Abstract
Recurrent mutations affecting the histone H3.3 residues Lys27 or indirectly Lys36 are frequent drivers of pediatric high-grade gliomas (over 30 % of HGGs). To identify additional driver mutations in HGGs, we investigated a cohort of 60 pediatric HGGs using whole-exome sequencing (WES) and compared them to 543 exomes from non-cancer control samples. We identified mutations in SETD2, a H3K36 trimethyltransferase, in 15 % of pediatric HGGs, a result that was genome-wide significant (FDR = 0.029). Most SETD2 alterations were truncating mutations. Sequencing the gene in this cohort and another validation cohort (123 gliomas from all ages and grades) showed SETD2 mutations to be specific to high-grade tumors affecting 15 % of pediatric HGGs (11/73) and 8 % of adult HGGs (5/65) while no SETD2 mutations were identified in low-grade diffuse gliomas (0/45). Furthermore, SETD2 mutations were mutually exclusive with H3F3A mutations in HGGs (P = 0.0492) while they partly overlapped with IDH1 mutations (4/14), and SETD2-mutant tumors were found exclusively in the cerebral hemispheres (P = 0.0055). SETD2 is the only H3K36 trimethyltransferase in humans, and SETD2-mutant tumors showed a substantial decrease in H3K36me3 levels (P < 0.001), indicating that the mutations are loss-of-function. These data suggest that loss-of-function SETD2 mutations occur in older children and young adults and are specific to HGG of the cerebral cortex, similar to the H3.3 G34R/V and IDH mutations. Taken together, our results suggest that mutations disrupting the histone code at H3K36, including H3.3 G34R/V, IDH1 and/or SETD2 mutations, are central to the genesis of hemispheric HGGs in older children and young adults.
Electronic supplementary material
The online version of this article (doi:10.1007/s00401-013-1095-8) contains supplementary material, which is available to authorized users.
Keywords: High-grade glioma, H3K36 methylation, SETD2, Epigenetic, Pediatric, Young adult
Introduction
Malignant primary brain and central nervous system (CNS) tumors occur at an age-adjusted incidence rate of 7.3 out of 100,000 people across all ages and are the leading cause of cancer-related death in children [9]. High-grade gliomas [HGG; grade III and grade IV astrocytomas/glioblastoma (GBM)] are highly aggressive and deadly brain tumors [9, 32] and are more commonly diagnosed in adults. GBM remains essentially incurable despite decades of concerted therapeutic efforts [5]. One impediment to treatment is that GBM is diagnosed as a single pathological entity, which cannot discriminate potential genetic drivers and molecular subtypes. This impacts the design and outcome of clinical trials and likely contributes to the apparent inherent resistance of GBM to adjuvant therapies. Because of the similar histology, current treatments for GBM in children are driven by adult studies and show, as in adults, little therapeutic success.
We and others recently identified two recurrent mutations in H3F3A, which encodes the replication-independent histone 3 variant H3.3, in over 30 % of pediatric and young adult GBM [32, 42]. The mutations, K27M and G34R/G34V, occur at positions in the histone tail that are critical for post-translational modifications involved in the histone code, which determines chromatin structure and gene expression. H3.3 K27M mutations were also identified in over 70 % of pediatric diffuse intrinsic pontine glioma (DIPG), a fatal HGG of the brainstem [18, 42] as well as K27M mutations in the canonical H3.1 in 18 % of samples [42]. H3.3 mutations significantly overlapped with mutations in TP53 and in ATRX (α-thalassemia/mental-retardation syndrome-X-linked) [13, 40] and less frequently with the ATRX hetero-dimer DAXX, which encode subunits of a chromatin remodeling complex required for H3.3 incorporation at pericentric heterochromatin and telomeres [8, 13]. H3.3 mutations represent the pediatric counterpart of the recurrent hotspot mutations in isocitrate dehydrogenase 1 or 2 (IDH1/2) [27, 44]. IDH1 R132 mutations are gain-of-function, causing the enzyme to produce 2-hydroxyglutarate (2-HG) [7, 27] and IDH1-mutant tumors display distinct DNA methylation profiles with global hypermethylation, termed a glioma-CpG island methylator phenotype (G-CIMP) [26, 35, 38]. Interestingly, similar to pediatric GBM, IDH1 mutations were shown to occur in association with TP53 [3, 27] and ATRX mutations in adult diffuse astrocytic tumors [14, 16, 25], illustrating an important constellation of mutations in the development of pediatric and secondary GBM. In the present study, we sought to identify drivers of HGG in pediatric samples that did not carry H3.3 or IDH mutations. We investigated a cohort of 60 pediatric HGGs utilizing statistical analysis of whole-exome sequencing (WES) on a genome-wide ranking scale and validated results in an independent validation cohort of 123 gliomas from all ages and grades. Herein, we present data showing the importance and functional impact of mutations in the H3K36 trimethyltransferase SETD2 in HGGs of the cerebral hemispheres.
