Background: Fumarate nitrate reduction (FNR) regulator, the master switch between aerobic and anaerobic respiration, responds in vivo to nitric oxide (NO).
Results: [4Fe-4S] FNR reacts rapidly with eight NO molecules in a complex, multistep reaction.
Conclusion: FNR nitrosylation is remarkably similar to that of WhiB-like proteins.
Significance: A common mechanism of nitrosylation exists for phylogenetically unrelated iron-sulfur regulatory proteins.
Keywords: Biophysics, Gene Regulation, Iron-Sulfur Protein, Nitric Oxide, Nitrosylation, Oxidative Stress, FNR
Abstract
The Fumarate nitrate reduction (FNR) regulator from Escherichia coli controls expression of >300 genes in response to O2 through reaction with its [4Fe-4S] cluster cofactor. FNR is the master switch for the transition between anaerobic and aerobic respiration. In response to physiological concentrations of nitric oxide (NO), FNR also regulates genes, including the nitrate reductase (nar) operon, a major source of endogenous cellular NO, and hmp, which encodes an NO-detoxifying enzyme. Here we show that the [4Fe-4S] cluster of FNR reacts rapidly in a multiphasic reaction with eight NO molecules. Oxidation of cluster sulfide ions (S2−) to sulfane (S0) occurs, some of which remains associated with the protein as Cys persulfide. The nitrosylation products are similar to a pair of dinuclear dinitrosyl iron complexes, [Fe(I)2(NO)4(Cys)2]0, known as Roussin's red ester. A similar reactivity with NO was reported for the Wbl family of [4Fe-4S]-containing proteins found only in actinomycetes, such as Streptomyces and Mycobacteria. These results show that NO reacts via a common mechanism with [4Fe-4S] clusters in phylogenetically unrelated regulatory proteins that, although coordinated by four Cys residues, have different cluster environments. The reactivity of E. coli FNR toward NO, in addition to its sensitivity toward O2, is part of a hierarchal network that monitors, and responds to, NO, both endogenously generated and exogenously derived.
Introduction
Nitric oxide (NO) is a gaseous lipophilic radical molecule that functions in eukaryotes both as a signal molecule (at nanomolar concentrations) and as a cytotoxic agent (at micromolar concentrations) (1). The latter arises from the ability of NO to react readily with a variety of cellular targets leading to thiol S-nitrosation (2), amino acid N-nitrosation (3), and nitrosative DNA damage (4). Iron-sulfur proteins, which are vital for many important cellular functions, can also be highly reactive toward NO (5).
Bacteria that carry out anaerobic respiration with nitrate/nitrite as terminal electron acceptors can produce NO endogenously (6, 7). Furthermore, many Gram-positive bacteria contain NO synthases, and the NO they generate is proposed to play a protective role against antibiotics (8). Pathogenic bacteria will encounter exogenous NO generated by the host as a defense mechanism. To survive and thrive, bacteria, both pathogenic and nonpathogenic, must be able to respond to fluctuations in NO concentrations, spanning a wide range, and to remove it when it reaches cytotoxic levels (9).
Several of the regulatory proteins known to respond to NO contain iron-sulfur clusters that function as the sensory module (10). Until recently, little was known about the reactions of iron-sulfur clusters with NO in regulatory proteins. Wbl family proteins, found exclusively in the actinomycetes, contain a [4Fe-4S] cluster that undergoes a remarkably rapid and complex multistep reaction involving a total of eight NO molecules per cluster (11). The products of the reaction have the stoichiometry of [Fe(I)2(NO)4(SR)2], similar to RRE2 complexes. Similar species have recently been recognized as a major product of cluster nitrosylation in a Rieske-type [2Fe-2S] protein (12). The products of the NO reaction are EPR-silent (most likely with electron spin S = 0), unlike the tetrahedral dinitrosyl iron complexes (DNICs), [Fe(I)(NO)2(SR)2]−, previously identified as products of iron-sulfur cluster nitrosylation, that do give EPR signals (S = ½).
The FNR regulator, which contains a [4Fe-4S] cluster, senses O2 and controls the switch between anaerobic and aerobic respiration in many bacteria (13, 14). FNR has sequence homology with cyclic-AMP receptor protein and, like the cyclic-AMP receptor protein, consists of two distinct domains that provide DNA binding and sensory functions (15). FNR becomes activated under anaerobic conditions by the insertion of an O2-labile [4Fe-4S]2+ cluster, coordinated by four Cys residues (Cys-20, -23, -29, and -122), into the N-terminal sensory domain (14). Cluster incorporation promotes dimerization, enabling the C-terminal DNA binding domain to recognize specific binding sites located in FNR-regulated promoters (16).
Upon exposure to O2, the [4Fe-4S]2+ cluster undergoes conversion to a [2Fe-2S]2+ cluster (17) via a [3Fe-4S]1+ intermediate (18). Cluster conversion causes the FNR dimer to dissociate into a transcriptionally inactive monomeric form (14). In this way, FNR regulates >300 genes, most of which are associated with anaerobic respiration, including nitrate and nitrite reductases, that are a major source of endogenous NO in bacteria (7, 19, 20).
In addition to its response to O2, FNR also responds to NO. Anaerobic exposure of E. coli cells to physiologically relevant concentrations (∼5 μm) of NO led to up-regulation of multiple FNR-repressed genes and down-regulation of FNR-activated genes, suggesting that NO inactivates FNR (20, 21). Among those genes up-regulated by FNR in the presence of NO is hmp, encoding a flavohemoglobin (21) that constitutes a principal NO detoxification pathway in E. coli and a wide range of other bacteria. It also plays an important role in the establishment of infection by pathogens (22). Reaction of [4Fe-4S] FNR with NO in vitro was shown to generate iron-nitrosyl species associated with FNR, which bound DNA with reduced affinity (21).
Here we report detailed kinetic and thermodynamic investigations that reveal a rapid, multistep reaction of NO with the [4Fe-4S] cluster of FNR that, in terms of both rate and mechanism, is remarkably similar to that previously observed for Streptomyces and Mycobacterium Wbl proteins (11). These results suggest a common mechanism for the nitrosylation of the tetra-Cys ligated iron-sulfur clusters of different regulator proteins.
EXPERIMENTAL PROCEDURES
RNA Isolation and RT-PCR
Escherichia coli JRG6348, which lacks a chromosomal copy of the fnr gene and carries a single-copy FNR-dependent lacZ reporter gene, was transformed with pBAD-fnr and cultured at 37 °C under anaerobic conditions in M9 minimal medium supplemented with 5% L-broth, 0.4% glycerol, 20 mm trimethylamine-N-oxide, and 20 mm fumarate (7) until the optical density at 600 nm was 0.175–0.200. For the time-course experiments, anaerobic supplements were then added to the cultures as follows: water; 10 mm sodium nitrite; 10 μm each NOC-5 and NOC-7 (Enzo Life Sciences). In addition, one culture was exposed to air with shaking. Samples after supplementation were taken at t = 0, 5, 10, and 15 min. For dose dependence experiments, anaerobic NOC-5 and NOC-7 mixtures were added to the cultures such that the indicated amounts of NO were released at the 15-min sampling point. All samples were mixed with 0.4 volumes of ice-cold 95% ethanol, 5% phenol, pH 4.5, to rapidly stabilize the mRNA. Total RNA was prepared using the RNeasy RNA purification kit (Qiagen) according to the manufacturer's instructions and quantified on a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Relative lacZ RNA quantities were determined on a Mx3005P Thermocycler using SYBR® Green detection of amplification in a one-step protocol. RNA (100 ng) was converted to cDNA and subsequently PCR-amplified using Brilliant III Ultra-Fast SYBR® Green quantitative RT-PCR master mix (Agilent Technologies) following the manufacturer's instructions (reverse transcription, 50 °C for 10 min; annealing temperature, 60 °C; elongation time, 20 s) using 5 pmol each of primers specific to lacZ (CGTGACGTCTCGTTGCTG, GTACAGCGCGGCTGAAAT) and gyrA (ACCTTGCGAGAGAAATTACACC, AATGACCGACATCGCATAATC). The housekeeping gene gyrA was used to control for differences in starting material, whereas a genomic DNA dilution series was used to correct for any differences in primer amplification efficiencies. All experiments were done in triplicate.
