lipids as the optimal). The detailed formulation of liposome composition is presented in Table 1.
Typical changes of R18 fluorescence are shown in Figure 3A. The fluorescence signal of solution containing unlabeled POPC liposomes (6 mM) mixed with an equal volume of R18-labeled liposomes (200 μM) at pH 8 was taken as 0% of fusion. After equilibrating at pH 8 for ~ 2 min, the pH was dropped from 8 to 4, and increase of fluorescence indicating on fusion was observed only for liposomes containing pHLIP in coat. Non-physiological pH 8 and pH 4 in this model assay on liposomes were used to ensure that all pHLIP molecules exist in either state II or state III, respectively. Fluorescence signal measured after 5 freeze-thaw cycles was taken as 100% of fusion. Furthermore, we investigated dependence of fluorescence increase on amount of unlabeled POPC liposomes (Figure 3B). Mixing of R18-liposomes containing pHLIP with increasing concentrations of unlabeled POPC liposomes led to the enhancement of de-quenching, indicating that more inter-liposomal fusion had occurred. On the contrary, control liposomes (without pHLIP) showed no fluorescence change as the concentration of unlabeled POPC increased. Thus, inter-liposomal fusion is strongly dependent on the presence of pHLIP.
To confirm that increase of R18 fluorescence is associated with fusion, not just flip-flopping of R18 we performed another fusion assay. 50 nm DOPC liposomes of ‘5:5’ formulation encapsulated with fluorescent dye ANTS were mixed with 200 nm POPC liposomes encapsulated with DPX, quencher of ANTS fluorescence. The fluorescence signal of ANTS was decreased only when pH was reduced from 8 to 4 (Figure 3C). The decrease of ANTS emission could occur only in the result of fusion of liposomes and mixing of ANTS with DPX.
pHLIP-mediated inter-liposomal association (‘sticking’) takes place at pH 8, since binding of pHLIP to lipid bilayer at pH8 (without insertion into membrane) is associated with energy release of about 6 kcal/mol [23]. At the end of the equilibration period of 2 min at pH 8, most of pHLIP peptides could be associated with an inter-liposomal POPC membrane, and the subsequent pH drop triggers membrane fusion, since pHLIP inserts into bilayer and adopts TM configuration within about 60 sec after changes of pH [22]. The peptide inserted state is stabilized by release of additional 2 kcal/mol of energy [23]. Our data indicate that most of pHLIPs prefer the inter-liposomal POPC membrane surface over its own intra-liposomal DOPE or DOPC surface, which is perhaps because adequate level of PEGylation effectively shields the intra-liposomal membrane from association with pHLIP. However, we cannot exclude that some amount of peptides inserts in membranes of their own liposomes as well.
The effect of PEGylation level (1–10 mol %), pHLIP content (1–10 mol %), and liposome type (DOPC vs. DOPE) on the efficiency of inter-liposomal fusion was further investigated (Figure 3D). We established that 5 mol % of PEG2000 is an optimal level of PEGylation. This level of PEGylation roughly corresponds to the transition between the tight packing ‘brush’ conformation and the loose ‘mushroom’ conformation of the liposomal PEG shield [26]. A lower level of PEGylation at 1 mol % led to the reduction of fusion for both DOPC and DOPE pHLIP-liposomes. In this case, we suspect that the membrane surface is not adequately shielded by PEG and too many pHLIP molecules associate and insert into the intra-liposomal DOPC/DOPE membrane at pH 8 and 4, respectively. However, more PEGylation at 10 mol % also led to lesser inter-liposomal fusion, especially for DOPC pHLIP-liposomes, perhaps. because shielding of the DOPC surface is too complete and membrane fusion becomes difficult. At 10 mol % PEGylation, DOPE-based liposomes show higher percentage of fusion than DOPC-based liposomes (35% vs. 12 %), in agreement with the fusogenic nature of PE lipids. However, when the amount of PEGylation is reduced to 5 mol %, DOPC-based pHLIP-liposomes are just as adapt at membrane fusion as DOPE-based liposomes. Given the same overall amount of PEGylated lipids, (1, 5, or 10 mol %), the presence of more pHLIP molecules always correlate with higher levels of membrane fusion, except of DOPC liposomes containing 10% of PEG-pHLIP (compare to DOPC liposomes containing just 5% of PEG-pHLIP), which most probably is associated with pure stability of this particular formulation. Therefore all experiments on cells were performed with liposomes containing no more than 5% of PEG-pHLIP.
Cellular uptake of liposomes
Cellular toxicity of liposomes with (‘5:5’) or without (‘0:10’) pHLIP was tested on A549 cells at both pH 6.2 and pH 7.4. Cells were treated with increasing concentrations of liposomes (up to 400 μM of lipids) for 1 and 2 days followed by standard MTS assay to check cells viability. No any cellular toxicity was observed (data not shown).