Materials and methods
Sample characteristics and pathological review
Samples were obtained with informed consent following approval of the Institutional Review Board of individual hospitals. Samples were reviewed by senior neuropathologists (S.A., A.K.) according to WHO guidelines. Fifty-one pediatric grade IV astrocytomas (glioblastoma, GBM) patients and nine pediatric grade III astrocytomas from patients aged 1–20 years were analyzed by whole-exome sequencing (44 previously published in [32]). An additional 123 adult and pediatric gliomas of diverse histology and grade were also included for targeted sequencing of SETD2, IDH1 and H3F3A. Available clinical and relevant mutational characteristics are detailed in Table S1. Tissues were obtained from the London/Ontario Tumor Bank, the Pediatric Cooperative Health Tissue Network, the Children’s Oncology Group, The Montreal Children’s Hospital and from collaborators in Poland, Hungary and Germany.
DNA extraction
Genomic DNA was extracted from frozen tumor tissue utilizing the Qiagen DNeasy Blood and Tissue kit according to instructions from the manufacturer (Qiagen).
Alignment and variant calling for whole-exome sequencing
Standard instructions from the manufacturer were used for target capture with the Illumina TruSeq exome enrichment kit and 100 bp paired-end sequencing reads on the Illumina HiSeq platform with bioinformatic processing and variant annotation as previously described [32]. For the selected genes of interest shown in Table S1, variants in these genes that were private to tumor samples are shown, i.e. those variants not seen within the 1000 genomes (http://www.1000genomes.org/) or NHLBI exome (http://evs.gs.washington.edu/EVS/) databases, or in any of our 543 control exomes. Missense mutations were highlighted if they occurred within highly conserved residues in vertebrates, assessed utilizing the UCSC Genome Browser (http://genome.ucsc.edu/) conservation track tool [17]. To assess significance of mutations in our tumor dataset, we used a case–control approach to compare the frequency of private mutations in each gene in the 60 tumor exomes to 543 control exomes, which were from constitutional DNA of patients with Mendelian diseases also sequenced at the McGill University and Genome Quebec Innovation Centre (Table S2). We controlled for false discovery rate using the Benjamini–Hochberg procedure. All variants in these genes are detailed in Table S2, whereas only private variants, likely to be somatic, and in highly conserved residues (likely to impact function), are highlighted in Table S1 and discussed in this study.
Targeted next-generation sequencing of SETD2
Coding regions of SETD2 were amplified using the Fluidigm. Access Array system (http://www.fluidigm.com/access-array-system.html) and sequenced on a half run of the GS FLX Titanium system from Roche 454. Forty pairs of primers were designed to cover all coding regions of the 21 exons of SETD2. Primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) [28]. The parameters were set to achieve melting temperatures ranging from 57 to 59 °C. Lengths of PCR products are between 197 and 394 bp. The UCSC Genome Browser (http://genome.ucsc.edu/) was used to download target genomic regions prior to design and identify variants (based on dbSNP135: http://www.ncbi.nlm.nih.gov/projects/SNP/) [17]. PCRs were performed on 48 × 48 IFC (Integrated Fluidic Circuit) chips. On each chip, 40 regions were amplified in 48 samples. Amplification of target regions and addition of 454 sequencing adapters and individual bar codes occur in the same PCR performed on the Fluidigm FC1 cycler. All samples were individually bar coded and sequenced in one half-region of a GS FLX Titanium run. Validation of variants was done with Sanger sequencing. Following this, statistical analyses of Fisher’s exact test for contingency comparisons were performed utilizing GraphPad Prism 5 software.
Immunoblotting analysis of H3K36me3 levels in patient tumors
Fresh-frozen tumor tissues with adequate material and known SETD2, H3F3A and IDH1 mutational status were lysed using the EpiQuik Total Histone Extraction Kit (Epigentek, USA). Lysates were quantified utilizing standard BioRad protein assay (BioRad) and loaded onto 15 % acrylamide gels and run for 2 h at 100 V. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes at room temperature for 5 min, using the Trans-Blot Turbo transfer system (BioRad) at LOW MW setting, blocked and immunoblotted with the following conditions overnight at 4 °C: rabbit polyclonal anti-H3K36me3 (Abcam #9050) at 1:1,000 in 5 % skim milk and rabbit polyclonal anti-H3 (Abcam #1791) at 1:1,000 in 5 % skim milk. Membranes were subsequently washed thrice with tris-buffered saline-Tween 20 (TBS-T) and incubated with horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG secondary antibody (GE Healthcare #NA934V) at 1:5,000 with Precision Protein StrepTactin-HRP conjugate at 1:10,000 (BioRad #161-0380) in 5 % skim milk for 1 h at room temperature and revealed utilizing Amersham ECL detection (Amersham Biosciences). H3K36me3 bands from four independent blots were quantified utilizing ImageQuant TL v2003.02 (Amersham Biosciences), normalized to total H3, and normalized ratios were compared statistically using two-tailed T test for significance.