Purification of FNR Proteins
A GST-FNR fusion protein was overproduced in aerobically grown E. coli BL21λDE3 harboring pGS572. Proteins were purified as described previously using assay buffer (25 mm Hepes, 2.5 mm CaCl2, 100 mm NaCl, 100 mm NaNO3, pH 7.5) (23, 24). FNR was cleaved from the fusion protein using thrombin and, where necessary, the [4Fe-4S] cluster was reconstituted, in vitro, as described previously (25). Protein concentrations were determined using the method of Bradford (Bio-Rad) (26), with bovine serum albumin as the standard. The iron and sulfide content of proteins was determined as described previously (11), and the [4Fe-4S]2+ cluster concentration was determined using ϵ406 = 16.22 (±0.14) mm−1 cm−1 (23) (Table 1).
TABLE 1.
Sulfide (S2−) and sulfur (S0) content of [4Fe-4S] FNR pre- and post-NO treatment
S2− measurements were made by the method of Beinert (45).
| Sample | Concentration | Before treatment with NO |
After treatment with NO (∼530 μm)a |
|||||
|---|---|---|---|---|---|---|---|---|
| [4Fe-4S] | S2− | S2−/[4Fe-4S] | S2− | S2−/[4Fe-4S] | S0 | S0/[4Fe-4S] | ||
| μm | μm | μm | μm | μm | ||||
| 1 | 37 | 33 | 134 (± 7) | 4.0 (± 0.2) | 7.6 (± 0.5) | 0.23 (± 0.01) | 76 (± 2) | 2.3 (± 0.1) |
| 2 | 39 | 35 | 139 (± 7) | 4.0 (± 0.2) | 24.5 (± 1.2) | 0.70 (± 0.04) | 50 (± 7) | 1.4 (± 0.2) |
| 3b | 26 | 23 | 92 (± 5) | 4.0 (± 0.2) | 17.6 (± 0.2) | 0.77 (± 0.10) | 43 (± 10) | 1.9 (± 0.4) |
a Samples had a final [NO]:[4Fe-4S] ratio of ∼15.
b Low molecular weight reactants and products were removed via gel filtration (PD10, GE Healthcare).
Spectroscopy
UV-visible absorbance measurements were made with a JASCO V500 spectrometer, and CD spectra were measured with a JASCO J810 spectropolarimeter. X-band EPR spectra were recorded with a Bruker EMX spectrometer equipped with an ESR-900 helium flow cryostat (Oxford Instruments). Spin intensities of paramagnetic samples were estimated by double integration of EPR spectra using 1 mm Cu(II), 10 mm EDTA as the standard.
Rapid Reaction Kinetics
UV-visible stopped-flow experiments were performed with a Pro-Data upgraded Applied Photophysics Bio-Sequential DX.17 MV spectrophotometer, with a 1-cm path length cell. Absorption changes were detected at a single wavelength (360 or 420 nm), as described previously (11). All stopped-flow experiments were carried out in assay buffer using gas tight syringes (Hamilton). Prior to use, the stopped-flow system was flushed with ∼30 ml of anaerobic assay buffer. All solutions used for stopped-flow experiments were stored and manipulated inside an anaerobic cabinet (Belle Technology). Fitting of the overall multiphase kinetic data at 360 and 420 nm (separately and together) was performed using DynaFit (BioKin) (27), which employs numerical integration of simultaneous first order differential equations, and verified by fitting individual phases to single or double exponential functions using Origin (version 8, OriginLab). Observed rate constants (kobs) obtained from the fits were plotted against the corresponding initial concentration of nitric oxide to obtain the apparent second order rate constant.
Analytical Methods
Stock solutions of the NO donor PROLI-NONOate (t½ = 1.5 s; Cayman Chemicals) were prepared in 25 mm NaOH, quantified optically (ϵ252 nm 8400 m−1 cm−1), and calibrated as described previously (21). For kinetic experiments, an aliquot of PROLI-NONOate was combined with assay buffer (25 mm Hepes, 2.5 mm CaCl2, 100 mm NaCl, 100 mm NaNO3, pH 7.5) and allowed to decompose in a gas-tight syringe (Hamilton) to achieve the desired NO concentration before addition to FNR samples. Elemental sulfur (S0) was determined by Sörbo's method, as described previously (11), in which the addition of cyanide ion is used to generate SCN− that yields an absorption band at 460 nm on the addition of Fe3+ ions. For LC-MS analyses, samples were diluted into 10% acetonitrile, 0.1% formic acid at a concentration of 0.5–1 pmol μl−1, and 10 μl were loaded onto a ProSwift RP-1S monolithic column (1.0 × 50 mm) (Dionex, Leeds, UK) on a U3000 HPLC System (Dionex). Proteins were eluted with three gradient steps of acetonitrile (in 0.05% formic acid) from 12 to 35% in 5 min, then from 35 to 60% in 10 min, and from 60 to 70% in 2 min. The eluent was continuously infused into a SYNAPT® G2 High Definition MSTM mass spectrometer (Waters, Manchester, UK) using electrospray ionization (ESI), and full MS spectra were recorded with Masslynx 4.1 (Waters) under standard conditions over the whole time of the LC run. The mass spectrometer was calibrated using sodium iodide. Spectra under the LC protein peak (at half-height) were combined, background-subtracted (5/20%), and smoothed (Savitzky-Golay, 3/2), and a mass range of approximately m/z = 650–1100 was selected for processing with MaxEnt1 in Masslynx 4.1 (Waters) until convergence. Gel filtration was carried out under anaerobic conditions using assay buffer and a Sephacryl S-100HR 16/50 column (GE Healthcare), at a flow rate of 1 ml min−1.
RESULTS
Escherichia coli FNR Responds to Exogenous and Endogenous NO in Vivo
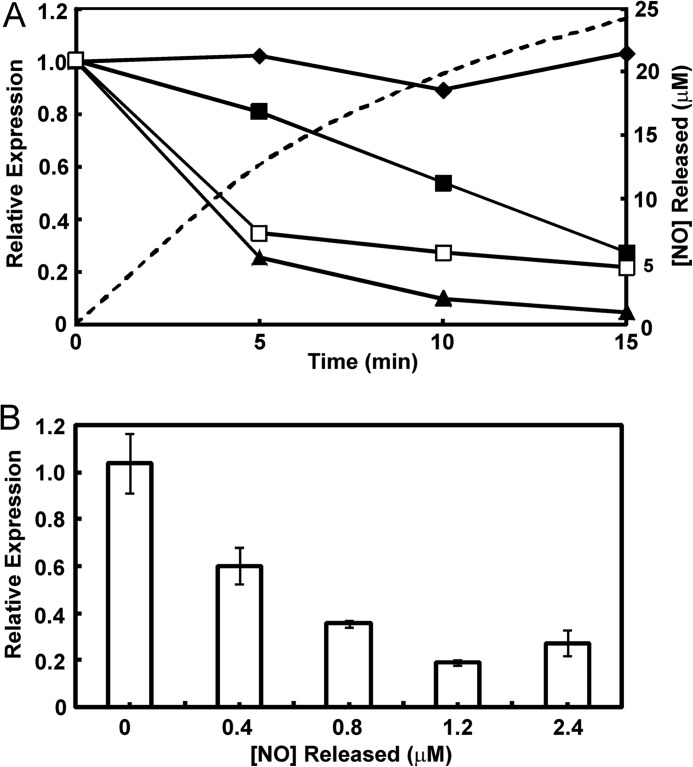
Exposure of E. coli cultures to micromolar concentrations of exogenous NO results in changes in FNR-regulated gene expression and nitrosylation of the FNR iron-sulfur cluster (20, 21, 28). However, it was recently reported that exogenous NO, at a concentration similar to the highest measured in vivo (∼10 μm), did not invoke an FNR-dependent transcriptional response (7). Because the dedicated NO-sensor NsrR failed to respond, it seems that in these experiments, NO did not reach the cytoplasm. Therefore, we compared in vivo transcription from a semisynthetic FNR-dependent reporter gene after exposure to exogenous and endogenous NO with the response to O2. FNR-dependent transcription decreased ∼4-fold after a 15-min exposure to a controlled release of exogenous NO provided by a mixture of NOC-5 and NOC-7 (10 μm each) (Fig. 1A). A more rapid FNR response (∼3-fold decrease after 5 min and ∼5-fold after 15 min) was observed when the cultures were supplemented by nitrite (10 mm; Fig. 1A), a source of endogenous NO (6, 7); anaerobic suspensions of E. coli contained 28 μm NO 15 min after the addition of 12.5 mm nitrite (6). The FNR response to aeration was initially similar to that observed upon the addition of nitrite (∼4-fold after 5 min) but was ultimately more pronounced (∼20-fold after 15 min; Fig. 1A). We conclude that in vivo, FNR responds to exogenous and endogenous NO and that the amplitude of the NO response is ∼4-fold lower than that observed for O2, suggesting that FNR is partially protected by actions initiated by the dedicated NO sensors NorR and NsrR.