Membrane fusion between liposomes and cells was evaluated in a cell suspension assay. A549 cells in suspension were treated with fluorescently labeled pHLIP-liposomes (“DOPE+pHLIP” or “DOPC+pHLIP”) or control-liposomes without pHLIP (“DOPE” or “DOPC”) under various conditions. After washing, fluorescence of individual cells was counted using a cellometer. The average fluorescence of ~ 2000 treated cells was calculated and normalized to that of untreated cells. First, we investigated whether pHLIP can induce membrane fusion between liposomes and cells in a pH-dependent fashion. A549 cells were treated with R18-labled DOPC or DOPE liposomes with or without pHLIP at pH 7.4 or 6.5. Incubation with cells was either 15 min in PBS (Figure 4A) or 60 min in serum-free DMEM (Figure 4B). For DOPC liposomes at pH 7.4, no difference in fluorescence intensity was observed between cells treated with pHLIP-liposomes vs. cells treated with control liposomes. However, at pH 6.5, the presence of pHLIP increased the amount of lipid mixing between liposomes and cell. The difference in cellular uptake of DOPE-based liposomes with and without pHLIP was observed already at pH 7.4, while the effect became more pronounced at pH 6.5. The results support the idea that pHLIP insertion into cellular membrane at low pH can facilitate uptake of liposomes by cells. Due to the fusogenic nature of DOPE lipids, DOPE-based liposomes show much higher cellular uptake than DOPC-based liposomes in general, which is in agreement with the data of inter-liposomal fusion assay discussed previously. The different incubation time of 15 min vs. 60 min might reflect different pathways of liposome-cell interaction. During short incubation period, predominantly direct liposomal fusion with plasma membrane occurs; while during long incubation period, both fusion and cellular internalization through endocytosis (most probably, macropinocytosis) could happen. Our data show that the fluorescence signal is significantly higher at longer incubation time, especially at low pH, suggesting that endocytosis plays an important role in pHLIP-mediated cellular uptake of liposomes.
Figure 4. Cellular uptake of liposomes.
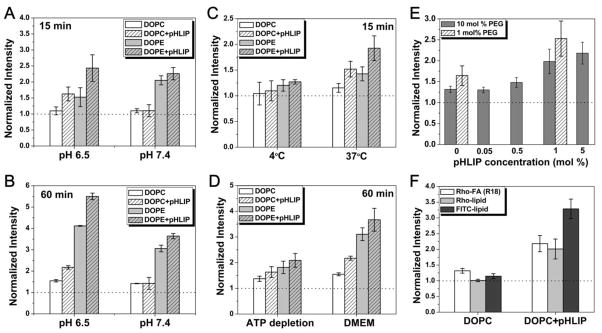
A549 cells in suspension were treated with fluorescently labeled liposomes (DOPC or DOPE) with or without pHLIP in liposome coat under various conditions. Cellular uptake of fluorescent liposomes was accessed by counting of fluorescent cells on a cellometer. All data were normalized to the fluorescence intensity of untreated cells (dotted line). (A–B) pH- and pHLIP-dependent cellular uptake of liposomes is shown. Cells were incubated with R18-labeled liposomes with (‘5:5’) or without (‘0:10’) pHLIP in liposome coat at pH 7.4 or 6.5 in PBS for 15 min (A) or in serum-free DMEM for 60 min (B). (C–D) Cells were incubated with R-18 labeled liposomes at pH 6.5 in PBS at 4°C or 37°C for 15 min (C) and in serum-free DMEM or ATP depletion medium at 37°C for 60 min (D). Low temperature and ATP depletion medium are used to reduce endocytotic uptake. (E) Cellular uptake of DOPC liposomes (pH 6.5, DMEM, 37°C, 60 min) depends on amount of pHLIP and PEG in liposome coat. (F) The pH- and pHLIP-dependent cellular uptake does not depend on choice of fluorescent lipids. Liposomes (‘5:5’ DOPC) containing different fluorescent lipids—Rho-FA (R18), Rhodamine-PE (Rho-lipid), or Fluorescein-DHPE (FITC-lipid)—were incubated with cells (pH 6.5, DMEM, 37°C, 60 min), and pHLIP-dependent fluorescence increase was observed in each case.
Next, cell treatment was carried out at low temperature or in an ATP-depletion medium to reduce energy dependent endocytic uptake. The decrease of temperature significantly (~ 80%) reduced overall liposome uptake (Figure 4C). Regardless of the route of entry (being endocytosis or direct interaction with plasma membrane), first, pHLIP needs to insert into the cellular membrane in an environment of low pH. Our kinetics studies indicate that the rate of insertion is reduced with decrease of temperature [22]. Therefore, we assume that experiments at low temperature would affect both fusion, and endocytotic pathways of liposomal uptake. Results from the experiments of cells treated at pH 6.5 for 60 min in the ATP depletion medium or serum-free DMEM (Figure 4D), indicate that endocytosis is responsible for ~ 50% of pHLIP-mediated cellular uptake of liposomes.
How does the amount of pHLIP and PEG present on liposome coat influence the level of cellular uptake of liposomes? To investigate this question, cells were incubated with DOPC-liposomes of various compositions in serum-free DMEM at pH 6.5 for 60 min. When the total amount of PEG-lipids was held constant, the presence of more pHLIP led to higher cellular uptake (Figure 4E, dark grey bars within the 10 mol % total PEG-lipid series). Furthermore, less amount of PEGylation was more favorable for membrane fusion, resulting in higher cellular uptake (Figure 4E, 1 mol % vs. 10 mol %). The results are consistent with the data obtained in inter-liposomal fusion assay.
The uptake of liposomes was accessed using the fluorescence of Rho-FA (R18). In place of R18, we also incorporated rhodamine-PE or fluorescein-DHPE into the liposomes as alternative fluorescent probes. Cells were incubated with DOPC-liposomes with or without pHLIP in serum-free DMEM at pH 6.5 for 60 min. A similar pattern of pHLIP-dependent cellular uptake of liposomes was observed regardless of fluorescent lipid probe used (Figure 4F). Therefore, pH- and pHLIP-dependent cellular uptake of liposomes is not influenced by the choice of the fluorescent lipid.