Methylation array profiling
DNA extracted from a subset of pediatric HGGs demonstrating defects in SETD2, IDH1, or H3F3A at G34 and wild-type tumors (n = 36) was analyzed for genome-wide DNA methylation patterns utilizing the HumanMethylation450 BeadChip according to the manufacturer’s instructions (Illumina, San Diego, USA) at the McGill University and Genome Quebec Innovation Centre. Of the >480,000 probes on the methylation chip, we discarded probes with ≥90 % sequence similarity to multiple genomic locations, with sequence variants in the probe binding region (1000 Genomes Project, SNPs with a minor allele frequency ≥2/120), and probes located on sex chromosomes, leaving 392,904 autosomal probes for further analysis. Subset-quantile within array normalization was performed on beta values using the SWAN method [23]. For unsupervised hierarchical clustering, the top 8,000 most variable probes (by standard deviation) were utilized with average linkage and Pearson correlation algorithms across the dataset. Consensus clustering was performed utilizing the k-means algorithm with 1,000 iterations on the top 8,000 most variable probes in the dataset. Methylation analysis was performed utilizing R (R version 2.14.2, http://cran.r-project.org/) with Minfi and ConsensusClusterPlus loaded packages.
Results and discussion
SETD2 mutations affect a significant proportion of pediatric HGGs
To identify genetic drivers in samples not carrying mutations in IDH1 and H3F3A, we analyzed 60 pediatric HGG tumors [grades III (n = 9) and IV (n = 51)] using WES (44 previously reported; Table S1) [32]. As matched normal DNA was unavailable for the majority of tumors, we identified private mutations that were present in tumors but were absent from public databases (1000 genomes project [24], NHLBI exomes) and from our set of 543 control exomes, and considered these as candidate somatic mutations. We compared the frequency of private mutations in each gene between the 60 tumors and 543 controls using Fisher’s exact test and used a false discovery rate threshold (FDR) of 0.05 to correct for multiple tests. Our case–control approach effectively corrects for the background rate of mutations in each gene (which implicitly includes the length of the gene and the mutability). We filtered out variants that were predicted to be tolerated/benign/unknown by both SIFT and PolyPhen-2 [1], and identified private mutations that we considered as candidate somatic mutations. The top genes by mutation frequency are shown in Table S2. As expected, four genes previously associated with pediatric HGG showed a highly significantly number of mutations (TP53, H3F3A, ATRX, NF1) [32] (Table S2). In addition, two genes not previously reported in HGG, SETD2 and CSMD3, achieved genome-wide significance (FDR = 0.029 and 0.031, respectively). For SETD2, this significance was more striking when only truncating mutations were considered (FDR = 0.0017), as no truncating mutations were seen in 543 controls, but tumor samples had frameshift (3), nonsense (1), and splicing (1) variants. In addition, the three missense variants in tumor samples occurred at highly conserved residues and were computationally predicted as damaging by both SIFT and Polyphen scores (Fig. 1a, b). In contrast, of the seven private variants in the control samples (all missense), only one is predicted damaging by both SIFT and Polyphen. H3.3 mutations occur at two positions within the histone tail involved in key regulatory post-translational modifications, K27 (directly) and K36 (indirectly). Driver loss-of-function SETD2 mutations have recently been identified in two high-grade cancers, renal cell carcinoma [6, 12] and early T-cell precursor acute lymphoblastic leukemia [37]. The other candidate gene, CSMD3 is expressed in adult and fetal brains; however, its functions are yet unclear [33]. Hence, we focused our next efforts on SETD2 as the top candidate gene.