FIGURE 1.
In vivo sensitivity of FNR to NO. A, dynamics of FNR-dependent in vivo transcription when E. coli cultures were exposed to NO, nitrite, or O2. Anaerobic cultures of E. coli with a chromosomal FNR-dependent lacZ reporter were exposed to anaerobic water (filled diamonds); a mixture of the NO-releasing compounds (NOC-5 and NOC-7, 10 μm each; release of NO is indicated by the dashed line) as a source of exogenous NO (filled squares); nitrite, 10 mm, as a source of endogenous NO (open squares); or aeration (filled triangles). The cultures were sampled at the indicated times, and the relative transcription of lacZ, which is entirely dependent on FNR in this experiment, was quantified by quantitative RT-PCR. The abundance of the lacZ mRNA at t = 0 min was set at 1.0 and normalized to the housekeeping gyrA mRNA. Data from a typical experiment are shown. B, effects of increasing NO concentrations on FNR-dependent in vivo transcription. The experimental conditions were as described above except that cultures were amended with mixtures of NOC-5 and NOC-7 (0, 0.33, 0.66, 1, and 2 μm of each NOC) for 15 min at which point the cultures were sampled for the lacZ quantitative RT-PCR measurements. The concentration of NO released from the NOC mixtures after 15 min was calculated from the measured half-lives of NOC-5 and NOC-7. Data are mean values (n = 3) with standard deviations indicated.
The dose dependence of the FNR response to NO was determined by measuring the response of the FNR-dependent lacZ reporter after anaerobic cultures were exposed for 15 min to different concentrations of NO, generated by the decay of mixtures of NOC-5 and NOC-7 (Fig. 1B). These experiments showed that FNR responded progressively to increases in NO concentration, with a maximum response (∼5.5-fold decrease in FNR activity) observed in the presence of 1.2 μm NO.
The [4Fe-4S] Cluster of FNR Reacts with Eight NO Molecules
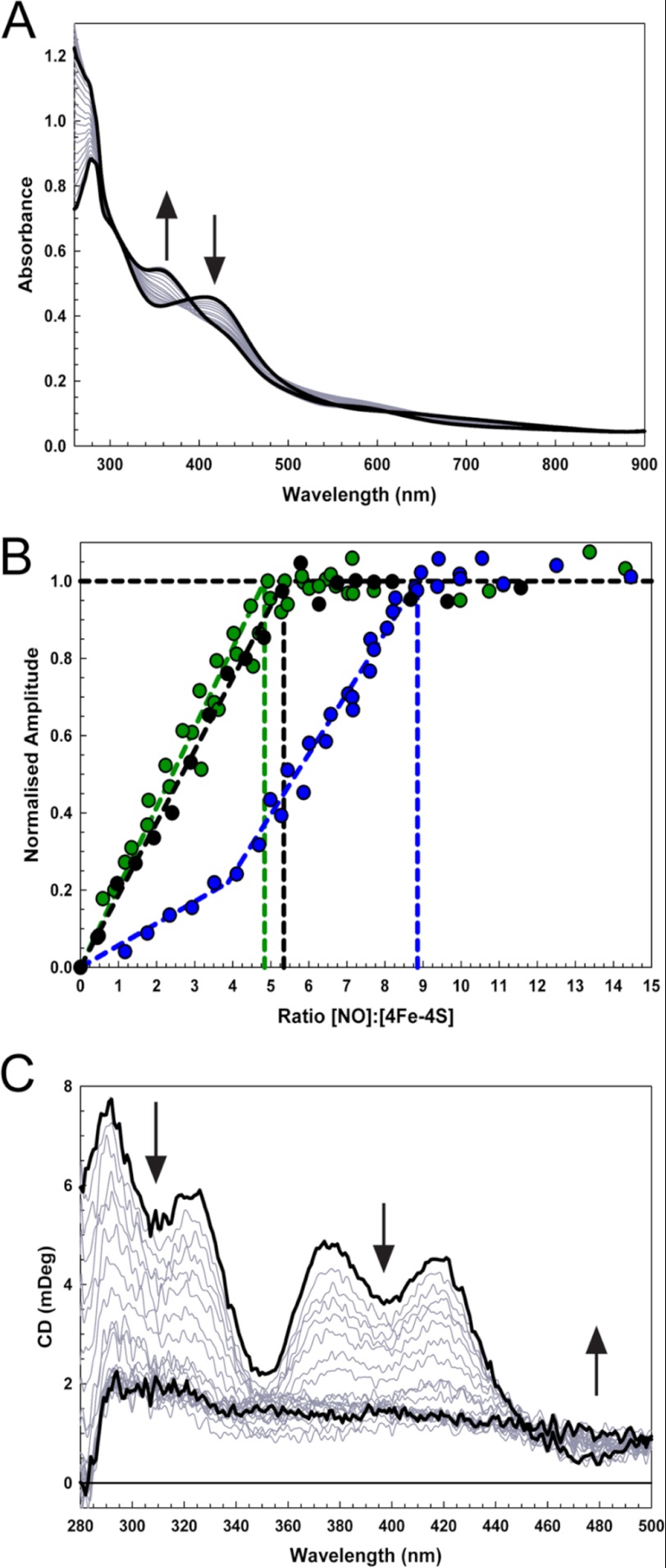
Having established that FNR responds to NO in vivo, the stoichiometry of the reaction of the FNR cluster with NO was investigated by measuring absorbance changes following sequential additions of NO to anaerobic FNR (Fig. 2A). A progressive decrease in A406 nm, an increase in A362 nm, and a lesser increase at 500–700 nm were observed. These changes are consistent with previous observations (21) and indicate the formation of iron-nitrosyl species (11). A plot of ΔA362 nm versus [NO]:[4Fe-4S] (Fig. 2B) revealed that the reaction was complete at a stoichiometry of 8–9 NO molecules per cluster, with a clear inflection point at ∼4 NO. A plot of ΔA406 nm versus [NO]:[4Fe-4S] (Fig. 2B) was quite different, with changes complete at 4–5 NO. The different stoichiometries and lack of isosbestic points reveal a complex reaction. The final UV-visible spectrum (Fig. 2A), with a principal absorption band at 362 nm and a shoulder at ∼430 nm, was consistent with that of an RRE complex. Based on a model RRE extinction coefficient (ϵ362 nm = 8530 m−1 cm−1 (29), the spectrum indicated the presence of two RREs per FNR cluster, consistent with the observed reaction stoichiometry and previous observations with Wbl proteins (11). The reaction was also studied using near UV-visible CD spectroscopy. Signals arising from the [4Fe-4S] cluster decreased almost to zero as the reaction with NO proceeded (Fig. 2C). A plot of intensity at 418 nm against the ratio [NO]:[4Fe-4S] gave a reaction stoichiometry of approximately five NO molecules per [4Fe-4S] cluster, similar to that obtained at ΔA406 nm (Fig. 2B).