Fluorescence microscopy
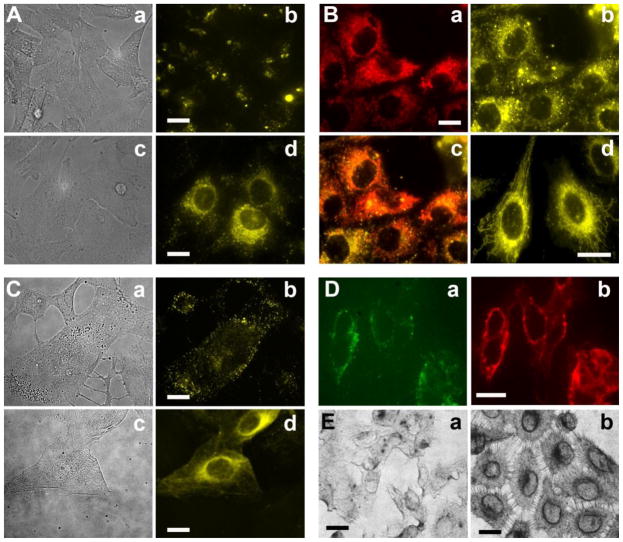
Fluorescent lipids incorporated in liposomes allow visualization of liposome-cell interactions using fluorescence microscopy. A549 cells in suspension were incubated with DOPE liposomes with or without pHLIP in serum-free DMEM medium at pH 6.5 for 1 hr. After washing, treated cells were grown on collagen-coated dishes and became adherent. The light (Figure 5A-a, -c) and fluorescent images (Figure 5A-b, -d) were taken 4 days after cell treatment with liposomes. Cells treated with pHLIP-containing liposomes show much stronger fluorescence than cells treated with liposomes containing no pHLIP. It is evident that R18 of liposomes containing pHLIP targeted specific cellular compartment. To figure out what organelle was marked by R18 fluorescence, cells treated with pHLIP-liposomes containing R18 (Figure 5B-b) were labeled with Mito-tracker and ER-tracker to stain mitochondria (Figure 5B-a) and endoplasmic reticulum (data not shown), respectively. The co-localization study reveals that the R18 (Figure 5B-b) was co-localized in mitochondria (Figure 5B-c, 5B-d). R18 contains a hydrophobic lipid tail and rhodamine ester, which is positively charged at physiological pH. It is well known that such amphiphilic molecules specifically target mitochondria in living cells [27–30]. It is important to outline that effective mitochondria targeting by R18 was observed mostly in case of pHLIP-mediated uptake of liposomes by cells.
Figure 5. Cellular localization of liposomes.
(A) A549 cells in suspension were incubated with R18 DOPE liposomes with (c & d: ‘5:5’) or without pHLIP (a & b: ‘0:10’) in liposome coat in serum-free DMEM medium at pH 6.5 for 1 hr. The light (a, c) and fluorescent images (b, d) were taken 4 days after cell treatment with liposomes. (B) Mito-tracker fluorescent dye was used to stain mitochondria of cells treated with R18 DOPE liposomes containing pHLIP (a). pHLIP mediated uptake of R18 liposomes show specific staining of mitochondria (b). An overlay of (a) & (b) is shown in panel (c) to be certain that the staining pattern of R18 is truly due to mitochondrial localization. On the panel (d) a zoom-in picture of mitochondria stained with R18 liposome is shown. (C) Adherent A549 cells were treated with Rho-PE DOPE liposomes with (c & d: ‘5:5’ formulation) or without pHLIP (a & b: ‘0:10’) in PBS at pH 6.3 for 30 min. Both light (a, c) and fluorescent images (b, d) were taken immediately after washing. (D) Adherent A549 cells were treated with fluorescein DOPC liposomes with 5 mol % of pHLIP-PEG-lipids (PBS, pH 6.2, 1 hr). Staining of plasma membrane by FITC-lipids of liposomes shown on panel (a) is confirmed by staining of the plasma membrane of the same cells by Texas Red-labeled wheat germ agglutinin, as shown on panel (b). (E) Adherent A549 cells were treated with nanogold-containing DOPE liposomes with or without pHLIP in serum-free DMEM (pH 6.5, 15 min). After fixation and silver enhancement, cells were visualized under the light microscope (a: no pHLIP; b: with pHLIP). Liposomes containing 10 mol % of pHLIP delivered more nanogold to cells (image b) than control liposomes without pHLIP (image a). The scale bar is 20 μm in all images.
Liposomes containing a different rhodamine lipid probe, Rhod-PE, were also investigated. In this case, the rhodamine portion of the molecule is zwitterionic and neutral, (while the overall molecule is −1 negatively charged due to the phosphate anion in the lipid headgroup). Adherent A549 cells were incubated with Rhod-PE labeled liposomes with or without pHLIP in PBS at pH 6.3 for 30 min, and distribution of fluorescent signal was monitored under microscope immediately after cell washing. Compared to cells treated with control liposomes without pHLIP (Figure 5C-b), cells treated with pHLIP-containing liposomes (Figure 5C-d) were more fluorescent and have different distribution of fluorescent signals. Unlike R18, Rhod-PE lipids do not stain mitochondria, but instead accumulate in perinuclear region and plasma membranes (Figure 5C-d). By contrast, punctate staining was observed for cells treated with liposomes without pHLIP (Figure 5C-b), indicating endosomal trapping of liposomes.
In addition, liposomes containing fluorescein-DHPE probes (5 mol %) were also tested. Adherent A549 cells were treated with fluorescein-labeled pHLIP-containing DOPC liposomes in PBS, pH 6.2, for 1 hr (Figure 5D-a) and the same cells were stained with wheat germ agglutinin conjugated with Texas Red to visualize plasma membrane (Figure 5D-b). It is clearly seen that the plasma membrane of cells treated with pHLIP-containing liposomes was stained with fluorescein lipids, revealing that pHLIP induced fusion between liposome and plasma membrane.