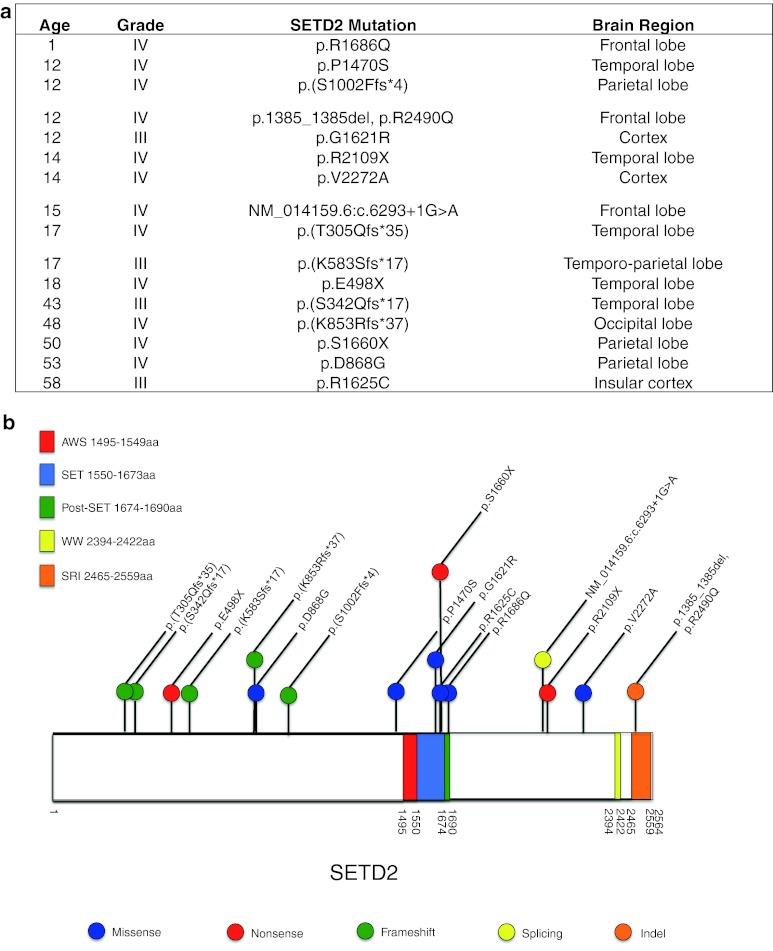
Fig. 1.
Missense/truncating mutations of the H3K36 trimethyltransferase SETD2 identified in pediatric and adult high-grade gliomas. a Patient age, tumor grade, and affected brain region of tumors with SETD2 mutation. b Schematic mapping type and distribution of missense/truncating mutations in SETD2 in 183 gliomas included in the study
SETD2 mutations affect pediatric and adult HGGs of the cerebral hemispheres
We expanded our sequencing analysis and next sequenced SETD2 in 123 additional gliomas of various ages and grades (Table 1; Table S1). Combining the discovery and the validation datasets, SETD2 mutations were identified in a total of 15 % of pediatric HGG (11/73) and 8 % of adult HGG (5/65), and were not seen in low-grade diffuse gliomas (0/45) (P = 0.0133; Table 1; Table S1). Except for one sample, all mutations occurred in children above the age of 12, in adolescents and in younger adults, mirroring the age range of H3.3 G34R/V and IDH1 mutations in HGG (Fig. 1a; Figure S1) [18–21, 32, 35]. Notably, all tumors carrying SETD2 mutations were localized in the cerebral hemispheres (P = 0.0055). SETD2 mutations were mutually exclusive with H3F3A mutations (P = 0.049) in HGGs (0/70), but showed partial overlap with IDH1 R132 mutations (4/14), TP53 (4/8) and ATRX (3/9) mutations (Table S1).
Table 1.
Frequencies of SETD2 mutations in 183 pediatric and adult gliomas
| Glioma | Mutated | Wild type | Total | Frequency (%) |
|---|---|---|---|---|
| Grade IV | 12 | 85 | 97 | 12.37 |
| Pediatric | 9 | 51 | 60 | 15 |
| Adult | 3 | 34 | 37 | 8.11 |
| Grade III | 4 | 37 | 41 | 9.76 |
| Pediatric | 2 | 11 | 13 | 15.38 |
| Adult | 2 | 26 | 28 | 7.14 |
| Grade II | 0 | 45 | 45 | 0 |
| Pediatric | 0 | 23 | 23 | 0 |
| Adult | 0 | 22 | 22 | 0 |
| Overall gliomas | 16 | 167 | 183 | 8.7 |
Missense/truncating mutations in SETD2 impair trimethyltransferase activity of the enzyme and confer distinct global DNA methylation signatures
SETD2 encodes the only H3K36 trimethyltransferase in humans [11, 41]. To support computational predictions of the damaging nature of SETD2 mutations, we assessed H3K36 trimethyltransferase activity in histone acidic extractions of patient tissue samples through Western blotting for H3K36me3 levels, an indicator of SETD2 activity [11]. Immunoblot analysis revealed a significant decrease in total H3K36me3 levels in SETD2-mutant gliomas (Fig. 2a), as well as a significantly decreased normalized ratio of H3K36me3 to total H3 levels in SETD2-mutant tumors (P < 0.001; Fig. 2b) showing loss-of-function as a result of SETD2 missense/truncating mutations.
Fig. 2.