FIGURE 2.
Titration of [4Fe-4S] FNR with NO. A, absorbance spectra of [4Fe-4S] FNR (28.2 μm) following sequential additions of NO up to a [NO]/[4Fe-4S] ratio of 12. B, ΔA360 nm (blue circles), ΔA406 nm (green circles), and ΔCD418 nm (black circles) values were normalized and plotted versus the concentration ratio [NO]:[4Fe-4S]. C, CD spectra obtained during a titration equivalent to that in A. Start and end-point spectra are in black. The sample buffer was 25 mm HEPES, 2.5 mm CaCl2, 100 mm NaCl, 100 mm NaNO3, pH 7.5. mDeg, millidegrees.
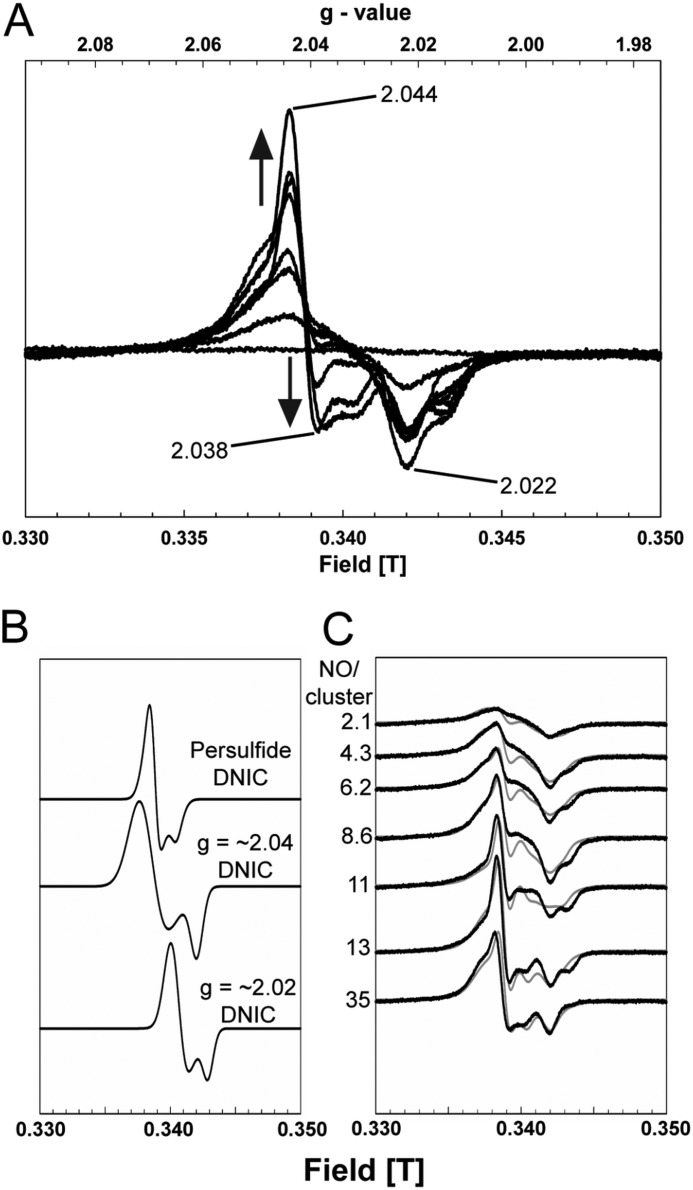
An EPR titration of wild-type [4Fe-4S] FNR with NO was conducted under conditions identical to those of the UV-visible absorption titration above, with spectra recorded at 15 (not shown) and 74 K at increasing levels of NO (Fig. 3A). Complex signals centered on g(∼) 2.03 were observed, similar to those previously reported for nitrosylated FNR in vivo (30). Spectra could be deconvoluted into signals arising from three distinct S = ½ species: two thiol-ligated DNIC species with g⊥ = 2.045, g= = 2.023 and g⊥ = 2.023, g= = 2.016, respectively, and a third, giving rise to the sharp feature observed at g(∼) 2.04 (g⊥ = 2.044, g= = 2.032) (Fig. 3, B and C). This latter species is similar to that previously assigned as an FNR-derived persulfide ligated DNIC (31). Quantification of the final signal envelope, including all of the EPR-detectable species, obtained with excess NO gave a concentration corresponding to ∼16% of the original cluster (or ∼4% of the original iron). We previously reported a signal due to a small but variable amount of a [3Fe-4S]1+ species, in addition to the DNIC signal, upon nitrosylation of wild-type FNR (21); this was not observed in the samples studied here.
FIGURE 3.
EPR analysis of [4Fe-4S] FNR nitrosylation. A, EPR spectra following the addition of NO to [4Fe-4S] WT-FNR (35.8 μm). The [NO]:[4Fe-4S] ratios were 0.0, 2.1, 4.3, 6.2, 8.6, 11.0, 13.0, and 35.0. The signal shape is indicative of the presence of more than one S = ½ species, as observed previously (11). Spectra were recorded at 74 K. Microwave power and frequency were 2 milliwatts and 9.68 GHz, respectively, and field modulation amplitude was 1 millitesla. The sample buffer was 25 mm HEPES, 2.5 mm CaCl2, 100 mm NaCl, 100 mm NaNO3, pH 7.5. B, simulated EPR spectra of persulfide-ligated DNIC (g⊥ = 2.044, g= = 2.032) (31, 46) and two thiol-coordinated DNICs at g(∼) 2.04 (g⊥ = 2.045, g= = 2.023) (29, 44) and g(∼) 2.02 (g⊥ = 2.023, g= = 2.016) (11, 47) that were used to deconvolute the recorded (74 K) EPR spectra in A. C, EPR spectra for nitrosylated FNR (black lines), together with simulations of the experimental data (R2(∼) 0.90) (gray lines) obtained by the combination of varying amounts of the spectra shown in B.
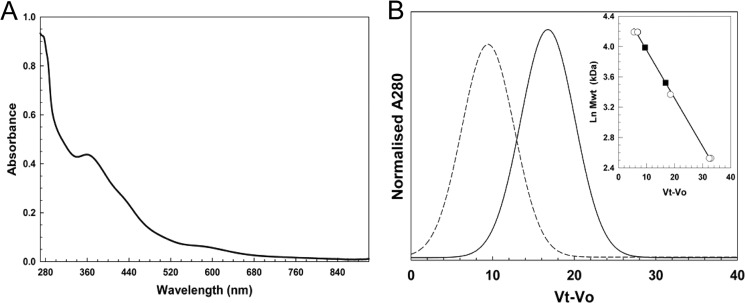
The RRE-like species remained associated with the protein following passage down a gel filtration column (Fig. 4A). Analysis by gel filtration of molecular mass changes upon nitrosylation of [4Fe-4S] FNR revealed a decrease in mass from 54 to 34 kDa (Fig. 4B). Given that wild-type FNR has an actual mass of 28 kDa but has previously been observed by gel filtration to run at ∼30 kDa following reaction with O2 (32), we conclude that nitrosylation, like reaction with O2, results in monomerization. In combination, the spectroscopic and gel filtration data indicate that the addition of NO results in FNR monomers containing two protein-bound RREs, similar to recent observations of the reactivity of NO with Wbl proteins (11).
FIGURE 4.