Finally, instead of fluorescent probes, nanogold particles (1.4 nm) conjugated to fatty acid, were introduced in liposome coat. Adherent A549 cells were treated with nanogold-containing DOPE liposomes with or without pHLIP in serum-free DMEM at pH 6.5, for 15 min, followed by fixation and silver enhancement to enlarge nanogold particles up to micron size particles and visualize them under light microscope. Liposomes containing pHLIP delivered more nanogold to cells (Figure 5E-b) than liposomes without pHLIP (Figure 6E-a). Similar to the localization of Rhod-PE probe, nanogold particles are clearly visible at perinuclear region and plasma membranes of treated cells.
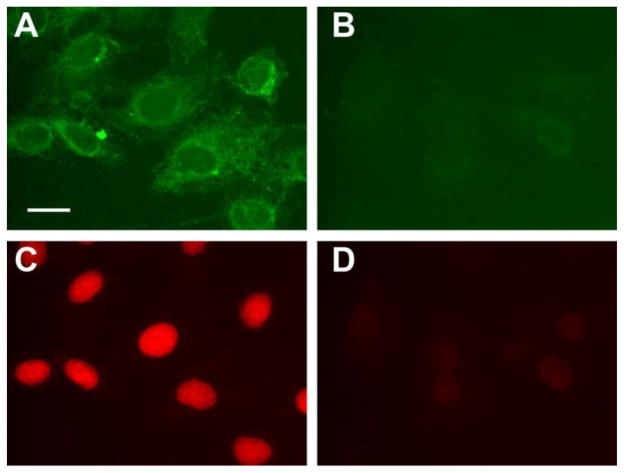
Figure 6. pHLIP-mediated liposomal delivery of propidium iodide (PI).
Adherent A549 cells were incubated in serum free DMEM (pH 6.5, 1 hr) with fluorescein-labeled DOPE liposomes loaded with PI, with or without pHLIP (‘5:5’ or ‘0:10’ formulation) in liposomal coating. Fluorescent images of cells treated with pHLIP-containing liposomes (A & C) or control liposomes without pHLIP (B & D) are shown side by side. The fluorescent images of the same cells were taken with two filter settings: green - fluorescein fluorescence; red - PI fluorescence. The exposure time is 172 ms for A & B and 80 ms for C & D. The scale bar is 20 μm.
Delivery of an encapsulated cargo
The main goal in effective use of liposomes, as a delivery agent, is to deliver and release polar payload into cells. Therefore, we investigated pHLIP-mediated liposomal delivery of an encapsulated cargo. Propidium iodide (PI), a membrane-impermeable DNA staining dye, was encapsulated in liposomes to serve as the model payload. Adherent A549 cells were incubated in serum free DMEM at pH 6.5, for 1 hr with fluorescein-labeled DOPE liposomes loaded with PI, with or without pHLIP in liposomal coating (Figure 6). Fluorescein (Figure 6A–B) and PI (Figure 6C–D) fluorescent images, taken after 1 hour treatment, evidently demonstrate that pHLIP-containing liposomes delivered much more PI to the nuclei than control liposomes. Further, more staining of cellular membranes was observed on cells treated with pHLIP-containing liposomes. According to the data presented on Figure 4, endocytosis is responsible only for ~ 50% of the pHLIP-mediated cellular uptake of liposomes and the other 50% are due to the pHLIP-induced direct fusion with plasma membrane (Figure 4D). Fusion is much faster than endocytosis. Our study shows that the internalization of liposomes was observed even after 15 min treatment (Figure 4). Thus, we assume that the release of PI encapsulated in liposomes occurred as a result of fusion of liposomes with plasma membrane. These results are consistent with the notion that pHLIP can promote membrane fusion and lipids exchange, leading to enhancement of payload release.
Delivery of liposomal ceramide inhibites proliferation of cancer cells
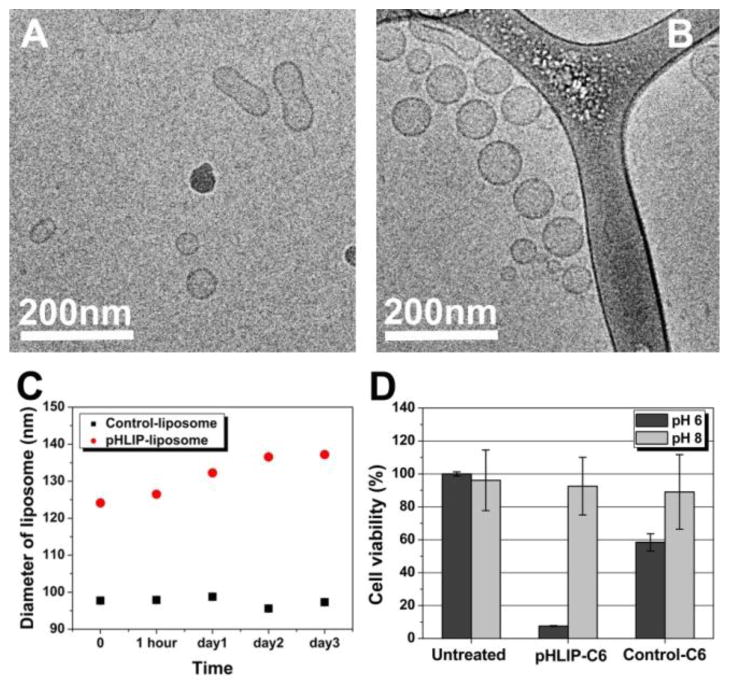
Our data suggest that pHLIP can enhance cellular delivery of liposomes. Can this approach be applied toward the delivery of cytotoxic agents of therapeutic potential? It was demonstrated that PEGylated liposomes containing short chain C6 ceramide can induce apoptosis in human breast cancer cells [31–33]. We prepared pHLIP-containing liposomes with 30 mol % of C6 ceramide to investigate whether pHLIP can enhance the cellular delivery of this cytotoxic signaling lipid. Cryo-TEM images confirmed the integrity of the ceramide- and pHLIP-containing liposomes (Figure 7A, B). The size of the liposomes were measured immediately after extrusion (Figure 7C) and then monitored for 3 days (Figure 7D). Ceramide-containing liposomes with pHLIP were slightly larger (~ 125 nm in diameter) than liposomes without pHLIP (~ 95 nm in diameter). Nonetheless, aggregation of pHLIP-containing liposomes was not significant during the 3-day follow-up period. These data confirmed that the ceramide liposomes were uniform in size and stable in solution.