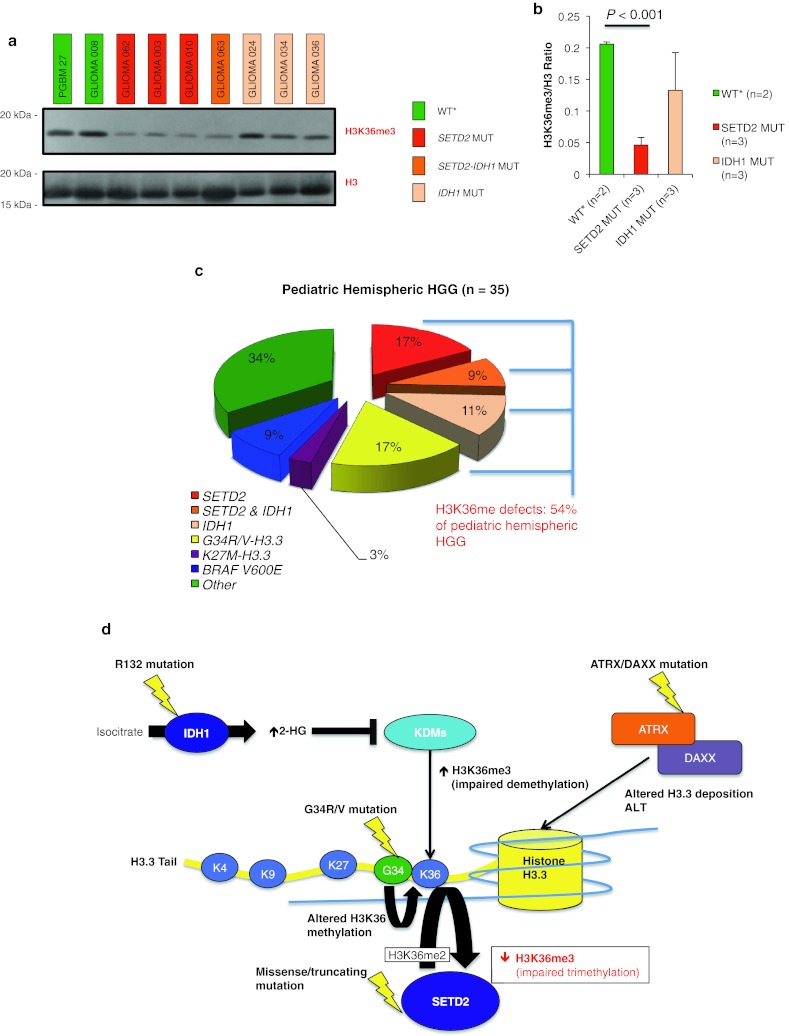
Missense/truncating mutations in SETD2 impair H3K36 trimethyltransferase activity of the enzyme. a Western blot analysis of histone acidic extracts of SETD2-mutant tumor samples demonstrating a significant decrease in H3K36me3 levels, indicating impaired H3K36 trimethyltransferase activity of the enzyme. b Densitometric quantification of H3K36me3 levels assessed in four independent blots demonstrating a significant decrease in H3K36me3/Total H3 normalized ratios in SETD2-mutant tumors. WT* = WT for SETD2, IDH1 and H3F3A. c Pie representation of mutations directly or indirectly affecting H3K36 methylation (H3K36me) in pediatric HGGs of the cerebral hemispheres (n = 35) indicating that approximately half of these tumors display defects, pointing to H3K36 dysregulation as a critical mechanism of hemispheric high-grade gliomagenesis. d Schematic representation of major genetic and epigenetic defects leading to altered H3K36 methylation in hemispheric HGGs
GBMs with epigenetic driver mutations such as H3.3 K27M or G34R/V, as well as those with IDH1 mutations, display distinct DNA methylation profiles and clinical characteristics. They also arise in distinct anatomic compartments, with IDH1- and H3.3 G34R/V-mutant tumors being restricted to areas of the cerebral hemispheres [18–21, 32, 35]. We and others have previously described the distinct heterogeneity of epigenetic profiles underlying HGGs including GBM [2, 35, 36]. We thus sought to characterize the DNA methylation profiles of 36 pediatric HGG tumors with mutations likely to affect K36 methylation status, using the Illumina 450K array platform as previously described [35]. SETD2 mutations yielded global DNA methylation patterns distinct from tumors with H3.3 G34R/V mutations, but which partly overlapped with IDH1-mutant methylation patterns (Fig. 3a–f). Notably, promoters at OLIG1/2 loci, characteristically hypermethylated in G34R/V-mutated samples, were not hypermethylated in SETD2 mutants (Figure S2) [2, 35].
Fig. 3.
