Abstract
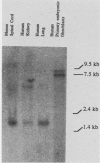
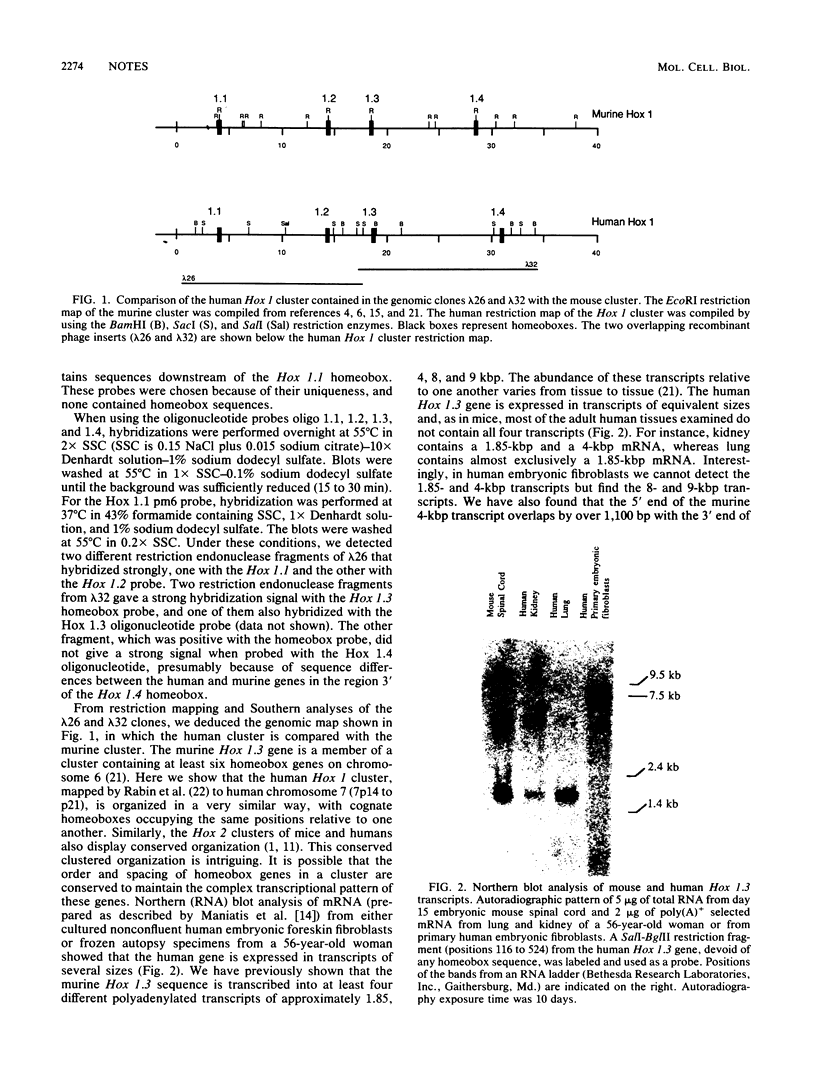
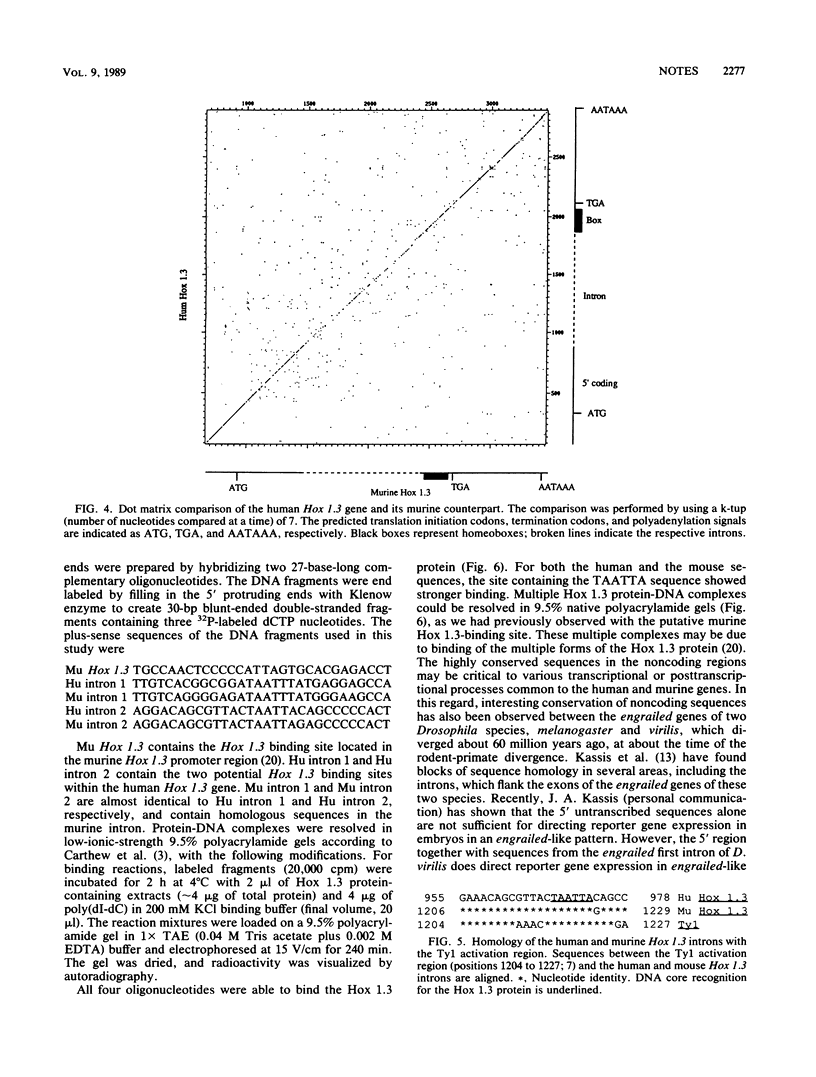
A high degree of conservation exists between the Hox 1.3 homeobox genes of mice and humans. The two genes occupy the same relative positions in their respective Hox 1 gene clusters, they show extensive sequence similarities in their coding and noncoding portions, and both are transcribed into multiple transcripts of similar sizes. The predicted human Hox 1.3 protein differs from its murine counterpart in only 7 of 270 amino acids. The sequence similarity in the 250 base pairs upstream of the initiation codon is 98%, the similarity between the two introns, both 960 base pairs long, is 72%, and the similarity in the 3' noncoding region from termination codon to polyadenylation signal is 90%. Both mouse and human Hox 1.3 introns contain a sequence with homology to a mating-type-controlled cis element of the yeast Ty1 transposon. DNA-binding studies with a recombinant mouse Hox 1.3 protein identified two binding sites in the intron, both of which were within the region of shared homology with this Ty1 cis element.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acampora D., Pannese M., D'Esposito M., Simeone A., Boncinelli E. Human homoeobox-containing genes in development. Hum Reprod. 1987 Jul;2(5):407–414. doi: 10.1093/oxfordjournals.humrep.a136559. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Colberg-Poley A. M., Voss S. D., Chowdhury K., Stewart C. L., Wagner E. F., Gruss P. Clustered homeo boxes are differentially expressed during murine development. Cell. 1985 Nov;43(1):39–45. doi: 10.1016/0092-8674(85)90010-8. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D., Baron A., Mähl P., Galliot B. A new homeo-box is present in overlapping cosmid clones which define the mouse Hox-1 locus. EMBO J. 1986 Aug;5(8):1973–1980. doi: 10.1002/j.1460-2075.1986.tb04452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Company M., Ferchak J. D., Hutchison C. A., 3rd, Yarnell W. S. Activation regions in a yeast transposon have homology to mating type control sequences and to mammalian enhancers. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5423–5427. doi: 10.1073/pnas.82.16.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone M. S., Baron A., Gaunt S. J., Mattei M. G., Duboule D. Hox-5.1 defines a homeobox-containing gene locus on mouse chromosome 2. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4760–4764. doi: 10.1073/pnas.85.13.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibi M., Zink B., Kessel M., Colberg-Poley A. M., Labeit S., Lehrach H., Gruss P. Coding sequence and expression of the homeobox gene Hox 1.3. Development. 1988 Feb;102(2):349–359. doi: 10.1242/dev.102.2.349. [DOI] [PubMed] [Google Scholar]