Figure 7. pHLIP-mediated delivery of liposomal ceramide inhibits proliferation of cancer cells at low pH.
(A–B) Liposomes containing 30 mol % of C-6 ceramide were characterized using Cryo-TEM: control liposomes without pHLIP and liposomes containing pHLIP are shown on panels A and B, respectively. (C) DLS was used to investigate the changes of liposome size with time. The size of the liposomes was measured immediately after liposome extrusion and monitored for 3 days. (D) A549 cells in suspension were incubated with ceramide liposomes (with or without pHLIP) at pH 8.0 or 6.1 for 60 min in a phosphate buffer. After washing, treated cells were grown in 96-well plates for 3 days before viable cell populations were estimated via MTS assays.
The cytotoxic effects of ceramide-containing liposomes were evaluated in anti-proliferation assay. First, A549 cells in suspension were incubated with ceramide-liposomes with or without pHLIP, at pH 8.0, or pH 6.1, for 60 min in a phosphate buffer. pH 8 was used to enhance pH-dependent effect of pHLIP-coated liposomes. After washing, treated cells were grown on 96-well plates (at ~ 5000 cells per well) for 3 days before viable cell populations were counted by MTS colorimetric assay. When the liposome treatment was carried out at pH 8, control liposomes exhibit some toxicity to the cells, while pHLIP-coated liposomes show practically no toxicity (Figure 7F). By contrast, when the treatment was carried out at pH 6.1, significant differences in cell viability was observed between cells treated with pHLIP- and ceramide-containing liposomes (< 10% of cell survival), and cells treated with control liposomes containing ceramide, but no pHLIP (~ 60% of cell survival) (Figure 7E). Compared to untreated cell population, the treatment with ceramide-containing liposomes at low pH alone inhibited <40% of cell proliferation, whereas the presence of pHLIP in liposome coat contributed to an additional 50% of inhibition, presumably by elevating the intracellular concentration of C6 ceramide. Thus, pHLIP enhances delivery of liposomal ceramide to cells at low pH, which induces cell death.
DISCUSSION
pH-sensitive liposomes were designed to mimic the highly efficient intracellular delivery systems utilized by viruses and pathogenic organisms. There are two main pathways of viruses entry into cells: direct fusion with plasma membrane and endocytotic uptake by host cells [34–41]. Membrane fusion also occurs in membrane trafficking, including neurotransmitter release in synaptic transmission, gamete formation in sexual reproduction, and myotube formation in organ development [42]. In all these cases, two distinct membranes have to come close to each other, merge, mix, which leads to the fusion. This process is triggered and regulated by fusion proteins. The entire fusion machinery of viruses is a single fusion protein within their own membrane, while intracellular fusion machines consist of several proteins located in both membranes to be fused. Despite the differences between viral and intracellular fusion proteins, their main role is to bring two individual membranes together [34, 37, 42–45]. Only upon activation, often by low pH in endosomes, does the fusion peptides (which are a part of the fusion protein) become exposed and available for insertion into the target membrane [42]. The process of membrane fusion mediated by fusion proteins can be as fast as 8 ms [46].
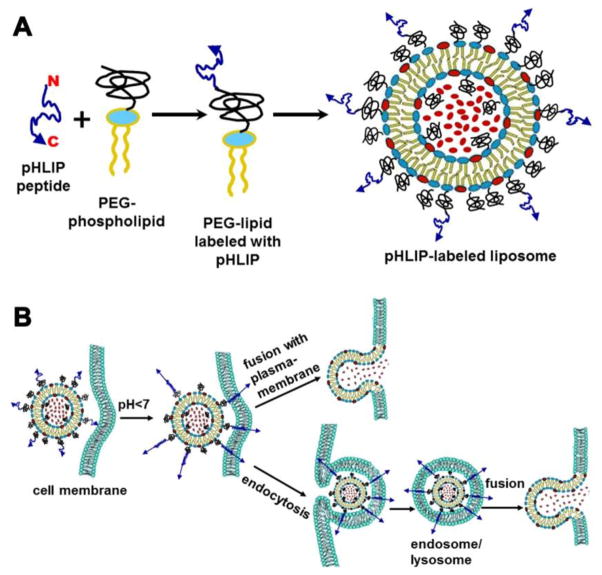
Inspired by the action of fusion peptides, which insert into the target membrane to induce fusion, we decorated PEG-coated liposomes with pHLIP (Figure 8A). Our data indicate that pHLIP adopts random coil configuration in aqueous solution at normal and high pH [21]. We assume that on the surface of liposomes coated with PEG polymer, pHLIP would adopt coil configuration as well. Our estimation of volume occupied by single pHLIP coil according to the freely-jointed model of polymer physics and the surface area on the liposomes available for the coating indicates that about 50% of liposome surface is coated with pHLIP peptides in the case of ‘5:5’ liposome formulation. At pH<7.0 Asp/Glu residues are protonated, which increases peptide hydrophobicity and affinity to membrane and leads to the peptide insertion into lipid bilayer and formation of a stable transmembrane helix. The biophysical studies indicate that process on model membrane could be completed within 60 sec [47]. Our biophysical inter-liposomal assays clearly demonstrates that indeed pHLIP induces fusion of both fusogenic (DOPE-based) and non-fusogenic (DOPC-based) PEG-liposomes with POPC vesicles at low pH. The content of liposomes is mixing in a result of the fusion. Experiments on cultured cells further confirmed pHLIP’s ability to induce fusion and lipid mixing between PEG-liposomes and biological membranes, evidenced by the following: