Abstract
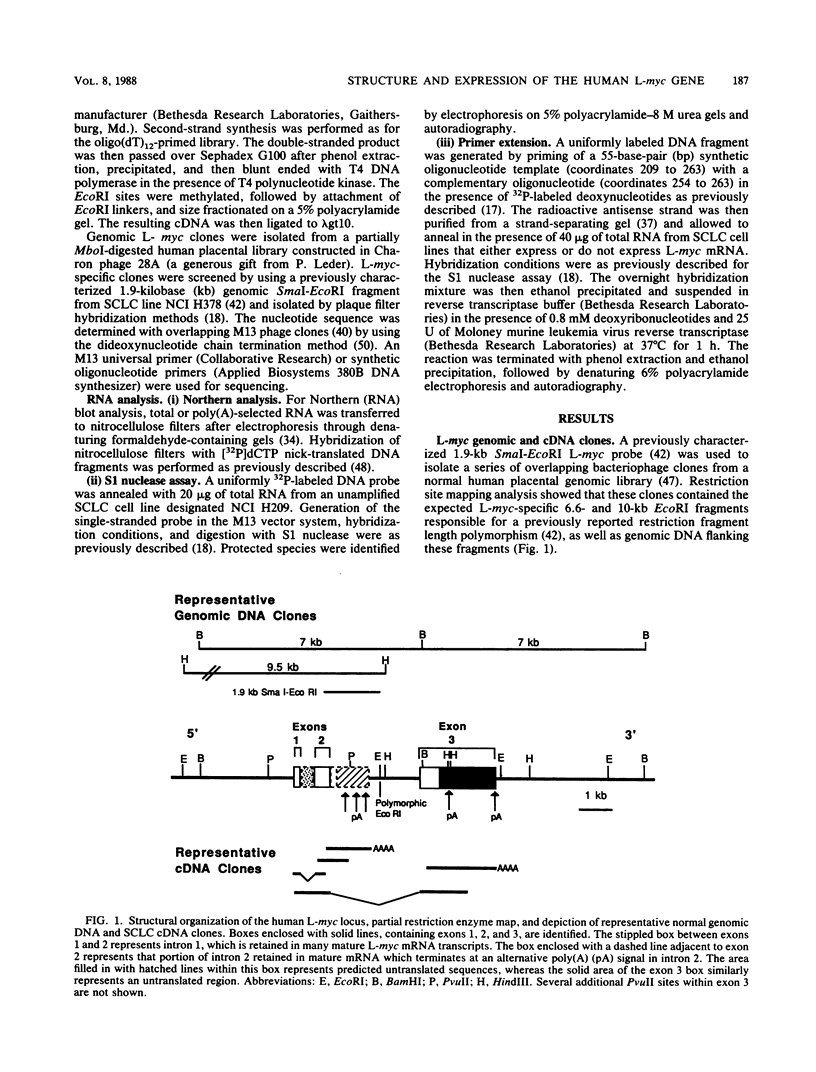
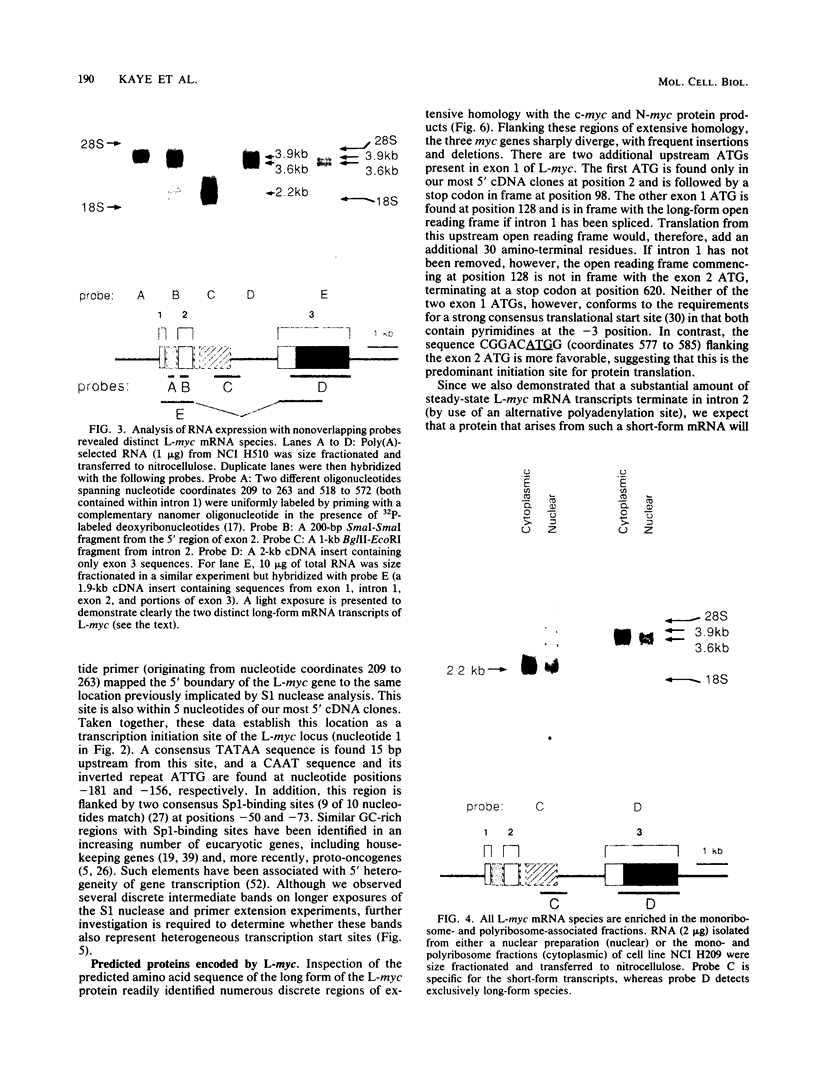
We analyzed in detail the structure of the L-myc gene isolated from human placental DNA and characterized its expression in several small-cell lung cancer cell lines. The gene is composed of three exons and two introns spanning 6.6 kilobases in human DNA. Several distinct mRNA species are produced in all small-cell lung cancer cell lines that express L-myc. These transcripts are generated from a single gene by alternative splicing of introns 1 and 2 and by use of alternative polyadenylation signals. In some mRNAs there is a long open reading frame with a predicted translated protein of 364 residues. Amino acid sequence comparison with c-myc and N-myc demonstrated multiple discrete regions with extensive homology. In contrast, other mRNA transcripts, generated by alternative processing, could encode a truncated protein with a novel carboxy-terminal end.