Properties of nitrosylated [4Fe-4S] FNR probed by gel filtration. A, absorbance spectrum of nitrosylated [4Fe-4S] FNR after rapid gel filtration (PD10, GE Healthcare). The sample (which corresponds to Sample 3 of Table 1) contained ∼2.0 (51 μm) RRE per protein, based on an ϵ362 nm of 8530 m−1 cm−1 (29). B, chromatogram for FNR pre- (dashed line) and post-NO exposure (solid black line). The inset is a standard calibration curve for a Sephacryl S100HR column. Open circles correspond to standard proteins (BSA, carbonic anhydrase, cytochrome c). Black circles and gray triangles correspond to FNR pre-NO (54 kDa) and post-NO (34 kDa), respectively. Ln Mwt, natural log of molecular weight in kDa.
The [4Fe-4S] Cluster of FNR Reacts Rapidly with NO
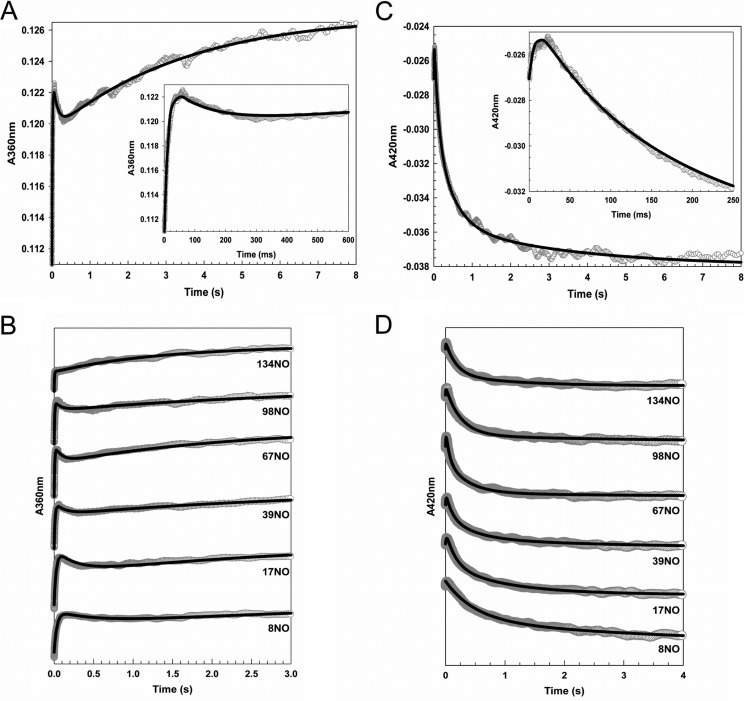
The reaction of [4Fe-4S] FNR with excess NO was followed using stopped flow by monitoring A360 nm and A420 nm, the maxima of the final nitrosylated product and the initial iron-sulfur cluster, respectively (Fig. 5). Three distinct kinetic phases were observed at both wavelengths, indicating a complex multistep process, with a minimum of three steps in the overall reaction, A → B → C → D. The data at the two wavelengths were fitted separately and simultaneously to exponential functions (Fig. 5D), as described previously (11). In both cases, the intermediate phase had quite different kinetic characteristics at the two wavelengths, indicating that they report on different processes in the intermediate part of the reaction. Therefore, the overall reaction was remodeled as a four step reaction, i.e. A → B → C → D → E, where the initial and final steps A → B and D → E, respectively, are detected at both wavelengths, whereas step B → C is detected at 360 nm and step C → D is detected at 420 nm. Data corresponding to step D → E at 420 nm were of very low amplitude as compared with 360 nm, consistent with the thermodynamic titrations that showed relatively little change at 420 nm during the second half of the titration. Overall, these observations are remarkably similar to those made previously with the Wbl proteins Streptomyces coelicolor WhiD and Mycobacterium tuberculosis WhiB1 (11).
FIGURE 5.
Stopped-flow measurements of the reaction of [4Fe-4S] FNR with NO. A–D, ΔA at 360 nm (A and B) and 420 nm (C and D) following the addition of NO to FNR (∼7.6 μm). A and C show data at 360 and 420 nm, respectively, for the addition of ∼33 NO molecules per cluster. B and D show data at 360 and 420 nm, respectively, for a range of other NO additions, as indicated. Insets in A and C show early events in the reaction time course. Fits to each of the observed phases are drawn in black lines.
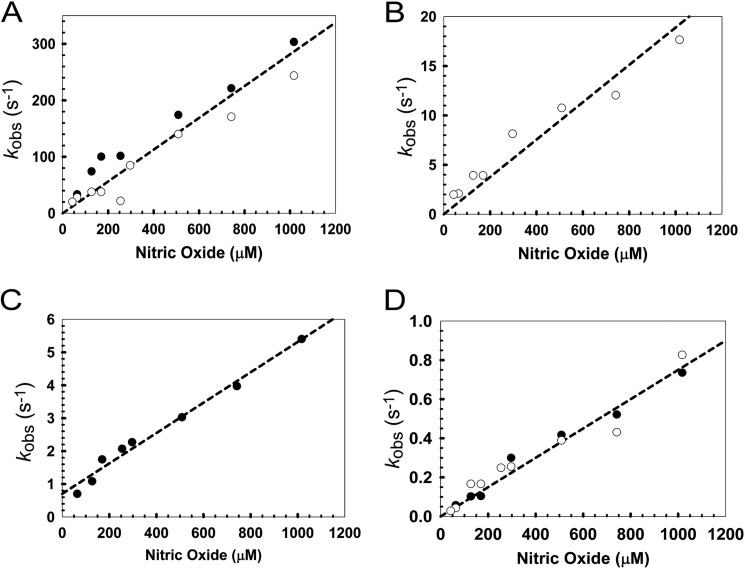
Plots of the observed pseudo-first order rate constants (kobs) against NO concentration for FNR were linear for each step, indicating a first order dependence on NO for each step (Fig. 6, A–D, and Table 2), consistent with the stepwise addition of NO. The rate constant for the slowest step of the NO reaction is at least ∼3 orders of magnitude greater than that for the slowest step with O2, indicating that FNR is actually much more sensitive to NO than it is to O2. The rate constants for each step are of the same order of magnitude as those previously observed for Wbl proteins (11) (Table 2).
FIGURE 6.
Dependence of the rate of each step of nitrosylation on NO. A–D, plots of the observed (pseudo-first order) rate constants (kobs) from fits of the kinetic data at 360 (white circles) and 420 nm (black circles), over a range of NO concentrations. Note that panels A–D correspond to steps 1–4, respectively (see under “Results”). Least squares linear fits are shown giving apparent second order rate constants (Table 2). The buffer was 25 mm HEPES, 2.5 mm CaCl2, 100 mm NaCl, 100 mm NaNO3, pH 7.5.
TABLE 2.
Rate constants for the nitrosylation of [4Fe-4S] FNR with NO in comparison with Wbl proteins
| Phase | Step | Rate constanta |
||
|---|---|---|---|---|
| FNR | WhiD | WhiB1 | ||
| m−1 s−1 | ||||
| 1 | A → B | 2.81 ± 0.10 × 105 | 6.50 × 105 | 4.40 × 105 |
| 2 | B → C | 1.89 ± 0.10 × 104 | 2.13 × 104 | 1.38 × 104 |
| 3 | C → D | 4.61 ± 0. 25 × 103 | 5.36 × 103 | 8.34 × 103 |
| 4 | D → E | 0.75 ± 0.03 × 103 | 1.00 × 103 | 0.90 × 103 |
FNR Cluster Sulfide Is Oxidized to S0 and Retained as Persulfide during Nitrosylation
The addition of NO to a [4Fe-4S]2+ cluster to generate DNIC/RRE species entails reduction of the mixed valence, iron core cluster [2Fe(III)-2Fe(II)]10+ to the [4Fe(I)]4+ state. The six electrons required could be derived from oxidation of cluster sulfide ion, S2−, to sulfane, S0, as shown previously in the case of WhiD (11). After reaction with excess NO, FNR was indeed found to contain ∼0.5 S2− and ∼1.9 S0 per [4Fe-4S] (Table 1). Although only ∼2–3 of the original sulfides could be detected following nitrosylation, the results demonstrate that S0 is generated through oxidation of S2− (11). Importantly, ∼2 S0 per cluster remained associated with the protein following gel filtration, whereas the majority of S2− was lost (Fig. 4A and Table 1).