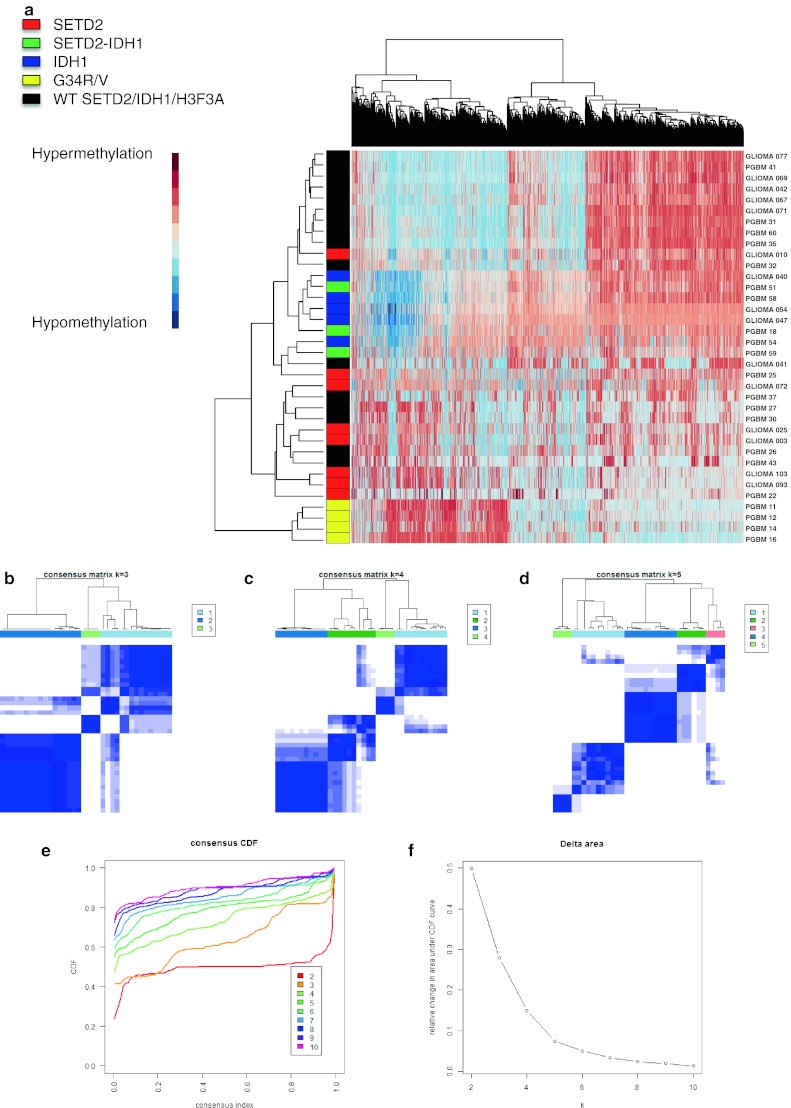
Mutations affecting H3K36 methylation confer distinct global DNA methylation signatures. a Unsupervised hierarchical clustering of methylation Beta-values representing the top 8,000 most variable probes between samples mutated for SETD2, IDH1 or H3.3 G34R/V and high-grade gliomas wild-type (WT) for these genes (n = 36). b k-means consensus matrices for k = 3 (b), k = 4 (c) or k = 5 (d) for the top 8,000 most variable probes. e Empirical cumulative distribution function (CDF) plot and delta area differences (f) for indicated numbers of clusters (k = 2 to k = 10)
Mutations identified in candidate oncogenic drivers and other genes involved in histone post-translational modifications in HGGs
We further investigated our dataset for mutations in other genes affecting PTM of H3K27 or H3K36 but which did not reach the statistically significant mutation levels. Eight distinct mammalian enzymes methylate H3K36 and share the catalytic SET domain, but have varying preferences for K36 residues in different methylation states (reviewed in [41]). SETD2 is the only enzyme in humans to catalyze H3K36 tri-methylation [11], while its mono- and/or di-methylation is catalyzed by NSD1, NSD2, NSD3, SETMAR, ASH1L, SMYD2 or SETD3 (reviewed in [41]). We identified one missense and one nonsense mutation in ASH1L (concurrently with SETD2 mutation) and SETD3 (1 missense mutation concurrently with SETD2 mutation). One PGBM mutant for SETD2 also had a missense mutation in UTX/KDM6A (H3K27 demethylase). This same PGBM had a missense mutation in PBRM1, a gene frequently mutated in renal cell carcinoma in association with SETD2 [39]. We also identified two mutations in KDM5C (H3K4 demethylase) (Table S1). Interestingly, no mutations in the cancer-implicated histone methyltransferase EZH2 were identified. Mutations in these genes were not prevalent enough to be statistically associated with HGGs in our sample set; however, it remains possible that they contribute to pathogenesis in a small fraction of HGG cases.
Further investigation of the exome dataset revealed previously described mutations in BRAF (V600E [30, 31], 5/60 pediatric HGG), which did not overlap with the epigenetic driver mutations we identify (Table S1; Fig. 2c). Other alterations also previously described in GBM, which may provide pathways alternative or complementary to epigenomic dysregulation, included PTEN mutations (two samples) which overlapped with H3.3 K27M while EGFR mutation or amplification (three samples) and CDKN2A mutation/loss (five samples) partially overlapped with SETD2 mutations (Table S1). Truncating mutations in the mismatch repair genes [10] MSH6 (three samples) and MSH2 (one sample) were identified and were concurrent with IDH1 (two samples) and SETD2 (three samples) mutations. Of note, SETD2 mutations were absent in a large cohort of 125 cases of medulloblastoma [15] another major group of pediatric brain tumors.
