Abstract
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) is frequently mutated in a variety of human cancers. However, the cell signaling pathways regulated by PTPN14 largely remain to be elucidated. Here, we identify a list of potential substrates of PTPN14 using a phospho-proteomic approach. We show that p130Cas is a direct substrate of PTPN14 and that PTPN14 specifically regulates p130Cas phosphorylation at tyrosine residue 128 (Y128) in colorectal cancer (CRC) cells. We engineered CRC cells homozygous for a p130Cas Y128F knock-in mutant and found that these cells exhibit significantly reduced migration and colony formation, impaired anchorage-independent growth, slower xenograft tumor growth in nude mice, and have decreased phosphorylation of AKT. Furthermore, we demonstrate that SRC phosphorylates p130Cas Y128 and that CRC cell lines harboring high levels of pY128 Cas are more sensitive to SRC family kinase inhibitor Dasatinib. These findings suggest that p130Cas Y128 phosphorylation may be exploited as a predictive marker for Dasatinib response in cancer patients. In aggregate, our studies reveal a novel signaling pathway that plays an important role in colorectal tumorigenesis.
Keywords: PTPN14, p130Cas, tumorigenesis, colorectal cancer
Introduction
Reversible tyrosine phosphorylation governs numerous signaling pathways that regulate cell proliferation, apoptosis, adhesion and migration (1). Protein tyrosine phosphorylation is coordinately controlled by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs) (1). Many PTKs have been identified to be oncogenes that are crucial to tumorigenic processes (2). Increasing evidence indicates that PTPs also play important roles in tumorigenesis (3). In a systematic mutational analysis of the PTP gene family in colorectal cancers (CRCs), protein tyrosine phosphatase non-receptor type 14 (PTPN14) was identified as one of the six PTPs that are mutated in CRCs (4). Recent studies revealed that PTPN14 is also mutated in breast, head and neck, kidney, liver, ovarian and skin cancers (5–10), suggesting that PTPN14 may play an important role in tumorigenesis of multiple types of cancers.
PTPN14, also known as Pez, PTP36 and PTPD2, consists of an N-terminal FERM (four-point-one, ezrin, radixin, moesin) domain and a C-terminal phosphatase domain (11). It has been shown that PTPN14 may regulate cell-cell adhesion, cell-matrix adhesion, cell migration and cell growth (12–14). Interestingly, a recent study indicates that PTPN14 also regulates TGFβ gene expression, thereby modulating epithelial-mesenchymal transition (15). Knockdown of the PTPN14 homolog in zebrafish results in developmental defects (15). Although PTPN14 was shown to regulate tyrosine phosphorylation of β-catenin (12), it remains to be determined how this regulation impacts tumorigenesis. Given that PTPN14 is mutated in various cancers including CRCs, it is important to identify more substrates of PTPN14 in CRC cells. In this study, we identified a list of putative substrates of PTPN14 using a phospho-proteomic approach. We further validated p130 Crk-associated substrate (p130Cas) as a critical PTPN14 substrate.
p130Cas was originally identified as a Crk-associated protein (16, 17). It is also known as breast-cancer anti-estrogen resistance protein 1 (BCAR1), because p130Cas overexpression renders breast cancer cells resistant to Tamoxifen treatment (18). Moreover, p130Cas is overexpressed in a subset of breast cancers (19). Transgenic mice overexpressing p130Cas shorten the latency of Her2/neu-induced breast cancer development (19). Furthermore, p130Cas is required for SRC-mediated transformation (20, 21). Together, these studies suggest that p130Cas is a key player in tumorigenesis. p130Cas is an adaptor protein that mediates integrin and growth factor signaling (22). It contains several conserved domains for protein-protein interactions. At the N-terminus, there is a substrate domain with 15 YXXP repeats which can be heavily phosphorylated (22). Here, we report that PTPN14 dephosphorylates the tyrosine 128 (Y128) residue, one of the YXXP repeats, on p130Cas. We further demonstrated that regulation of Y128 p130Cas phosphorylation plays a critical role in CRC tumorigenesis.
Results
Identification of p130Cas as a substrate of PTPN14 using a phospho-proteomic approach
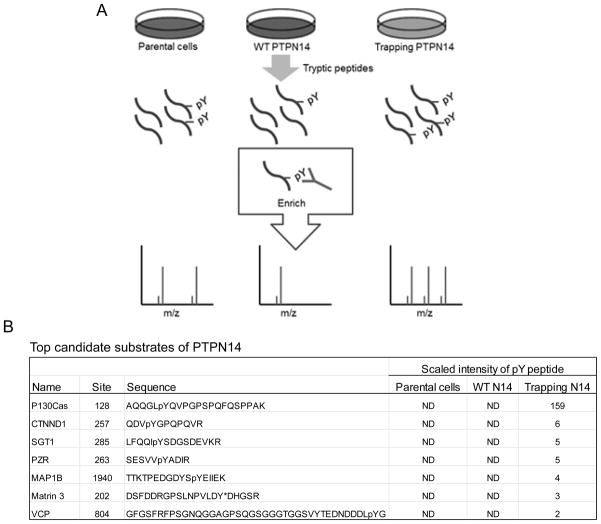
To identify potential substrates of PTPN14, we constructed two stable SW480 CRC cell lines expressing either a wild-type (WT) PTPN14 PTP domain or a PTPN14 D1079A trapping mutant PTP domain. It is well documented that PTP trapping mutants can trap phospho-substrates in their catalytic pockets and protect the substrates from dephosphorylation, thereby potentially enriching the phospho-substrates (23, 24). Proteins were extracted from these cell lines as well as the parental SW480 cells and digested with trypsin. Phospho-tyrosine (pY) containing peptides were enriched using anti-pY antibody columns. Peptides from each cell line were profiled by mass spectrometry analyses (Fig 1A). pY peptides were quantified as described by Rush at al (25). Those pY containing peptides that were either down-regulated in the cell lines overexpressing the WT phosphatase or up-regulated in the cell line overexpressing the trapping mutant compared to the parental cells are potential candidate substrates (Fig. 1B and Table S1). Of these candidate substrates, we chose to further characterize p130Cas because the pY128 containing peptides of p130Cas were enriched over 150 fold in the cell line overexpressing the PTPN14 trapping mutant phosphatase (Fig. 1B).