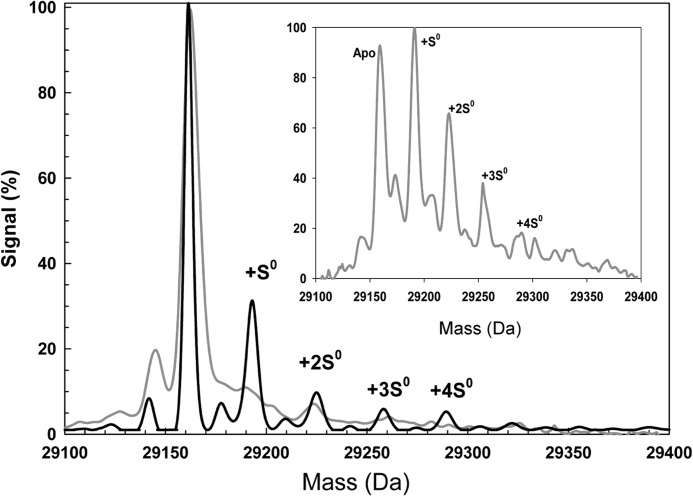
Although ESI-MS of acidified iron-sulfur proteins leads to loss of the cluster, cysteine persulfides/polysulfides remain attached to the protein (33, 34), thus providing a means to detect S0 associated with FNR following nitrosylation. The mass spectrum of [4Fe-4S] FNR (Fig. 7) was dominated by the FNR monomer peak at 29,163 Da (theoretical mass of 29,165 Da). Exposure of [4Fe-4S] FNR to excess NO prior to analysis by MS resulted in significant additional peaks at +32, +64, +96, and +128 Da (Fig. 7). These peaks correspond to apo-FNR containing between 1 and 4 S0 adducts per protein.
FIGURE 7.
Detection of persulfide species of FNR by mass spectrometry. ESI-TOF mass spectra of [4Fe-4S] FNR (891 nm) before (gray line) and after the addition of NO (black line) are shown. The non-NO-treated sample was maintained entirely under anaerobic conditions until dilution in the MS solvent. The peak at 29,163 Da corresponds to the monomer molecular ion peak of FNR, and the peaks at +32, +64, +96, and +128 Da correspond to the addition of one, two, three, and four covalently bound sulfur atoms, respectively, as indicated. The inset is the ESI-TOF mass spectrum of wild-type [4Fe-4S] (34 μm in 25 mm HEPES, 2.5 mm CaCl2, 100 mm NaCl, 100 mm NaNO3, pH 7.5) after exposure to O2 (102 μm) for 60 s at 20 °C prior to dilution in the MS solvent. Peaks at +32, +64, +96, and +128 Da again correspond to the addition of one, two, three, and four covalently bound sulfur atoms.
Recently, it was shown that the reaction of O2 with [4Fe-4S] FNR results in oxidation of cluster sulfide to S0, with FNR containing one or two S0 species per protein as the major species (34). The ESI-TOF MS spectrum of [4Fe-4S] FNR following the addition of oxygen (Fig. 7, inset) shows the same persulfide adducts, although more abundant following reaction with O2 as compared with NO. Although this suggests that more persulfide species are generated during reaction with O2 than with NO, our S0 analytical data are not consistent with this conclusion. This may arise from instability of the persulfide species under the conditions of the MS experiment following nitrosylation (35). In this case, one possibility is that persulfide species may become re-reduced to cysteine plus S2− and iron reoxidized from Fe(I) to Fe(II)/(III) upon disruption of iron-nitrosyl-FNR complex(es) during acidification in preparation for MS. Resonance Raman studies of O2-treated [4Fe-4S] FNR showed that the S0 is carried as persulfide that coordinates the [2Fe-2S] cluster (34). Therefore we propose that the S0 adduct species formed in FNR by the action of NO are also persulfides that ligate the Fe(I) cluster core.
DISCUSSION
The reactivity of O2/reactive oxygen species and NO toward protein-bound iron-sulfur clusters is a major cause of their toxicity, but also underlies the biological mechanisms through which such species are sensed, often as a first step of stress response (10, 13). Understanding the chemistry of the reaction between NO and iron-sulfur clusters is, therefore, an area of growing importance. A recent mechanistic study of the nitrosylation of the [4Fe-4S] clusters of Wbl proteins, some of which have been proposed to function as NO sensors (36, 37), revealed a rapid and complex multistep reaction involving a total of eight NO molecules per cluster (11).
E. coli FNR, which functions as a global O2 sensor that controls the switch between aerobic and anaerobic metabolism, is mechanistically the best characterized of the iron-sulfur cluster-containing regulators (13). Here and elsewhere it has also been shown to regulate gene expression in response to physiological concentrations of NO (20, 21). Previous in vitro studies of this reaction led to the conclusion that ∼3 NO molecules per cluster are involved in the reaction, resulting in the formation of a mixture of DNIC and RRE-like species (21). The data presented here demonstrate that the addition of ∼4–5 NO molecules results in loss of the [4Fe-4S] cluster, but 8 NO molecules are required for complete reaction.
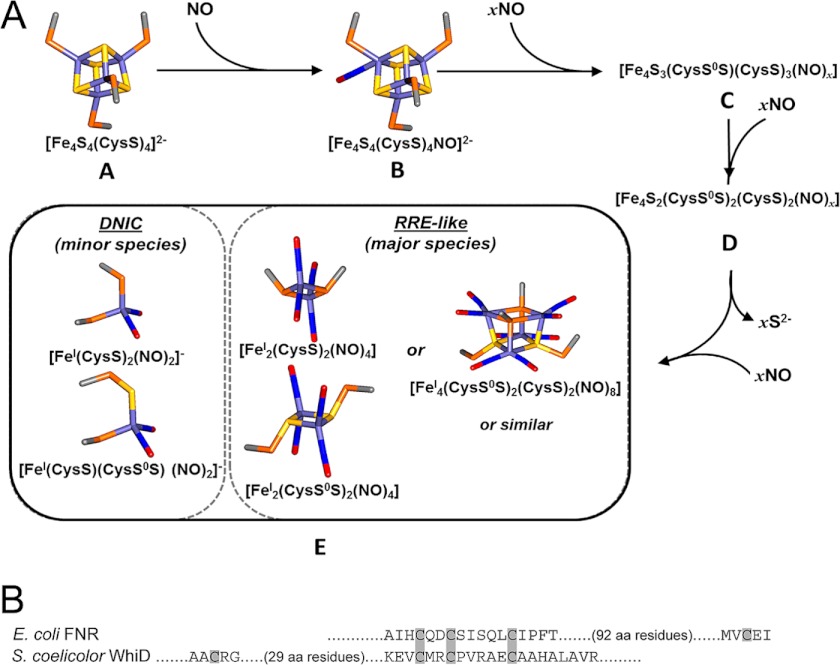
Stopped-flow kinetics showed that nitrosylation of the cluster proceeds via a multiphasic reaction. The first step of the reaction very probably corresponds to the binding of one NO molecule to the FNR [4Fe-4S] cluster (A → B; first order with respect to NO) (Fig. 8A). This most likely enhances accessibility to further NO binding in steps B → C (detected at 360 nm) and C → D (detected at 420 nm), which are both first order in NO, indicating either that a single NO is involved in each or that NO binding occurs independently to different irons of the cluster, giving an overall first order dependence. During the intermediate step C → D, a major decrease in absorbance occurs at 420 nm, suggesting that iron-sulfide and/or cysteine interactions (which give rise to the cluster absorbance) are significantly disrupted and may also be simultaneous with reduction of iron (to the +1 state) and oxidation of S2− (to S0). The optical titration data (absorbance and CD) revealed a stable intermediate at a stoichiometry of approximately four NO molecules per [4Fe-4S] cluster, and EPR showed that this is diamagnetic. Thus, intermediate D may be an EPR-silent tetra-nitrosylated multi-iron species. The last step of the reaction (D → E) resulted in a large increase in absorbance at 360 nm, consistent with significant further nitrosylation, and was again first order in NO, resulting in an EPR-silent protein-bound product composed of four irons and eight NO molecules. This could be a pair of EPR-silent RRE-like species, [FeI2(NO)4(Cys)2], with only minor amounts of EPR-active DNIC, as recently proposed for Wbl proteins (11).