Full text
PDF


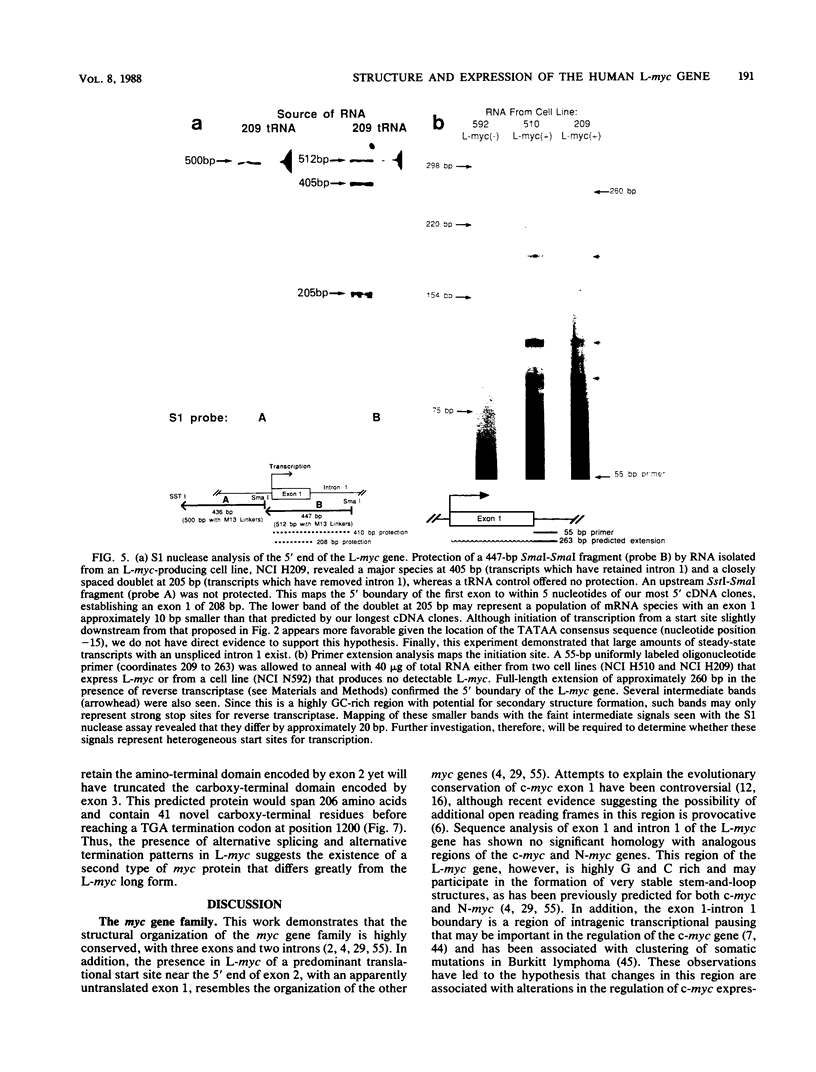
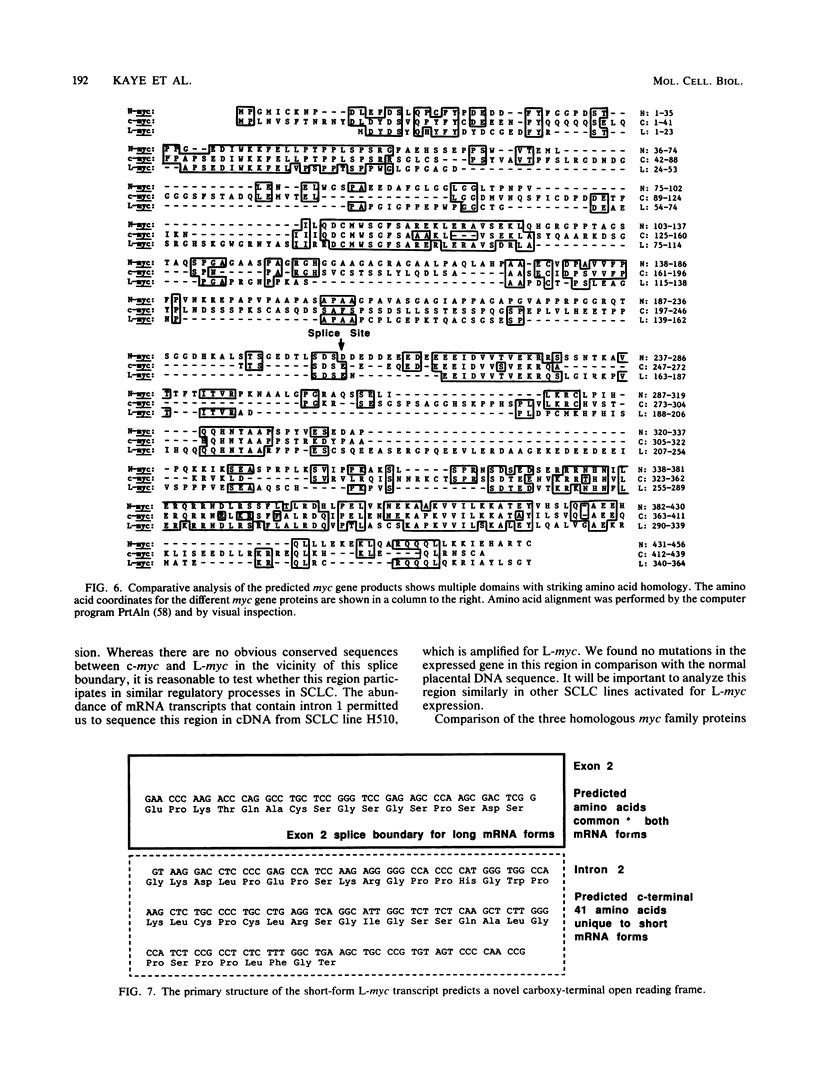



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Alt F. W., DePinho R., Zimmerman K., Legouy E., Hatton K., Ferrier P., Tesfaye A., Yancopoulos G., Nisen P. The human myc gene family. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 2):931–941. doi: 10.1101/sqb.1986.051.01.106. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bender T. P., Kuehl W. M. Murine myb protooncogene mRNA: cDNA sequence and evidence for 5' heterogeneity. Proc Natl Acad Sci U S A. 1986 May;83(10):3204–3208. doi: 10.1073/pnas.83.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986 Oct;6(10):3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Butnick N. Z., Miyamoto C., Chizzonite R., Cullen B. R., Ju G., Skalka A. M. Regulation of the human c-myc gene: 5' noncoding sequences do not affect translation. Mol Cell Biol. 1985 Nov;5(11):3009–3016. doi: 10.1128/mcb.5.11.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney D. N., Gazdar A. F., Bepler G., Guccion J. G., Marangos P. J., Moody T. W., Zweig M. H., Minna J. D. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985 Jun;45(6):2913–2923. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Martinotti S., Gallo R. C., Erikson J., Croce C. M. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983 Feb 25;219(4587):963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- Darveau A., Pelletier J., Sonenberg N. Differential efficiencies of in vitro translation of mouse c-myc transcripts differing in the 5' untranslated region. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2315–2319. doi: 10.1073/pnas.82.8.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Sazer S., Tjian R., Schimke R. T. Transcription factor Sp1 recognizes a DNA sequence in the mouse dihydrofolate reductase promoter. Nature. 1986 Jan 16;319(6050):246–248. doi: 10.1038/319246a0. [DOI] [PubMed] [Google Scholar]
- Gazdar A. F., Carney D. N., Nau M. M., Minna J. D. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res. 1985 Jun;45(6):2924–2930. [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Abrams H. D., Rohrschneider L. R., Eisenman R. N. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983 Oct;34(3):789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- Hann S. R., Eisenman R. N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984 Nov;4(11):2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney M. L., Pierce J., Parsons J. T. Site-directed mutagenesis of the gag-myc gene of avian myelocytomatosis virus 29: biological activity and intracellular localization of structurally altered proteins. J Virol. 