Figure 1. Identification of potential substrates of PTPN14 using a phospho-proteomic approach.
(A) Schematic diagram of a phospho-proteomic approach to identify potential substrates of PTPN14. Cell lysates were made from the parental SW480 CRC cells, stable clones expressing either a wild-type (WT) PTP domain or a PTPN14 D1079A trapping mutant PTP domain. Proteins were digested with trypsin and pY containing peptides were enriched by anti-pY antibody (pY100)-conjugated beads. Peptide mixtures were profiled by mass-spectrometry. (B) List of the top candidate substrates of PTPN14. ND, not detected.
PTPN14 regulates phosphorylation of p130 at the tyrosine 128 residue in CRC cells
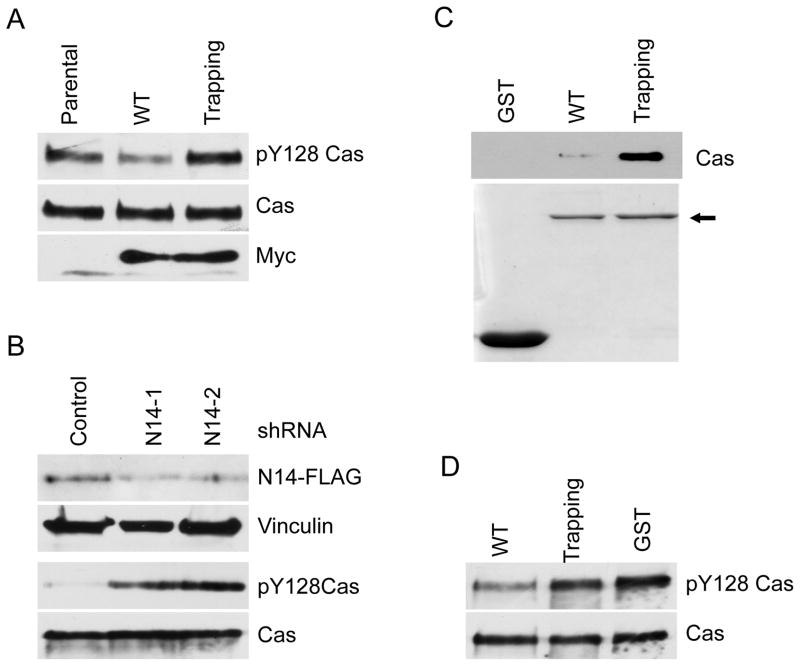
We set out to determine if PTPN14 indeed regulates p130Cas Y128 phosphorylation. To this end, we first generated a pY128 p130Cas specific antibody. As shown in Fig. S1, the antibody recognized a phospho-signal on the wild-type p130Cas. In contrast, it failed to detect any signal on p130Cas Y128F mutant protein (Fig. S1), indicating that this antibody specifically recognizes phosphorylated Y128 of p130Cas. We then performed Western blot analyses on the aforementioned SW480 CRC cell lines using this antibody. Consistent with the proteomic data, compared to parental cells, overexpression of WT PTPN14 phosphatase resulted in reduced levels of pY128 p130Cas (Fig. 2A), whereas overexpression of the trapping mutant led to increased levels of this phosphorylation (Fig. 2A). Conversely, knockdown of PTPN14 by two independent shRNAs resulted in increased levels of pY128 p130Cas in a genetically engineered DLD1 CRC cell line (Fig. 2B), in which the endogenous PTPN14 proteins are epitope-tagged with 3xFLAG (26). Taken together, these data demonstrated that PTPN14 regulates phosphorylation of p130Cas Y128 in CRC cells.
Figure 2. Validation of p130Cas as a substrate of PTPN14.
(A) Overexpression of WT PTPN14 PTP domain decreases p130Cas Y128 phosphorylation, whereas the trapping mutant enriches pY128 Cas in CRC cells. Cell lysates made from the cell lines used in Fig. 1 were blotted with the indicated antibodies. (B) Knock-down of PTPN14 leads to increased levels of pY128 Cas in CRC cells. Cell lysates from stable clones of DLD1 CRC cells expressing scramble shRNAs or shRNAs against PTPN14 were blotted with the indicated antibodies. N14-1 and N14-2 are two independent shRNAs against PTPN14. (C) A PTPN14 trapping mutant traps p130Cas protein. GST, GST-WT and GST-D1079A trapping mutant PTPN14 PTP domains were expressed in E. Coli, purified and resolved on a SDS-PAGE gel (bottom panel). Arrow indicates the GST fusion proteins. Equal amounts of the purified protein were mixed with DLD1 cell lysates in the phosphatase trapping assay. The trapped p130Cas proteins were detected by Western blot (top panel). (D). PTPN14 dephosphorylates pY128 Cas in vitro. Phosphorylated p130Cas proteins were incubated with the indicated GST fusion proteins. The pY128 Cas proteins were quantified by Western blot analysis.
PTPN14 directly dephosphorylates p130Cas
To determine whether PTPN14 directly regulates p130Cas Y128 phosphorylation, we first employed a substrate trapping assay developed by Flint et al. (23). WT or substrate trapping mutant (D1079A) of PTPN14 phosphatase domain was fused to GST and expressed in E. coli. GST fusion proteins were purified and attached on Glutathione beads. Equal amounts of WT and mutant GST-fusion proteins were used for a trapping assay (Fig. 2C). As shown in Fig. 2C and Fig S2, the trapping mutant fusion proteins pulled down p130Cas abundantly from cell lysates made from two different CRC cell lines DLD1 and SW480. In contrast, the WT GST-fusion proteins only pulled down minimal amount of p130Cas, whereas GST alone did not bind to p130Cas. Secondly, when mixed with phosphorylated p130cas substrates in vitro, the WT PTPN14 GST fusion proteins dephosphorylated p130Cas at the Y128 site in comparison with controls with equal amounts of GST or the enzymatically inactive mutant proteins (trapping mutant) (Fig. 2D). In aggregate, our results showed unequivocally that p130Cas is a direct substrate of PTPN14. However, we failed to detect protein-protein interaction between PTPN14 and p130Cas in DLD1 cell lysates by a co-immunoprecipitation assay (data not shown), suggesting that the two proteins only interact transiently when the enzymatic reaction occurs.