- Gehring W. J. Homeo boxes in the study of development. Science. 1987 Jun 5;236(4806):1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Hart C. P., Fainsod A., Ruddle F. H. Sequence analysis of the murine Hox-2.2, -2.3, and -2.4 homeo boxes: evolutionary and structural comparisons. Genomics. 1987 Oct;1(2):182–195. doi: 10.1016/0888-7543(87)90011-5. [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Gehring W. J. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987 Sep 11;50(6):963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R. Nomenclature for homoeobox-containing genes. Nature. 1987 Jan 1;325(6099):21–22. doi: 10.1038/325021b0. [DOI] [PubMed] [Google Scholar]
- Mavilio F., Simeone A., Giampaolo A., Faiella A., Zappavigna V., Acampora D., Poiana G., Russo G., Peschle C., Boncinelli E. Differential and stage-related expression in embryonic tissues of a new human homoeobox gene. Nature. 1986 Dec 18;324(6098):664–668. doi: 10.1038/324664a0. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Meijlink F., de Laaf R., Verrijzer P., Destrée O., Kroezen V., Hilkens J., Deschamps J. A mouse homeobox containing gene on chromosome 11: sequence and tissue-specific expression. Nucleic Acids Res. 1987 Sep 11;15(17):6773–6786. doi: 10.1093/nar/15.17.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto C., Smith G. E., Farrell-Towt J., Chizzonite R., Summers M. D., Ju G. Production of human c-myc protein in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1985 Oct;5(10):2860–2865. doi: 10.1128/mcb.5.10.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald W. F., Garbern J., Arnheiter H., Tournier-Lasserve E., Lazzarini R. A. The Hox-1.3 homeo box protein is a sequence-specific DNA-binding phosphoprotein. Genes Dev. 1989 Feb;3(2):158–172. doi: 10.1101/gad.3.2.158. [DOI] [PubMed] [Google Scholar]
- Odenwald W. F., Taylor C. F., Palmer-Hill F. J., Friedrich V., Jr, Tani M., Lazzarini R. A. Expression of a homeo domain protein in noncontact-inhibited cultured cells and postmitotic neurons. Genes Dev. 1987 Jul;1(5):482–496. doi: 10.1101/gad.1.5.482. [DOI] [PubMed] [Google Scholar]
- Rabin M., Ferguson-Smith A., Hart C. P., Ruddle F. H. Cognate homeo-box loci mapped on homologous human and mouse chromosomes. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9104–9108. doi: 10.1073/pnas.83.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Carroll S. B. The segmentation and homeotic gene network in early Drosophila development. Cell. 1987 Dec 4;51(5):689–698. doi: 10.1016/0092-8674(87)90092-4. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A., Mavilio F., Acampora D., Giampaolo A., Faiella A., Zappavigna V., D'Esposito M., Pannese M., Russo G., Boncinelli E. Two human homeobox genes, c1 and c8: structure analysis and expression in embryonic development. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4914–4918. doi: 10.1073/pnas.84.14.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]