1986 Oct;60(1):167–176. doi: 10.1128/jvi.60.1.167-176.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. T., Grollman A. P. Effects of aurintricarboxylic acid on ribosomes and the biosynthesis of globin in rabbit reticulocytes. Mol Pharmacol. 1972 Mar;8(2):111–127. [PubMed] [Google Scholar]
- Ishii S., Kadonaga J. T., Tjian R., Brady J. N., Merlino G. T., Pastan I. Binding of the Sp1 transcription factor by the human Harvey ras1 proto-oncogene promoter. Science. 1986 Jun 13;232(4756):1410–1413. doi: 10.1126/science.3012774. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl N. E., Legouy E., DePinho R. A., Nisen P. D., Smith R. K., Gee C. E., Alt F. W. Human N-myc is closely related in organization and nucleotide sequence to c-myc. Nature. 1986 Jan 2;319(6048):73–77. doi: 10.1038/319073a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H., Parada L. F., Weinberg R. A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983 Aug 18;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Little C. D., Nau M. M., Carney D. N., Gazdar A. F., Minna J. D. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983 Nov 10;306(5939):194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- Littman D. R., Thomas Y., Maddon P. J., Chess L., Axel R. The isolation and sequence of the gene encoding T8: a molecule defining functional classes of T lymphocytes. Cell. 1985 Feb;40(2):237–246. doi: 10.1016/0092-8674(85)90138-2. [DOI] [PubMed] [Google Scholar]
- Martinez R., Mathey-Prevot B., Bernards A., Baltimore D. Neuronal pp60c-src contains a six-amino acid insertion relative to its non-neuronal counterpart. Science. 1987 Jul 24;237(4813):411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- McGrogan M., Simonsen C. C., Smouse D. T., Farnham P. J., Schimke R. T. Heterogeneity at the 5' termini of mouse dihydrofolate reductase mRNAs. Evidence for multiple promoter regions. J Biol Chem. 1985 Feb 25;260(4):2307–2314. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau M. M., Brooks B. J., Battey J., Sausville E., Gazdar A. F., Kirsch I. R., McBride O. W., Bertness V., Hollis G. F., Minna J. D. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985 Nov 7;318(6041):69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- Nau M. M., Brooks B. J., Jr, Carney D. N., Gazdar A. F., Battey J. F., Sausville E. A., Minna J. D. Human small-cell lung cancers show amplification and expression of the N-myc gene. Proc Natl Acad Sci U S A. 1986 Feb;83(4):1092–1096. doi: 10.1073/pnas.83.