SRC phosphorylates p130Cas at Tyr128
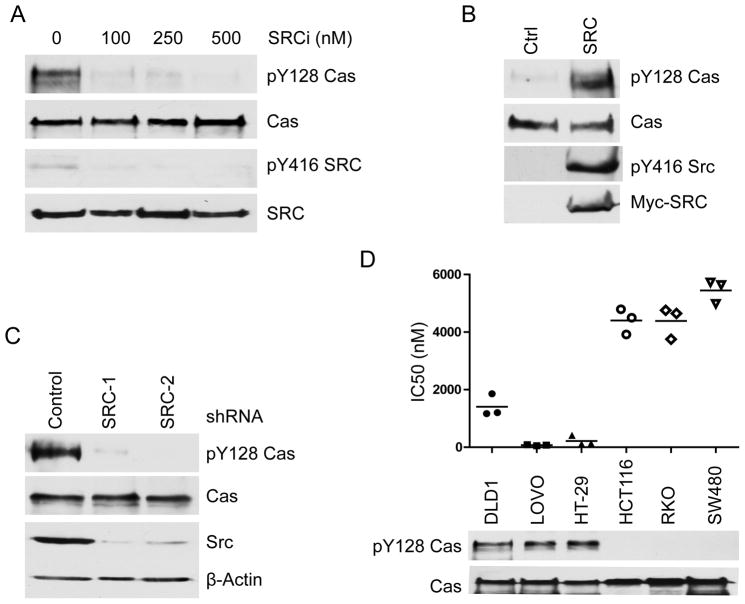
Having demonstrated that PTPN14 dephosphorylates Y128 of p130Cas, we set out to identify the kinase that phosphorylates this site. Because SRC kinase has been shown to phosphorylate p130Cas at several tyrosine residues other than Y128, we elected to test whether SRC could also phosphorylate Y128 on p130Cas. Three lines of evidence indicated that SRC indeed phosphorylates this site: (1) Treatment of DLD1 cells with a SRC inhibitor AZD0530 led to dephosphorylation of pY128 p130Cas (Fig. 3A); (2) Knockdown of SRC by two independent shRNAs also resulted in reduced levels of pY128 p130Cas in DLD1 cells (Fig. 3B); (3) Overexpression of SRC led to increased levels of pY128 on p130Cas (Fig. 3C).
Figure 3. SRC phosphorylates the Y128 residue of p130Cas.
(A) SRC kinase inhibitor treatment down-regulates pY128 p130Cas. DLD1 cells were treated with the indicated concentrations of SRC inhibitor AZD0530. Cell lysates were blotted with the indicated antibodies. (B) Overexpression of SRC results in increased levels of pY128 p130Cas. HEK 293 cells were transfected with either a Myc-tagged SRC plasmid or an empty vector. Cell lysates were blotted with the indicated antibodies. (C) Knock-down of SRC down-regulates pY128 Cas in CRC cells. Cell lysates from stable clones of DLD1 cells expressing scramble shRNAs or shRNAs against SRC were blotted with the indicated antibodies. SRC-1 and SRC-2 are two independent shRNAs against SRC. (D) CRC cells harboring high levels of pY128 Cas are more sensitive to SRC inhibitor Dasatinib-induced growth inhibition. The indicated CRC cell lines are treated with a serial concentration of Dasatinib. The IC50s of each cell line from three independent experiments are plotted. The levels of pY128 Cas in each cell line are quantified by Western blot analysis.
Colon cancer cell lines harboring higher levels of pY128 p130Cas are more sensitive to the killing by SRC family kinase inhibitor Dasatinib
SRC family kinase inhibitor Dasatinib is used in clinic to treat chronic myelogenous leukemia and there are several ongoing clinical trials using Dasatinib in combination with other drugs to treat colon cancers (27). To test if p130Cas Y128 phosphorylation associates with Dasatinib inhibition, we treated six colon cancer cell lines with various doses of Dasatinib and the IC50s of these cell lines were shown in up panel of Fig. 3D. Remarkably, colon cancer cell lines harboring higher levels of pY128 p130Cas were more sensitive to growth inhibition by Dasatinib (Fig. 3D).
Phosphorylation of p130Cas Y128 can be stimulated by EGF
Given that we have identified the kinase and phosphatase that regulate p130Cas Y128 phosphorylation, it is of interest to identify stimuli that may activate this phosphorylation in CRC cells. To this end, we treated DLD1 cells with various growth factors and cytokines including EGF, FGF, PDGF-AA, Il-6, and VEGF. Among them, EGF could robustly stimulate p130Cas Y128 phosphorylation after 30 minute treatment (Fig S3A). In contrast, other growth factors and cytokines, including PDGF-AA, failed to activate p130Cas Y128 phosphorylation, although PDGF was able to activate ERK1/2 (Fig S3B). Furthermore, EGF also induced phosphorylation of p130Cas Y128 in multiple CRC cells including RKO cells (Fig. S3A). Given that EGF signaling plays a critical role in CRC tumorigenesis and that anti-EGFR antibodies are approved by the FDA to treat CRC patients (28), we chose to study p130Cas Y128 phosphorylation in the context of EGF stimulation.