Alteration of H3K36 post-translational modifications characterize hemispheric adolescent and younger adult HGG
Post-translational modification of resident histones modulates the properties of chromatin, impacting cell state and differentiation and determining the outcome of virtually all DNA processes in eukaryotes. Methylation of H3K36 is a key histone mark and has been widely associated with active chromatin but also with transcriptional repression, alternative splicing, DNA replication and repair, DNA methylation and the transmission of memory of gene expression from parents to offspring during development (reviewed in [41]). We identify loss-of-function mutations in SETD2, in 15 % of pediatric and 8 % of adult high-grade gliomas in a cohort of 183 samples from all ages and grades II–IV of glioma (Fig. 1a; Table S1). We further show SETD2 mutations to be specific to high-grade tumors (P = 0.013), to HGGs located within the cerebral hemispheres (P = 0.005), and to be mutually exclusive with H3.3 mutations we [18, 32] and others [42] previously identified in pediatric high-grade astrocytomas (P = 0.049). SETD2 alterations overlapped with IDH1 mutations in 4 of 14 tumors (Table S1). Strikingly, the oncometabolite produced by IDH1 mutations inhibits a plethora of histone demethylases (KDMs) causing aberrant histone methylation at defined residues including K27 and K36 and a block to cell differentiation [4, 22, 29, 43]. We [21] and others [14] have previously shown the association of ATRX and TP53 mutations in IDH1-mutant diffuse astrocytic gliomas, and others have pointed to mutations in CIC and 1p19q loss in IDH1-mutant oligodendroglial tumors. Thus, IDH1 mutations may require other key genetic events in a specific context for full-blown tumorigenesis, which may include SETD2 mutations as suggested by our cohort. H3.3K36 methylation can be thus disrupted by H3.3 G34R/V mutation, IDH mutations and the SETD2 mutations we report herein (Fig. 2a, b). Furthermore, our current analysis suggests that this histone mark is specifically altered in hemispheric adolescent and younger adult HGG (Fig. 2c, d) [2, 18, 19, 25, 32, 34, 35], and that the functional effect differs between SETD2 and H3.3 mutations (Fig. 3; Figure S2). Future studies directed towards elucidating the importance of H3K36 methylation in cortical astrocytes and neural progenitor cells, and its dysregulation in tumorigenesis may lend insight into the regional specificity of these defects, while improved understanding of the consequences of altered chromatin remodeling induced by these mutations will help guide alternative therapeutic avenues for these deadly cancers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to acknowledge the excellent staff of the McGill University and Genome Quebec Innovation Centre High-Throughput sequencing platform for performing the exome capture and sequencing. This work was supported by the Cole Foundation, and was funded by Genome Canada and the Canadian Institute for Health Research (CIHR) with co-funding from Genome BC, Genome Quebec, CIHR-ICR (Institute for Cancer Research) and C17, through the Genome Canada/CIHR joint ATID Competition [project title: The Canadian Paediatric Cancer Genome Consortium: Translating next-generation sequencing technologies into improved therapies for high-risk childhood cancer (NJ)], the Hungarian Scientific Research Fund (OTKA) Contract No. T-04639, TAMOP-4.2.2.A-11/1/KONV-2012-0025, the National Research and Development Fund (NKFP) Contract No. 1A/002/2004 (PH, MG, LB), the PedBrain project contributing to the International Cancer Genome Consortium funded by the German Cancer Aid (109252) and the CNS tumor tissue bank within the priority program on tumor tissue banking of the German Cancer Aid (108456), the BMBF, the Samantha Dickson Brain Tumor Trust, and a grant from the National Cancer Center Heidelberg (“Paediatric Brain Tumor Preclinical Testing”). A. Fontebasso and X-Y. Liu are the recipients of studentship awards from CIHR. DA Khuong-Quang is the recipient of a studentship from the Fonds de la recherche en santé du Québec (FRSQ). S. Pfister is the recipient of the Sybille Assmus Award for Neurooncology in 2009 and N. Jabado is the recipient of a Chercheur Clinicien Award from Fonds de Recherche en Santé du Québec and an award from the Canadian Gene Cure Foundation.
Conflict of interest
The authors declare no competing financial interests.
Footnotes
A. M. Fontebasso and J. Schwartzentruber contributed equally to this work.
S. M. Pfister, N. Jabado, J. Majewski are co-senior and co-corresponding authors.
Contributor Information
Stefan M. Pfister, Email: s.pfister@dkfz-heidelberg.de
Nada Jabado, Email: nada.jabado@mcgill.ca.
Jacek Majewski, Email: jacek.majewski@mcgill.ca.