i) pHLIP-induced cellular uptake of DOPE- and DOPC-based PEG-liposomes are dependent on acidic extracellular pH, ii) fluorescent lipid probes (or nanogold-lipid conjugates) brought in by the liposomes are distributed throughout the cell. Important that lipid mixing occurs not in a result of flip-flopping of fluorescent lipids and fatty acids, but in a result of membrane fusion, which leads to the efficient cytoplasmic delivery of liposomal payload. In addition to delivering probes and model cargos we also showed that, at low extracellular pH (pH 6.0), pHLIP can enhance the biological effect of PEGylated liposomes containing short chain C6 ceramide, which induces apoptosis and cell death [48, 49]. We concluded that the presence of up to 5 mol % of PEG polymer in the liposomal coatings favors pHLIP interaction with the inter-membrane surface over its own intra-liposomal surface. The amount of pHLIP peptide in the liposome coat correlates with the degree of fusion: the more pHLIP, the more fusion.
Figure 8. Schematic presentation of pHLIP/PEG coated liposomes (A) and their interaction with cellular membranes (B).
The encapsulated model payload molecules and the head-groups of fluorescent lipids are shown in red.
Our data indicate that there might be several routes of liposome entry into the cell: i) direct fusion with plasma membrane, and ii) endocytotic uptake of liposomes, and then, fusion with endosomal membrane (Figure 8B). In both cases, the first event is the insertion of pHLIP peptides into target membrane triggered by reduced pH, bringing liposomal and cellular membranes close to each other, which can induce fusion of liposome with cellular membrane. Since there are many pHLIP peptides on a liposome surface, their simultaneous insertion might lead to plasma membrane raffling and blebbing, which may trigger internalization of liposomes. It is well documented that pHLIP shows direct spontaneous insertion into membrane of liposomes and cells in vitro and in vivo, and this process does not involve receptor interaction [19, 50, 51]. Therefore, we assume that plasma membrane raffling and blebbing might promote macropinocytosis, which is an endocytic route of cellular internalization not associated with the activation of a particular receptor on the cellular surface [52–54]. More detailed investigation is needed to confirm that pHLIP-triggered endocytotic uptake of liposomes occur via macropinocytosis. When endosomes with encapsulated liposomes detach from the plasma membrane inside a cell, they undergo acidification. Low pH (5.0–5.5) inside endosomes would promote even more efficient insertion of pHLIP into the endosomal membrane, leading to lipid mixing and fusion. As a result, liposomal payload is released into the cytoplasm and liposomal lipids could be found in the membrane of various cellular compartments.
pHLIP-coated liposomes represent a novel type of pH-sensitive “fusogenic” liposomes. In pHLIP-coated liposomes, the fusogenic properties are determined not only by the choice of lipids, but also by the presence of membrane-inserting peptides on the liposome surface, as in some pathogens. The energy of membrane-associated folding of pHLIP is utilized to bring liposomal and cellular membranes close to each other to induce lipid mixing, or membrane blebbing and raffling. Since extracellular acidity is associated with the development of various pathological states, such as solid tumors, ischemic stroke, neurotrauma, epileptic seizure, inflammation, infection, wounds, cystic fibrosis and others, pHLIP-coated liposomes could be used to deliver and release various diagnostic and therapeutic agents to these sites of disease.
CONCLUSIONS
We introduced new formulation of pHLIP-PEG-coated liposomes. pHLIP promotes cellular uptake of both fusogenic (DOPE-based) and non-fusogenic (DOPC-based) liposomes at low pH. Our data indicate that about 50% of cellular uptake of pHLIP-PEG-coated liposomes at low pH could be attributed to the direct fusion with plasma membrane and about 50% due to the endocytotic pathway of cellular entry. Lipids and fatty acids conjugated with various reporters (such as fluorescent dyes or gold nanoparticles), when incorporated into pHLIP-PEG-coated liposomes, were distributed to various cellular compartments, including mitochondrial, nuclear and plasma membranes, at the same time, encapsulated in liposome polar cargo, such as propidium iodide, was delivered to nuclear DNA at low pH. We show that pHLIP-PEG-coated liposomes containing C6-ceramide induced cell death much more effectively (~90% inhibition) than PEG-liposomes containing no pHLIP in their coating (~40% inhibition) at low pH. Since low extracellular pH is a characteristic feature of many pathological states, we assume that pHLIP-PEG-coated liposomes could be used for selective delivery and cytoplasmic release of diagnostic and therapeutic agents.