Engineering p130Cas Y128F mutant knock-in (KI) CRC cells
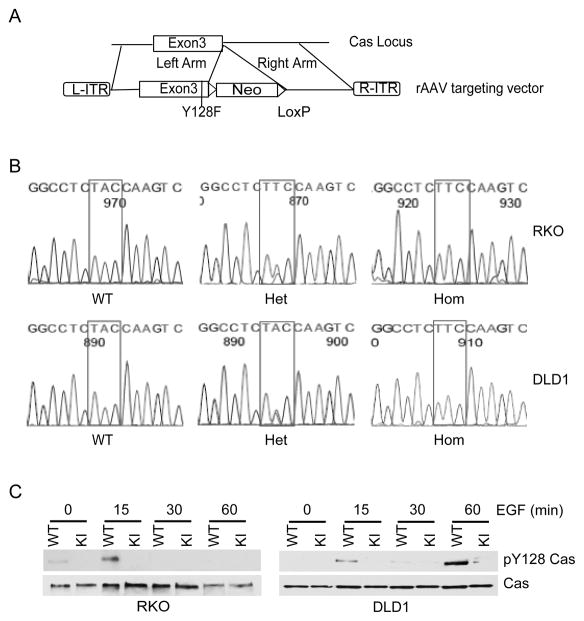
To rigorously test whether regulation of p130Cas Y128 phosphorylation is critical to colorectal tumorigenesis, we set out to engineer p130Cas Y128F knock-in (KI CRC) cell lines. The adeno-associated virus (AAV) targeting system was used to engineer the knock-in cell lines because of its high homologous recombination frequency in somatic cells (26, 29, 30). We first chose to knock in the p130Cas Y128F mutant allele into the human colon cancer cell line RKO, because we had shown that p130Cas Y128 phosphorylation can be activated by EGF in RKO cells as described above and that RKO cells are widely used for gene targeting (31). The targeting strategy is outlined in the schematic diagram in Fig 4A. After the first round of gene targeting, 7 targeted clones were identified out of 96 geneticin resistant clones screened. To ensure the presence of the mutant allele, genomic PCR products of exon 3, which encodes the Y128 residue, from the targeted clones were sequenced. Five of the 7 targeted clones harbored p130Cas Y128F mutant allele. Two clones were infected with adenovirus expressing Cre-recombinase to excise the neomycin resistance gene (Fig. 4A) and targeted for the second allele to generate homozygous KI clones. To confirm that there was no WT p130Cas allele expression in the homozygous p130Cas Y128F mutant clones, the genomic PCR products of p130Cas were sequenced. As expected, both WT p130Cas alleles were replaced by the p130Cas Y128F mutants in the homozygous KI clones (Fig 4B). Furthermore, Western blot analyses showed that p130Cas proteins were expressed in the homozygous KI cells but they remained unphosphorylated at residue 128 after EGF stimulation, while p130Cas proteins in the parental cells were heavily phosphorylated at Y128 post EGF stimulation (Fig. 4C). These data indicated that we had successfully engineered p130Cas Y128F mutant KI cells. We chose two independently derived heterozygous and homozygous KI clones for in-depth analyses and both clones behaved similarly in all the studies described below. To ensure that what we observe with RKO cells is not cell line specific, we used the same method to generate p130Cas Y128F mutant DLD1 cells (Fig. 4).
Figure 4. Generation of p130Cas Y128F mutant knockin (KI) CRC cells.
(A) Diagram of the KI construct. (B) Genomic sequences of the parental (WT), heterozygous (Het) and homozygous (Hom) KI cells. The rectangular boxes indicate codons for the Cas amino acid 128 position. (C) The parental (WT) and p130Cas Y128F homozygous KI CRC cells were serum-starved and stimulated with EGF. Cell lysates were blotted with the indicated antibodies.
In vitro, p130Cas Y128F mutant CRC cells are reduced in properties predicative of in vivo tumorigenicity
When grown under normal tissue culture conditions (McCoy’s 5A supplemented with 10%FBS), the average doubling times of the DLD1 p130Cas Y128F mutant clones increased by 1.5 hours in comparison to the parental cells (Fig. S4), whereas no doubling time difference was observed between RKO parental and the mutant clones (Fig. S4). Cell cycle profiling showed slightly increased G1 populations in the p130Cas Y128F homozygous KI clones derived from both DLD1 and RKO cells (Fig S5).
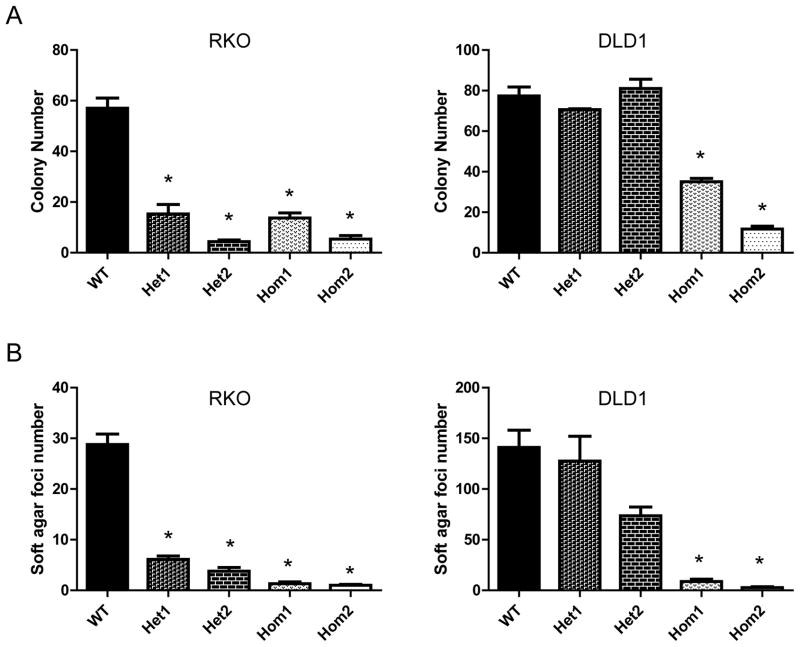
To test whether p130Cas Y128F mutant affects tumorigenicity correlated responses in vitro, we performed colony formation and soft agar assays with the p130Cas mutant KI cells. Compared to the parental cells, homozygous p130Cas Y128F KI RKO and DLD1 cells exhibited 3–6 fold (p < 0.001) reduced abilities to form colonies in colony-formation assays (Fig. 5A). Similarly, homozygous p130Cas mutant CRC cell clones formed ~25 fold (p < 0.001) less foci in soft agar assay than their wild-type counterparts (Fig. 5B). Interestingly, the RKO heterozygous KI clones, but not these of DLD1 heterozygous KI clones, displayed significant (p < 0.001) reduction in colony numbers and soft-agar foci with respect to wild-type cells (Fig. 5A and B).
Figure 5. p130Cas Y128F mutant CRC cells are less tumorigenic in vitro.