FIGURE 8.
A conserved mechanism for [4Fe-4S] cluster nitrosylation. A, a scheme illustrating proposed steps in the nitrosylation of [4Fe-4S] FNR. The first step of the reaction, A → B, is proposed to correspond to the binding of one NO molecule to the FNR [4Fe-4S] cluster to yield a mono-nitrosylated cluster, which may facilitate further NO binding in steps B → C and C → D. Intermediate D may be the stable, but EPR-silent, tetra-nitrosylated multi-iron species observed during optical titrations (Fig. 2). The last step of the reaction, D → E, results in further nitrosylation of species D to give a product resembling a pair of EPR-silent RRE-like species (∼96% of starting iron) and S = ½ DNIC species (total ∼4%). Models were made using Discovery Studio (Accelrys Software Inc., San Diego, CA) showing iron (pale blue), sulfide (yellow), Cys β-carbon (gray), Cys γ-sulfur (orange), nitrogen (blue), and oxygen (red). B, comparison of amino acid sequence in the cluster-coordinating regions of FNR and WhiD. Note that the spacing between the three coordinating Cys residues is conserved but the identity of the spacing amino acid residues themselves are not. Furthermore, the location of the fourth coordinating Cys residue, although necessarily close to the other three Cys residues in three-dimensional space, is located toward the N terminus in WhiD but is toward the C terminus in FNR.
Nitrosylation of the [4Fe-4S] cluster results in the oxidation of bridging sulfide ions to S0 and most likely addition to cysteine forming a persulfide. The O2 reaction of [4Fe-4S] FNR also leads to sulfide oxidation, resulting in a [2Fe-2S] cluster coordinated by one or two cysteine persulfides (17, 34). It is likely therefore that the iron-nitrosyl species formed upon cluster nitrosylation are coordinated by one or more cysteine persulfides (Fig. 8A). A possible overall reaction is given in Reaction 1. We note that, as described in the case of Wbl proteins (11), the two RRE-like species may dimerize to form a novel cluster, e.g. [FeI4(NO)8(S-Cys)4]0.
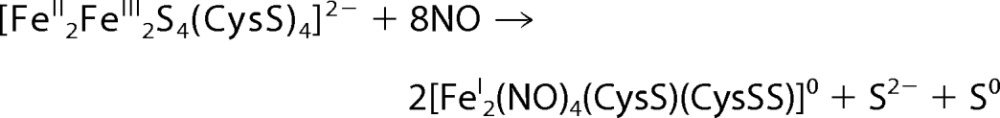 |
Nitrosylation of [4Fe-4S] FNR led to monomerization of the protein, consistent with previous studies that showed a significantly reduced DNA binding affinity following NO reaction (21) and consistent with the in vivo data reported here indicating that NO alleviates FNR-mediated transcriptional repression.
The kinetic and thermodynamic characteristics of FNR nitrosylation are similar to those recently reported by us for the Wbl proteins S. coelicolor WhiD and M. tuberculosis WhiB1 (11). Although both FNR and WhiD/WhiB1 contain [4Fe-4S]2+ clusters ligated by four cysteines (37, 38), they represent phylogenetically unrelated protein families that have different residues surrounding the cluster (Fig. 8B). Clearly, nitrosylation of these Cys-coordinated clusters is not strongly influenced by the protein environment, and we suggest that this pathway is likely to occur widely in proteins during the reaction of [4Fe-4S] clusters with NO.
FNR regulates the aerobic-anaerobic switch via its sensitivity toward O2; what, then, is the biological significance of its reactivity toward NO? Many transcriptional regulators respond to NO in E. coli. Principal among these are NorR, NsrR, and FNR (9, 21, 39). NorR is a non-heme iron-containing regulator that, when bound to NO, activates the transcription of norVW, which encodes the flavorubredoxin NorV and its associated oxidoreductase NorW that, together, detoxify NO to form nitrous oxide (N2O) under anaerobic conditions (40). NsrR regulates at least 60 genes involved in detoxification and/or repair, including nrfA and hmp, for which NO is a substrate. It is suggested that if these dedicated NO detoxification systems are unable to lower the NO concentration sufficiently to counteract the ensuing nitrosative stress, FNR will become nitrosylated (30), leading to lowered expression of the nar, nir, nrf, and nap operons (that require [4Fe-4S] FNR for activation) and derepression of hmp expression. We note that NarG is the most important source of endogenously derived NO during nitrate/nitrite respiration (7, 41–43). Thus, nitrosylation of FNR would serve to protect the cell by down-regulating the principal source of endogenous NO and by up-regulating Hmp, an important NO-detoxifying enzyme. Although Hmp is most active under aerobic conditions, where it catalyzes oxidation of NO to nitrate, under anaerobic conditions, it exhibits some activity, catalyzing reduction of NO to nitrous oxide (N2O) (44), and may have a role in NO detoxification under anaerobiosis (20).
Thus, the nitrosylation of FNR should only occur when the cellular NO detoxification systems are overwhelmed. This implies that FNR functions as a final safeguard against NO damage in a hierarchal network that has evolved to monitor, and respond to, NO both endogenously generated during anaerobic respiration with nitrate or nitrite and exogenously generated by the host immune system.
Acknowledgments
We thank Nick Cull and Dr. Gerhard Saalbach (John Innes Centre, Norwich, Norfolk, United Kingdom) for technical assistance and Drs. Myles Cheesman and Nick Watmough for access to instrumentation.
This work was supported by Grants BB/G018960/1 and BB/J003247/1 from the Biotechnology and Biological Sciences Research Council.
- RRE
- Roussin's Red
- DNIC
- dinitrosyl iron complex
- ESI-MS
- electrospray ionization-mass spectrometry
- FNR
- fumarate nitrate reduction regulator.