References
- 1.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310(5746):306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J, Kung HF. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46(1):200–208. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 4.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloughesy TF, Mischel PS. New strategies in the molecular targeting of glioblastoma: how do you hit a moving target? Clin Cancer Res Off J Am Assoc Cancer Res. 2011;17(1):6–11. doi: 10.1158/1078-0432.CCR-09-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O’Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, Ragozin S, Jeltsch A The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet 20(11):2195–2203. doi:10.1093/hmg/ddr107 [DOI] [PMC free article] [PubMed]
- 9.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neurooncology. 2012;14(Suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert A, Kloor M, Giersch A, Ahmadi R, Herold-Mende C, Hampl JA, Heppner FL, Zoubaa S, Holinski-Feder E, Pietsch T, Wiestler OD, von Knebel Doeberitz M, Roth W, Gebert J. Microsatellite instability in pediatric and adult high-grade gliomas. Brain Pathol. 2007;17(2):146–150. doi: 10.1111/j.1750-3639.2007.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, Shi Y. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol. 2011;18(7):769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Marie SK, Bettegowda C, Agrawal N, Lipp E, Pirozzi C, Lopez G, He Y, Friedman H, Friedman AH, Riggins GJ, Holdhoff M, Burger P, McLendon R, Bigner DD, Vogelstein B, Meeker AK, Kinzler KW, Papadopoulos N, Diaz LA, Yan H. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, McCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, Kastenhuber ER, Heguy A, Petrini JH, Chan TA, Huse JT. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, Bourgey M, Bourque G, Montpetit A, Bourret G, Lepage P, Fleming A, Lichter P, Kool M, von Deimling A, Sturm D, Korshunov A, Faury D, Jones DT, Majewski J, Pfister SM, Jabado N, Hawkins C. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khuong-Quang DA, Gerges N, Jabado N. Mutations in histone H3.3 and chromatin remodeling genes drive pediatric and young adult glioblastomas. Med Sci M/S. 2012;28(10):809–812. doi: 10.1051/medsci/20122810004. [DOI] [PubMed] [Google Scholar]
- 20.Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, Ellingson BM, Bowers CW, Soriano RH, Schmidt NO, Mohan S, Yong WH, Seshagiri S, Modrusan Z, Jiang Z, Aldape KD, Mischel PS, Liau LM, Escovedo CJ, Chen W, Nghiemphu PL, James CD, Prados MD, Westphal M, Lamszus K, Cloughesy T, Phillips HS. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol Off J Am Soc Clin Oncol. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, Dong Z, Siegel P, Albrecht S, Croul S, Jones DT, Kool M, Tonjes M, Reifenberger G, Faury D, Zadeh G, Pfister S, Jabado N. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 22.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maksimovic J, Gordon L, Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The 1000 Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature 467(7319):1061–1073. doi:10.1038/nature09534 [DOI] [PMC free article] [PubMed]
- 25.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. The Journal of biological chemistry. 2006;281(13):8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- 26.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, You-Ten A, Li WY, Schalm S, Su SM, Virtanen C, Reifenberger G, Ohashi PS, Barber DL, Figueroa ME, Melnick A, Zuniga-Pflucker JC, Mak TW. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiffman JD, Hodgson JG, VandenBerg SR, Flaherty P, Polley MY, Yu M, Fisher PG, Rowitch DH, Ford JM, Berger MS, Ji H, Gutmann DH, James CD. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70(2):512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, Schmieder K, Wesseling P, Mawrin C, Hasselblatt M, Louis DN, Korshunov A, Pfister S, Hartmann C, Paulus W, Reifenberger G, von Deimling A. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 32.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tönjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482((7384)):226–231. doi:10.1038/nature10833 [DOI] [PubMed]
- 33.Shimizu A, Asakawa S, Sasaki T, Yamazaki S, Yamagata H, Kudoh J, Minoshima S, Kondo I, Shimizu N. A novel giant gene CSMD3 encoding a protein with CUB and sushi multiple domains: a candidate gene for benign adult familial myoclonic epilepsy on human chromosome 8q23.3-q24.1. Biochem Biophys Res Commun. 2003;309(1):143–154. doi: 10.1016/S0006-291X(03)01555-9. [DOI] [PubMed] [Google Scholar]
- 34.So AY, Jung JW, Lee S, Kim HS, Kang KS. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS ONE. 2011;6(5):e19503. doi: 10.1371/journal.pone.0019503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Fruhwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Durken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Sudo T, Utsunomiya T, Mimori K, Nagahara H, Ogawa K, Inoue H, Wakiyama S, Fujita H, Shirouzu K, Mori M. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer. 2005;92(9):1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, Zhang HB, Liao YJ, Zheng F, Zhu W, Liu TH, Bian XW, Guan XY, Lin MC, Zeng MS, Zeng YX, Kung HF, Xie D. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31(5):583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 38.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villard L, Gecz J, Mattei JF, Fontes M, Saugier-Veber P, Munnich A, Lyonnet S. XNP mutation in a large family with Juberg-Marsidi syndrome. Nat Genet. 1996;12(4):359–360. doi: 10.1038/ng0496-359. [DOI] [PubMed] [Google Scholar]
- 41.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012 doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.