(A) p130Cas Y128F mutant CRC cells form fewer colonies. Cells from indicated clones were plated in 6-well plates in triplicates. Cells were grown for 14 days and stained with crystal violet. Colony numbers were counted and plotted for each of the clones. (B) p130Cas Y128F mutant CRC cells impair anchorage-independent growth. CRC cells of the indicated clones were mixed in 0.4% soft agar and plated in 6-well plates in triplicates. Cells were grown for 30 days. Colony foci were counted and plotted for each of the clones. * p < 0.0001, t test. Het and Hom indicate the heterozygous and homozygous KI clones respectively.
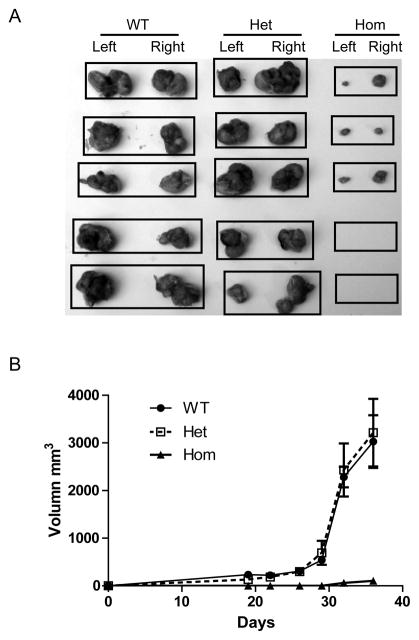
The p130Cas mutant RKO cells were less tumorigenic in vivo
Tumorigenicity of the KI cells was also tested in a more stringent in vivo model. For these studies, p130Cas Y128F homozygous, heterozygous clones or the parental RKO and DLD1 cells were injected subcutaneously into nude mice. After 35 days of growth, wild-type cells formed tumors in all mice injected, whereas the p130Cas Y128F homozygous RKO KI clones failed to form tumors in two of the five mice injected (Fig. 6A). The average tumor volumes of p130Cas Y128F homozygous RKO KI clones were 30-fold smaller than those produced by the parental cells (p < 0.001) (Fig. 6B). However, no significant difference in xenograft tumor growth was observed between the DLD1 homozygous KI clones and the parental (Fig. S6). Both RKO and DLD1 heterozygous KI clones formed similar sizes of tumors to those of parental cells (Fig. 6A and B, and Fig. S6).
Figure 6. The RKO p130Cas Y128F mutant cells are less tumorigenic in vivo.
Athymic nude mice were injected subcutaneously with cells from the indicated clones. Tumor sizes were measured weekly for 5 weeks. Mice were then sacrificed and tumors were harvested. (A) Tumors grown from the RKO clones. Each black rectangle indicates tumors harvested from a mouse. (B) Average sizes of the tumors formed by the indicated clones were plotted.
The p130Cas Y128F mutant CRC cells display defects in cell spreading and migration
Given that both PTPN14 and p130Cas are involved in cell adhesion and migration (11, 22), we set out to determine how p130Cas Y128 phosphorylation impacts cancer cell adhesion and migration. Boyden chamber cell migration assay showed that the p130Cas Y128F mutant cells exhibited significantly reduced ability in cell migration (Fig. S7). When grown on cover slips coated with fibronectin, the majority of parental RKO and DLD1 cells spread fully and displayed a fibroblast-like morphology (Fig. S8 A and B). In contrast, most of the p130Cas Y128F DLD1 mutant cells were not fully-spreading (Fig. 7A and B). The percentages of fully-spreading mutant RKO cells were also significantly reduced, although not as dramatic as the mutant DLD1 cells. However, no apparent focal adhesion defect was observed with the mutant cells (Fig. S8A).
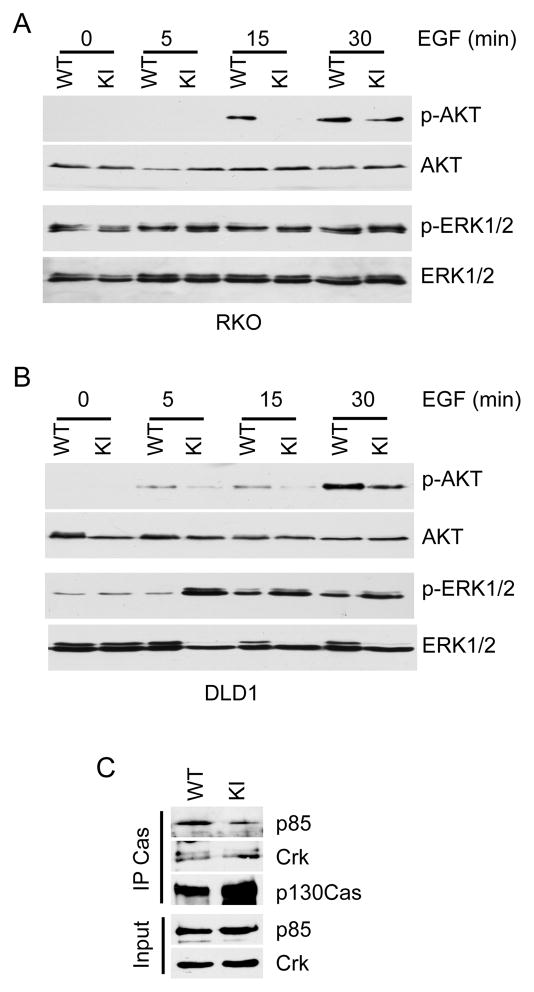
Figure 7. Reduced AKT phosphorylation in the p130Cas Y128F mutant cells.
(A) and (B) Parental and p130Cas Y128F mutant (KI) cells were serum-starved and stimulated with EGF for the indicated time. Cell lysates were blotted with the indicated antibodies. (C) The p130Cas Y128F mutant proteins show weaker binding affinity to p85. Cell lysates from either the parental or p130Cas Y128F mutant (KI) cells were immunoprecipitated with an anti-p130Cas antibody and blotted with the indicated antibodies.