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci P. G., Knowles D. M., 2nd, Magrath I., Dalla-Favera R. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt lymphoma. Proc Natl Acad Sci U S A. 1986 May;83(9):2984–2988. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G., Stanton L., Schwab M., Bishop J. M. Human proto-oncogene N-myc encodes nuclear proteins that bind DNA. Mol Cell Biol. 1986 Dec;6(12):4450–4457. doi: 10.1128/mcb.6.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch J. V., Siebenlist U., Korsmeyer S., Waldmann T., Leder P. Structure of the human immunoglobulin mu locus: characterization of embryonic and rearranged J and D genes. Cell. 1981 Dec;27(3 Pt 2):583–591. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sausville E. A., Lebacq-Verheyden A. M., Spindel E. R., Cuttitta F., Gazdar A. F., Battey J. F. Expression of the gastrin-releasing peptide gene in human small cell lung cancer. Evidence for alternative processing resulting in three distinct mRNAs. J Biol Chem. 1986 Feb 15;261(5):2451–2457. [PubMed] [Google Scholar]
- Sazer S., Schimke R. T. A re-examination of the 5' termini of mouse dihydrofolate reductase RNA. J Biol Chem. 1986 Apr 5;261(10):4685–4690. [PubMed] [Google Scholar]
- Schwab M., Ellison J., Busch M., Rosenau W., Varmus H. E., Bishop J. M. Enhanced expression of the human gene N-myc consequent to amplification of DNA may contribute to malignant progression of neuroblastoma. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4940–4944. doi: 10.1073/pnas.81.15.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Schwab M., Bishop J. M. Nucleotide sequence of the human N-myc gene. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1772–1776. doi: 10.1073/pnas.83.6.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. A., Pattengale P. K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984 Oct;38(3):627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Stone J., de Lange T., Ramsay G., Jakobovits E., Bishop J. M., Varmus H., Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987 May;7(5):1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D., Nisen P. D., Tesfaye A., Kohl N. E., Goldfarb M. P., Alt F. W. N-myc can cooperate with ras to transform normal cells in culture. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5455–5459. doi: 10.1073/pnas.82.16.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]