Acknowledgments
We thank Prof. Ming An (SUNY-Binghamton) for reading the manuscript and useful comments. The work was supported by the NIH grant CA133890 to OAA and YKR. Mass spectrometry and cellometer analysis was done in RI-INBRE core facility funded by NCRR/NIH P20RR016457.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Torchilin VP. Passive and active drug targeting: drug delivery to tumors as an example. Handb Exp Pharmacol. 2010:3–53. doi: 10.1007/978-3-642-00477-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Boomer JA, Qualls MM, Inerowicz HD, Haynes RH, Patri VS, Kim JM, Thompson DH, et al. Cytoplasmic delivery of liposomal contents mediated by an acid-labile cholesterol-vinyl ether-PEG conjugate. Bioconjug Chem. 2009;20:47–59. doi: 10.1021/bc800239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fattal E, Couvreur P, Dubernet C, et al. “Smart” delivery of antisense oligonucleotides by anionic pH-sensitive liposomes. Adv Drug Deliv Rev. 2004;56:931–946. doi: 10.1016/j.addr.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 4.Budker V, Gurevich V, Hagstrom JE, Bortzov F, Wolff JA, et al. pH-sensitive, cationic liposomes: a new synthetic virus-like vector. Nat Biotechnol. 1996;14:760–764. doi: 10.1038/nbt0696-760. [DOI] [PubMed] [Google Scholar]
- 5.Guo X, Szoka FC, Jr, et al. Steric stabilization of fusogenic liposomes by a low-pH sensitive PEG--diortho ester--lipid conjugate. Bioconjug Chem. 2001;12:291–300. doi: 10.1021/bc000110v. [DOI] [PubMed] [Google Scholar]
- 6.Simoes S, Moreira JN, Fonseca C, Duzgunes N, de Lima MC, et al. On the formulation of pH-sensitive liposomes with long circulation times. Adv Drug Deliv Rev. 2004;56:947–965. doi: 10.1016/j.addr.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 7.Yuba E, Kojima C, Harada A, Tana, Watarai S, Kono K, et al. pH-Sensitive fusogenic polymer-modified liposomes as a carrier of antigenic proteins for activation of cellular immunity. Biomaterials. 2010;31:943–951. doi: 10.1016/j.biomaterials.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez M, Aranda FJ, Teruel JA, Ortiz A, et al. New pH-sensitive liposomes containing phosphatidylethanolamine and a bacterial dirhamnolipid. Chem Phys Lipids. 2011;164:16–23. doi: 10.1016/j.chemphyslip.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Moghimi SM, Szebeni J, et al. Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog Lipid Res. 2003;42:463–478. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 10.Klibanov AL, Maruyama K, Beckerleg AM, Torchilin VP, Huang L, et al. Activity of amphipathic poly(ethylene glycol) 5000 to prolong the circulation time of liposomes depends on the liposome size and is unfavorable for immunoliposome binding to target. Biochim Biophys Acta. 1991;1062:142–148. doi: 10.1016/0005-2736(91)90385-l. [DOI] [PubMed] [Google Scholar]
- 11.Klibanov AL, Maruyama K, Torchilin VP, Huang L, et al. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 1990;268:235–237. doi: 10.1016/0014-5793(90)81016-h. [DOI] [PubMed] [Google Scholar]
- 12.Allen TM, Hansen C, et al. Pharmacokinetics of stealth versus conventional liposomes: effect of dose. Biochim Biophys Acta. 1991;1068:133–141. doi: 10.1016/0005-2736(91)90201-i. [DOI] [PubMed] [Google Scholar]
- 13.Park JW, Kirpotin DB, Hong K, Shalaby R, Shao Y, Nielsen UB, Marks JD, Papahadjopoulos D, Benz CC, et al. Tumor targeting using anti-her2 immunoliposomes. J Control Release. 2001;74:95–113. doi: 10.1016/s0168-3659(01)00315-7. [DOI] [PubMed] [Google Scholar]
- 14.Legendre JY, Szoka FC, Jr, et al. Delivery of plasmid DNA into mammalian cell lines using pH-sensitive liposomes: comparison with cationic liposomes. Pharm Res. 1992;9:1235–1242. doi: 10.1023/a:1015836829670. [DOI] [PubMed] [Google Scholar]
- 15.Sosunov EA, Anyukhovsky EP, Sosunov AA, Moshnikova A, Wijesinghe D, Engelman DM, Reshetnyak YK, Andreev OA, et al. pHLIP targets ischemic myocardim. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1220038110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daumar P, Wanger-Baumann CA, Pillarsetty N, Fabrizio L, Carlin SD, Andreev OA, Reshetnyak YK, Lewis JS, et al. Efficient (18)F-Labeling of Large 37-Amino-Acid pHLIP Peptide Analogues and Their Biological Evaluation. Bioconjug Chem. 2012;23:1557–1566. doi: 10.1021/bc3000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macholl S, Morrison MS, Iveson P, Arbo BE, Andreev OA, Reshetnyak YK, Engelman DM, Johannesen E, et al. In Vivo pH Imaging with (99m)Tc-pHLIP. Mol Imaging Biol. 2012;14:725–734. doi: 10.1007/s11307-012-0549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vavere AL, Biddlecombe GB, Spees WM, Garbow JR, Wijesinghe D, Andreev OA, Engelman DM, Reshetnyak YK, Lewis JS, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reshetnyak YK, Yao L, Zheng S, Kuznetsov S, Engelman DM, Andreev OA, et al. Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol Imaging Biol. 2011;13:1146–1156. doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreev OA, Dupuy AD, Segala M, Sandugu S, Serra DA, Chichester CO, Engelman DM, Reshetnyak YK, et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci U S A. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reshetnyak YK, Segala M, Andreev OA, Engelman DM, et al. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreev OA, Karabadzhak AG, Weerakkody D, Andreev GO, Engelman DM, Reshetnyak YK, et al. pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path. Proc Natl Acad Sci U S A. 2010;107:4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reshetnyak YK, Andreev OA, Segala M, Markin VS, Engelman DM, et al. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc Natl Acad Sci U S A. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafez IM, Cullis PR, et al. Roles of lipid polymorphism in intracellular delivery. Adv Drug Deliv Rev. 2001;47:139–148. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]