AKT signaling is impaired in the p130Cas Y128F mutant KI cells
We demonstrated that phosphorylation of the p130Cas Y128 residue plays an important role in colorectal tumorigenesis. To gain insights into the effects of this phosphorylation on downstream signaling, we examined how the p130Cas Y128F KI affects phosphorylation of signaling molecules in CRC cells after EGF stimulation. It is well-documented that EGF receptors, once they are engaged by their ligands, activate multiple well-characterized signaling pathways including Ras-MAPK, PI3K-AKT, PLC-γ and STATs (32). We tested the phosphorylation status of 27 sites on 16 proteins that could be potentially modulated by EGF signaling (Table S2). In both RKO and DLD1 CRC cells, phosphorylation of AKT Thr308 was significantly reduced in p130Cas Y128F KI cells in comparison with the parental cells (Fig. 7A and B). However, the kinetics of AKT activation appeared to be faster in RKO cells than in DLD1 cells (Fig. 7A and B). Although no difference in ERK1/2 phosphorylation was observed between the RKO parental and Y128F KI cells, ERK1/2 phosphorylation levels were elevated in DLD1 p130Cas Y128F mutant cells, suggesting that a compensatory effect occurred in the DLD1 mutant cells.
The p130Cas Y128F mutant proteins bind less p85 than the WT p130Cas
The above results indicate that AKT is a critical downstream mediator of the p130Cas pY128 signaling. It is well documented that AKTs are activated by phosphatidylinositol 3-kinase (PI3K) (33). PI3K, which consists of a p85 regulatory subunit and a p110 catalytic subunit (34), converts phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-trisphosphate (PIP3) (33, 34). PIP3 then recruits AKT the plasma membrane, which, in turn, activates AKT (34). PI3K becomes activated when it is recruited to the membrane by interaction between p85 and pY residues on membrane-bound receptors or adaptor proteins (34). Interestingly, several studies demonstrate that membrane-associated p130Cas activates PI3K through interaction with p85 (35, 36) and that this interaction is dependent on tyrosine phosphorylation of the substrate-binding domain on p130Cas (35). Given that the Y128 belongs to one of the YXXP repeats (22), we postulated that the reduced AKT activity in the p130Cas Y128F mutant cells might be due to weaker binding affinity of the mutant p130Cas protein to p85. Indeed, as shown in Fig. 7C, less p85 proteins associated with the p130Cas Y128F mutant compared to the WT p130Cas. In contrast, the p130Cas Y128F mutation did not affect its binding with CRK, a known p130Cas interacting protein (Fig. 7C) (16, 17). Under these conditions, we failed to detect protein interactions between p130Cas and focal adhesion kinase (FAK).
Discussion
Using a phospho-proteomic approach, we identified and validated p130Cas as a substrate of PTPN14. PTPN14 dephosphorylates an unappreciated Y128 residue on p130Cas protein. PTPN14 regulated p130Cas Y128 phosphorylation plays an important role in colorectal tumorigenesis, because the genetically engineered p130Cas Y128F mutant CRC cells exhibit reduced abilities in colony formation, anchorage-independent growth, cell migration and xenograft tumor growth.
PTPN14 is identified to be mutated in various human cancers including breast, colon, head and neck, kidney, liver, ovarian and skin cancers (4–10), thereby providing genetic evidence suggesting that this PTP plays a role in tumorigenesis. In a fraction of tumors, one allele of PTPN14 is mutated whereas the other allele is lost (4), consistent with a “two-hit” tumor suppressor model. Therefore we postulate that PTPN14 may function as a tumor suppressor. Consistent with this notion, PTPN14 negatively regulates an oncogenic target pY128 p130Cas, because the unphosphorylatable p130Cas Y128F KI DLD1 and RKO cells exhibit reduced colony formation ability and soft-agar growth (Fig. 6). Furthermore, the RKO p130Cas Y128F mutant cells grow much slower as xenograft tumors, although the p130Cas Y128F mutation in DLD1 cell does not affect xenograft tumor growth. We hypothesize that this discrepancy is due to a compensatory effect in the DLD1 p130Cas mutant cells. In support, we observed that ERK activities are up-regulated in the DLD1 p130Cas mutant KI cells, but not in the RKO mutant KI cells (Fig. 7A and B).
We also demonstrated that SRC is the kinase that phosphorylates p130Cas Y128 residue (Fig. 3). Interestingly, CRC cell lines harboring high levels of pY128 p130Cas are more sensitive to SRC inhibitor Dasatinib-induced growth inhibition than those cell lines with low levels of pY128 p130Cas (Fig. 3D). Dasatinib is used in the clinic to treat chronic myelogenous leukemia, and there are several ongoing clinical trials using Dasatinib in combination with other drugs to treat solid tumors including colon cancers (27). Our data suggest that pY128 p130Cas may be exploited as a prediction marker for Dasatinib response in cancer patients. It is worth noting that Src also phosphorylates other sites of the YXXP repeats on p130Cas including the Y253 that regulate transformed cell growth and migration (37, 38). As the future directions, it is of interest to determine if PTPN14 dephosphorylates the other sites and if there is a cross-talk between the Y128 phosphorylation and the other Y residues of the YXXP repeats.
The oncogenic signaling of p130Cas Y128 phosphorylation seems to be mediated by the PI3K/AKT pathway, as we showed that AKT T308 phosphorylation, a key to AKT activation, is reduced in both DLD1 and RKO p130Cas Y128 mutant KI cells (Fig. 7A and B). It has been reported previously that p130cas can activate PI3K through binding to the regulatory subunit p85 (35, 36). Here, we demonstrate that p130Cas pY128 phosphorylation is crucial for its binding to p85. Although the p130Cas interacting protein CRK is shown to activate PI3K through the FAK-p85 interaction (39), our results indicate that the p130Cas Y128F mutant does not affect its binding to CRK.
Lastly, the phospho-proteomic approach used in this study represents a novel method to globally identify potential substrates of PTPs. Identification of substrates of PTPs has been a challenge. Although phosphatase trapping pull-down has been widely used to validate PTP substrates, it has limited success in identification of new substrates (40). We show here that, coupling with global phospho-proteomic profiling, overexpression of the trapping mutant PTPN14 PTP domain can enrich the target pY containing peptides and provides a systematic approach to identify candidate substrates. We also successfully employed a similar method to identify substrates of protein tyrosine phosphatase receptor T (PTPRT) (41). We further validated that STAT3 and paxillin are the substrates of PTPRT (41, 42). Therefore, our studies suggest that the phospho-proteomic approach is a generally applicable method for identification of PTP substrates.