REFERENCES
- 1. Bruckdorfer R. (2005) The basics about nitric oxide. Mol. Aspects Med. 26, 3–31 [DOI] [PubMed] [Google Scholar]
- 2. Broillet M. C. (1999) S-Nitrosylation of proteins. Cell. Mol. Life Sci. 55, 1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwon Y. M., Weiss B. (2009) Production of 3-nitrosoindole derivatives by Escherichia coli during anaerobic growth. J. Bacteriol. 191, 5369–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiss B. (2006) Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J. Bacteriol. 188, 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drapier J. C. (1997) Interplay between NO and [Fe-S] clusters: relevance to biological systems. Methods 11, 319–329 [DOI] [PubMed] [Google Scholar]
- 6. Corker H., Poole R. K. (2003) Nitric oxide formation by Escherichia coli: dependence on nitrite reductase, the NO-sensing regulator FNR, and flavohemoglobin Hmp. J. Biol. Chem. 278, 31584–31592 [DOI] [PubMed] [Google Scholar]
- 7. Vine C. E., Purewal S. K., Cole J. A. (2011) NsrR-dependent method for detecting nitric oxide accumulation in the Escherichia coli cytoplasm and enzymes involved in NO production. FEMS Microbiol. Lett. 325, 108–114 [DOI] [PubMed] [Google Scholar]
- 8. Gusarov I., Shatalin K., Starodubtseva M., Nudler E. (2009) Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325, 1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spiro S. (2007) Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31, 193–211 [DOI] [PubMed] [Google Scholar]
- 10. Crack J. C., Green J., Hutchings M. I., Thomson A. J., Le Brun N. E. (2012) Bacterial iron-sulfur regulatory proteins as biological sensor-switches. Antioxid. Redox Signal. 17, 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crack J. C., Smith L. J., Stapleton M. R., Peck J., Watmough N. J., Buttner M. J., Buxton R. S., Green J., Oganesyan V. S., Thomson A. J., Le Brun N. E. (2011) Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 133, 1112–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tinberg C. E., Tonzetich Z. J., Wang H., Do L. H., Yoda Y., Cramer S. P., Lippard S. J. (2010) Characterization of iron dinitrosyl species formed in the reaction of nitric oxide with a biological Rieske center. J. Am. Chem. Soc. 132, 18168–18176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crack J. C., Green J., Thomson A. J., Le Brun N. E. (2012) Iron-sulfur cluster sensor-regulators. Curr. Opin. Chem. Biol. 16, 35–44 [DOI] [PubMed] [Google Scholar]
- 14. Kiley P. J., Beinert H. (1998) Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352 [DOI] [PubMed] [Google Scholar]
- 15. Green J., Scott C., Guest J. R. (2001) Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44, 1–34 [DOI] [PubMed] [Google Scholar]
- 16. Lazazzera B. A., Bates D. M., Kiley P. J. (1993) The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7, 1993–2005 [DOI] [PubMed] [Google Scholar]
- 17. Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P. J. (1997) Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. U.S.A. 94, 6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crack J. C., Green J., Cheesman M. R., Le Brun N. E., Thomson A. J. (2007) Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. U.S.A. 104, 2092–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Constantinidou C., Hobman J. L., Griffiths L., Patel M. D., Penn C. W., Cole J. A., Overton T. W. (2006) A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281, 4802–4815 [DOI] [PubMed] [Google Scholar]
- 20. Pullan S. T., Gidley M. D., Jones R. A., Barrett J., Stevanin T. M., Read R. C., Green J., Poole R. K. (2007) Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: Unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 189, 1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cruz-Ramos H., Crack J., Wu G., Hughes M. N., Scott C., Thomson A. J., Green J., Poole R. K. (2002) NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21, 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stevanin T. M., Read R. C., Poole R. K. (2007) The hmp gene encoding the NO-inducible flavohaemoglobin in Escherichia coli confers a protective advantage in resisting killing within macrophages, but not in vitro: links with swarming motility. Gene 398, 62–68 [DOI] [PubMed] [Google Scholar]
- 23. Crack J. C., Gaskell A. A., Green J., Cheesman M. R., Le Brun N. E., Thomson A. J. (2008) Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J. Am. Chem. Soc. 130, 1749–1758 [DOI] [PubMed] [Google Scholar]
- 24. Bates D. M., Popescu C. V., Khoroshilova N., Vogt K., Beinert H., Münck E., Kiley P. J. (2000) Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275, 6234–6240 [DOI] [PubMed] [Google Scholar]
- 25. Crack J. C., Le Brun N. E., Thomson A. J., Green J., Jervis A. J. (2008) Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli. Methods Enzymol. 437, 191–209 [DOI] [PubMed] [Google Scholar]
- 26. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 28. Vasilieva S. V., Strel'tsova D. A., Moshkovskaya E. Y., Vanin A. F., Mikoyan V. D., Sanina N. A., Aldoshin S. M. (2010) Reversible NO-catalyzed destruction of the Fe-S cluster of the FNR[4Fe-4S]2+ transcription factor: a way to regulate the aidB gene activity in Escherichia coli cells cultured under anaerobic conditions. Dokl. Biochem. Biophys. 435, 283–286 [DOI] [PubMed] [Google Scholar]
- 29. Costanzo S., Menage S., Purrello R., Bonomo R. P., Fontecave M. (2001) Re-examination of the formation of dinitrosyl-iron complexes during reaction of S-nitrosothiols with Fe(II). Inorg. Chim. Acta 318, 1–7 [Google Scholar]
- 30. Vasilieva S. V., Streltsova D. A., Vlaskina A. V., Mikoian V. D., Vanin A. F. (2012) Sources of divalent sulfur allow recovery of the Fnr [4Fe-4S]2+ center in Escherichia coli incubated with nitric oxide donors. Biophysics 57, 166–169 [Google Scholar]
- 31. Frolov E. N., Vanin A. F. (1973) [New type of paramagnetic nitrosyl complexes of non-heme iron]. Biofizika 18, 605–610 [PubMed] [Google Scholar]
- 32. Lazazzera B. A., Beinert H., Khoroshilova N., Kennedy M. C., Kiley P. J. (1996) DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271, 2762–2768 [DOI] [PubMed] [Google Scholar]
- 33. Smith A. D., Agar J. N., Johnson K. A., Frazzon J., Amster I. J., Dean D. R., Johnson M. K. (2001) Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123, 11103–11104 [DOI] [PubMed] [Google Scholar]
- 34. Zhang B., Crack J. C., Subramanian S., Green J., Thomson A. J., Le Brun N. E., Johnson M. K. (2012) Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 109, 15734–15739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zal F., Leize E., Lallier F. H., Toulmond A., Van Dorsselaer A., Childress J. J. (1998) S-Sulfohemoglobin and disulfide exchange: the mechanisms of sulfide binding by Riftia pachyptila hemoglobins. Proc. Natl. Acad. Sci. U.S.A. 95, 8997–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh A., Guidry L., Narasimhulu K. V., Mai D., Trombley J., Redding K. E., Giles G. I., Lancaster J. R., Jr., Steyn A. J. C. (2007) Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U.S.A. 104, 11562–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith L. J., Stapleton M. R., Fullstone G. J. M., Crack J. C., Thomson A. J., Le Brun N. E., Hunt D. M., Harvey E., Adinolfi S., Buxton R. S., Green J. (2010) Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 432, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crack J. C., den Hengst C. D., Jakimowicz P., Subramanian S., Johnson M. K., Buttner M. J., Thomson A. J., Le Brun N. E. (2009) Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry 48, 12252–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tucker N. P., Le Brun N. E., Dixon R., Hutchings M. I. (2010) There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 18, 149–156 [DOI] [PubMed] [Google Scholar]
- 40. Gomes C. M., Giuffrè A., Forte E., Vicente J. B., Saraiva L. M., Brunori M., Teixeira M. (2002) A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J. Biol. Chem. 277, 25273–25276 [DOI] [PubMed] [Google Scholar]
- 41. Rowley G., Hensen D., Felgate H., Arkenberg A., Appia-Ayme C., Prior K., Harrington C., Field S. J., Butt J. N., Baggs E., Richardson D. J. (2012) Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. Biochem. J. 441, 755–762 [DOI] [PubMed] [Google Scholar]
- 42. Ralt D., Wishnok J. S., Fitts R., Tannenbaum S. R. (1988) Bacterial catalysis of nitrosation: involvement of the nar operon of Escherichia coli. J. Bacteriol. 170, 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Metheringham R., Cole J. A. (1997) A reassessment of the genetic determinants, the effect of growth conditions, and the availability of an electron donor on the nitrosating activity of Escherichia coli K12. Microbiology 143, 2647–2656 [DOI] [PubMed] [Google Scholar]
- 44. Kim S. O., Orii Y., Lloyd D., Hughes M. N., Poole R. K. (1999) Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445, 389–394 [DOI] [PubMed] [Google Scholar]
- 45. Beinert H. (1983) Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 46. Bryar T. R., Eaton D. R. (1992) Electronic configuration and structure of paramagnetic iron dinitrosyl complexes. Can. J. Chem. 70, 1917–1926 [Google Scholar]
- 47. Pieper G. M., Halligan N. L., Hilton G., Konorev E. A., Felix C. C., Roza A. M., Adams M. B., Griffith O. W. (2003) Non-heme iron protein: a potential target of nitric oxide in acute cardiac allograft rejection. Proc. Natl. Acad. Sci. U.S.A. 100, 3125–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]