- 25.Arnhold J, Wiegel D, Hussler O, Arnold K, et al. Quenching and dequenching of octadecyl Rhodamine B chloride fluorescence in Ca(2+)-induced fusion of phosphatidylserine vesicles: effects of poly(ethylene glycol) Biochim Biophys Acta. 1994;1191:375–383. doi: 10.1016/0005-2736(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 26.Li SD, Huang L, et al. Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. J Control Release. 2010;145:178–181. doi: 10.1016/j.jconrel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson LV, Walsh ML, Chen LB, et al. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci U S A. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissig V, Boddapati SV, Cheng SM, D’Souza GG, et al. Liposomes and liposome-like vesicles for drug and DNA delivery to mitochondria. J Liposome Res. 2006;16:249–264. doi: 10.1080/08982100600851169. [DOI] [PubMed] [Google Scholar]
- 29.Boddapati SV, Tongcharoensirikul P, Hanson RN, D’Souza GG, Torchilin VP, Weissig V, et al. Mitochondriotropic liposomes. J Liposome Res. 2005;15:49–58. doi: 10.1081/lpr-64958. [DOI] [PubMed] [Google Scholar]
- 30.Biswas S, Dodwadkar NS, Sawant RR, Koshkaryev A, Torchilin VP, et al. Surface modification of liposomes with rhodamine-123-conjugated polymer results in enhanced mitochondrial targeting. J Drug Target. 2011;19:552–561. doi: 10.3109/1061186X.2010.536983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover T, Kester M, et al. Liposomal delivery enhances short-chain ceramide-induced apoptosis of breast cancer cells. J Pharmacol Exp Ther. 2003;307:468–475. doi: 10.1124/jpet.103.054056. [DOI] [PubMed] [Google Scholar]
- 32.Stover TC, Sharma A, Robertson GP, Kester M, et al. Systemic delivery of liposomal short-chain ceramide limits solid tumor growth in murine models of breast adenocarcinoma. Clin Cancer Res. 2005;11:3465–3474. doi: 10.1158/1078-0432.CCR-04-1770. [DOI] [PubMed] [Google Scholar]
- 33.Shabbits JA, Mayer LD, et al. Intracellular delivery of ceramide lipids via liposomes enhances apoptosis in vitro. Biochim Biophys Acta. 2003;1612:98–106. doi: 10.1016/s0005-2736(03)00108-1. [DOI] [PubMed] [Google Scholar]
- 34.Luo M, et al. Influenza virus entry. Adv Exp Med Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skehel JJ, Wiley DC, et al. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 36.Yates SP, Jorgensen R, Andersen GR, Merrill AR, et al. Stealth and mimicry by deadly bacterial toxins. Trends Biochem Sci. 2006;31:123–133. doi: 10.1016/j.tibs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Chen YA, Scheller RH, et al. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 38.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH, et al. Herpes virus fusion and entry: a story with many characters. Viruses. 2012;4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilen CB, Tilton JC, Doms RW, et al. Molecular mechanisms of HIV entry. Adv Exp Med Biol. 2012;726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 40.Mercer J, Helenius A, et al. Gulping rather than sipping: macropinocytosis as a way of virus entry. Curr Opin Microbiol. 2012;15:490–499. doi: 10.1016/j.mib.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Kielian M, Jungerwirth S, et al. Mechanisms of enveloped virus entry into cells. Mol Biol Med. 1990;7:17–31. [PubMed] [Google Scholar]
- 42.Tamm LK, Crane J, Kiessling V, et al. Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr Opin Struct Biol. 2003;13:453–466. doi: 10.1016/s0959-440x(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 43.Poschner BC, Fischer K, Herrmann JR, Hofmann MW, Langosch D, et al. Structural features of fusogenic model transmembrane domains that differentially regulate inner and outer leaflet mixing in membrane fusion. Mol Membr Biol. 2010;27:1–11. doi: 10.3109/09687680903362044. [DOI] [PubMed] [Google Scholar]
- 44.Li F, Pincet F, Perez E, Eng WS, Melia TJ, Rothman JE, Tareste D, et al. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 2007;14:890–896. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]
- 45.Markin VS, Albanesi JP, et al. Membrane fusion: stalk model revisited. Biophys J. 2002;82:693–712. doi: 10.1016/S0006-3495(02)75432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domanska MK, Kiessling V, Stein A, Fasshauer D, Tamm LK, et al. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J Biol Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karabadzhak AG, Weerakkody D, Wijesinghe D, Thakur MS, Engelman DM, Andreev OA, Markin VS, Reshetnyak YK, et al. Modulation of the pHLIP Transmembrane Helix Insertion Pathway. Biophys J. 2012;102:1846–1855. doi: 10.1016/j.bpj.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hannun YA, Luberto C, et al. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000;10:73–80. doi: 10.1016/s0962-8924(99)01694-3. [DOI] [PubMed] [Google Scholar]
- 49.Hannun YA, Obeid LM, et al. Ceramide and the eukaryotic stress response. Biochem Soc Trans. 1997;25:1171–1175. doi: 10.1042/bst0251171. [DOI] [PubMed] [Google Scholar]
- 50.Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM, et al. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci U S A. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreev OA, Engelman DM, Reshetnyak YK, et al. pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol Membr Biol. 2010;27:341–352. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercer J, Helenius A, et al. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 53.Swanson JA, Watts C, et al. Macropinocytosis. Trends Cell Biol. 1995;5:424–428. doi: 10.1016/s0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- 54.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]