Material and Methods
Cell Lines
DLD1, HCT116, HT29, RKO, SW480, LOVO and HEK 293T cells were obtained from the American Type Culture Collection. (Manassas. VA). All the CRC cells were maintained in McCoy’s 5A media plus 10% FBS. HEK 293T cells were maintained in DMEM media plus 10% FBS.
Establishment of Cell Line Stably Expressing PTPN14 Catalytic Domain
The catalytic domain of PTPN14 was PCR amplified using primers 5′-cgggatcccgggaagagaatcgagttga-3′ and 5′-ccgctcgaggagctggattggggtgatta-3′. The PCR products were cloned into the pcDNA4-Myc-HisB (Invitrogen) by BamHI and XhoI, which were incorporated into the primers. The PTPN14 D1079A mutant catalytic domain was made by site-directed mutagenesis with primers 5′-aatatactgactggccagctcacggctgtccagaaga-3′ and 5′-tcttctggacagccgtgagctggccagtcagtatatt-3′. Stable clones of SW480 cells were obtained according to the manufacturer’s instructions and were maintained in medium containing Zeocin (100 μg/ml) plus blasticidin (10 μg/ml). Protein expression was induced by addition of 1 μg/ml of doxycycline.
Profiling pY Peptides
Cell lysates were prepared under denaturing conditions in the presence of phosphatase inhibitors (20 mM HEPES, pH 8.0, 9 M urea, 1mM sodium vanadate). Following trypic digestion, peptides were concentrated and partitioned into three fractions by reversed-phase solid-phase extraction. The phosphopeptides from each fraction were bound to agarose beads conjugated with the phosphotyrosine-specific antibody pTyr-100. After thorough washing, peptides were eluted from the immobilized antibody with dilute acid and analyzed by nanoflow LC-MS/MS using an ion trap mass spectrometer. Lists of credible phosphopeptide sequence assignments were assembled.
Generation of Anti-p130Cas pY128 Antibody
Rabbits were immunized with a pY128 p130Cas peptide [KAQQGL (pY) QVPGP] conjugated to BSA. The anti-serum was first absorbed to a BSA-Sepharose 4B column and then passed twice through BSA-conjugated unphosphorylated peptide KAQQGLYQVPGP columns.
Stimulation of CRC Cells with Growth Factors
DLD-1 and RKO cells were serum-starved for 24 h and stimulated with EGF (200 ng/ml), PDGF-AA (10 ng/ml), FGF (10 ng/ml), VEGF (10 ng/ml) and IL-6 (10ng/ml) for various times.
Western Blot Analysis
Cells were lysed in lysis buffer (10mM Tris-HCl, pH 8.0; 100mM NaH2PO4; 8M Urea; 1mM Na3VO4; 20 mM NaF; 80uM beta-glycerophosphate; 20 mM Sodium pyrophosphate). Western blots were performed as previously described. Antibodies used include anti-FLAG (M2; Sigma, St. Louis, MO), anti-ERK1/2, anti-SRC, anti-pERK1/2, anti-AKT, anti-pAKT (Cell Signaling, Danvers, MA) and anti-p130Cas (BD Biosciences, San Jose, CA).
Phosphatase Substrate Trapping Assay
Substrate trapping was performed as described (41) and modified as follows. Ten million cells were treated with 100 μM pervanadate for 30 min and collected by centrifugation. The cell pellet was lysed with 1 ml of lysis buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 1x complete protease inhibitor cocktail, 1 mM EDTA, 1 mM Benzmidine), treated with 5 mM iodoacetic acid on ice for 5 min, neutralized by addition of 10 mM DTT for 15 min, and subjected to centrifugation at 16,000 × g for 30 min to remove debris. GST-PTPN14 bound beads were incubated with this lysate at 4°C for 1 h. The beads were pelleted and washed three times for 5 min with lysis buffer supplemented with 1 mM DTT. The beads were then boiled and aliquots analyzed by SDS-PAGE and Western blotting.
In Vitro Phosphatase Assay
HEK293T cells overexpressing FLAG-tagged p130Cas proteins were treated with 50 μM pervanadate for 30 min and lysed in RIPA buffer. Cas proteins were immunoprecipitated with anti-FLAG antibody-conjugated agarose beads. The immune complexes were washed twice in wash buffer (50 mM Tris·HCl pH 7.4, 150 mM NaCl, 5 mM EDTA,1% Nonidet P-40) with the phosphatase inhibitors (10 mM NaF and 2 mM Na3VO4), twice in the same buffer without the phosphatase inhibitors, once in ST buffer (50 mM Tris·HCl pH 7.4, 150 mM NaCl) and once in phosphatase assay buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM DTT). The p130 Cas immunocomplexes were then incubated with equal amounts of either GST alone, GST-PTPN14, or the trapping mutant at 37°C for 30 min. Western blots were performed to quantitate p130Cas pY128.
shRNA knockdown
shRNA lentiviral vectors were purchased from Sigma. Lentiviruses were packaged in HEK-293T cells with the transfection of lentiviral vectors together with pMD2.G VSVG and pCMV delta R8.74. 24 hrs after transfection, lentiviruses were harvested and purified and then were used to transduce DLD1 cells. Stable clones were then selected in medium containing 2.5 μg/ml of puromycin.
Targeted Knock-in of p130Cas Y128F mutant Allele
Somatic cell gene targeting was performed as described (26). Briefly, a 1.3 KB fragment from intron 2 to intron 3 of the p130Cas locus containing the exon 3 sequences was amplified 20 cycles from genomic DNA using primers 5′-gggaaag/deoxyU/gggtttgctgagggcgacggg -3′ and 5′-ggagaca/deoxyU/ggtgattgagggtggctgggccctt-3′. The coding sequences for Y128 were then mutated from TAC (Tyr) to TTC (Phe) by site-directed mutagenesis using primers 5′-ggctcagcaaggcctcttccaagtcccgggtcccag -3′ and 5′-aagaggccttgctgagcc -3′. This mutated fragment was used as the left homologous arm. Another 1.0 KB fragment from intron 3 of the p130Cas gene was also amplified 20 cycles from genomic DNA as the right arm using primers 5′-ggtccca/deoxyU/ggcattccaggagtggctgtg-3′ and 5′-ggcatag/deoxyU/gttcatctgtgtggggtgtggg-3′. The left and right homologous arms were cloned into pAAV-USER-Neo-LoxP vector using USER system as previously described. The targeting AAV viruses were packaged in 293T cells (a T75 flask at 70% confluence) by transfecting equal amounts of the targeting vector, pHelper and pRC plasmids (3 μg each). Viruses were harvested 72 hours post-transfection. DLD1 and RKO cells were infected with the p130Cas knock-in targeting viruses and selected with geneticin for 20 days. The geneticin resistant clones were then screened for homologous recombination by 35 cycles of genomic PCR with primers derived from the neomycin resistance gene 5′-gttgtgcccagtcatagccg-3′ and the upstream region of the left homologous arm 5′-gggccacatggagcacgtacc -3′. Confirmatory genomic PCR was also performed with positive clones identified using primers derived from the neomycin resistant gene (5′-tctggattcatcgactgtgg-3′) and the downstream region of the right homologous arm (5′-gcatgggtgttcgtatctgtgg-3′). The DNA fragments from the screening PCRs were then sequenced to ensure the presence of the mutant Y128F alleles. In order to target the second allele with the same targeting virus, correctly targeted clones were infected with adenoviruses expressing the Cre-recombinase to delete the drug selection marker. To select clones with successful deletion of the drug selection marker, 30 cycles of genomic PCR were performed to amplify a ~200 bp genomic fragment in which the Lox P site was inserted (using primers 5′-cacgcagctgggagggcacgaag-3′ and 5′-ctctgggtccattcacatccatc 3′). The heterozygous KI clones were infected with the same targeting virus to target the second allele and the neomycin resistance gene was excised as described above.
Colony formation assay
DLD1 and RKO cells were trypsinized, counted twice using a hemocytometer, and placed into 6-well plates at 200 cells per well. Cells were grown for 14 days before staining with Crystal Violet (Sigma, St. Louis, MO). The experiment was repeated three times with two replicates each. Average numbers of colonies from each experiment were plotted.
Focus Formation Assay in Soft agar
Soft agar assays were performed as described (43). Briefly, DLD1 and RKO clones were trypsinized, counted twice using a hemocytometer, and plated at 5000 cells/ml in top plugs consisting of 0.4% SeaPlaque agarose (FMC Bioproducts, Rockland, Maine) and McCoy’s 5A medium. After 30 days, the colonies were photographed and counted. The experiment was repeated three times with two replicates each. Average numbers of colonies from each experiment were plotted.
Xenografts
Five million cells were injected subcutaneously and bilaterally into 4- to 6- week old female nude mice (5 nude mice in each group). Tumor formation and size were assessed by weekly caliper measurements of the length and width of the tumors. Tumor volumes were calculated using the formula: Volume = (width)2 × length/2. After 35 days, the mice were sacrificed and tumors were harvested.
Focal Adhesion Assay
Focal adhesion assay was performed using the Acitn Cytoskeleton and Focal Adhesion Staining Kit (Millipore) according to the manufacturer’s instructions. Briefly, cells were seeded on coverslips treated with Fibronectin. The next day, cells were fixed with 4% paraformaldehyde in 1X PBS for 15 mins and washed once. Cells were then permeabilized with 0.1% Triton X-100 in 1XPBS for 5 mins and washed twice. Cells were incubated in anti-Vinculin antibody (1:200 dilution) for 1 hour and followed by three washes. Cells were then incubated in Alexa 488-conjugated secondary antibody (1:400) and TRITC-conjugated Phalloidin (1:250) for 1 hour and were washed three times. Coverslips were mounted on a slide and were visualized under Zeiss LSM 510 Confocal Microscope.
Boyden Chamber Cell Migration Assay
Transwell membranes (pore size 8.0 μm; Corning Incorporated., Corning, NY) were coated with 50 μg/ml fibronectin. Cells were detached with 2 mM EDTA and re-suspended in serum-free medium with 0.1% of BSA. Five hundred thousand cells were added to the upper compartment of the transwell chamber in the wells of a 24-well plate and allowed to migrate to the underside of the inserts for 24 hours. Non-migrating cells on the upper membrane were removed with a cotton swab, and cells that had migrated and become attached to the bottom surface of the membranewere fixed and stained with crystal violet. Migrated cells were counted microscopically. The experiments were repeated three times with two replicates for each cell line. Average numbers of migrated cells from each experiment were plotted.
Statistical Analysis
We applied the t test to compare the means between two groups assuming unequal variances. For xenograft-growth, we performed MANOVA analysis.
Supplementary Material
Acknowledgments
We thank Yueting Chen for excellent technical assistance, Drs. Susann Brady-Kalnay, Hua Luo and CK Qu for helpful discussions, and Anthony Scott for critical reading of the manuscript. This research was supported by grants from the National Institutes of Health Grant R01-CA127590, R01-HG004722 and a pilot grant from the Case Comprehensive Cancer Center.
Footnotes
Conflict of Interest:
The authors declare no conflict of interest.
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001 May 17;411(6835):355–65. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Brognard J, Hunter T. Protein kinase signaling networks in cancer. Curr Opin Genet Dev. 2011 Feb;21(1):4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julien SG, Dubé N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11(1):35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004 May 21;304(5674):1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 5.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 30;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006 Oct 13;314(5797):268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011 Aug 26;333(6046):1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011 Sep;43(9):828–9. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009 May;41(5):521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011 May;43(5):442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt L, Khew-Goodall Y. PTP-Pez: a novel regulator of TGFbeta signaling. Cell Cycle. 2008 Aug;7(15):2290–5. doi: 10.4161/cc.6443. [DOI] [PubMed] [Google Scholar]
- 12.Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol Biol Cell. 2003 Jun;14(6):2520–9. doi: 10.1091/mbc.E02-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogata M, Takada T, Mori Y, Oh-hora M, Uchida Y, Kosugi A, et al. Effects of overexpression of PTP36, a putative protein tyrosine phosphatase, on cell adhesion, cell growth, and cytoskeletons in HeLa cells. J Biol Chem. 1999 Apr 30;274(18):12905–9. doi: 10.1074/jbc.274.18.12905. [DOI] [PubMed] [Google Scholar]
- 14.Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. Translocation of protein tyrosine phosphatase Pez/PTPD2/PTP36 to the nucleus is associated with induction of cell proliferation. J Cell Sci. 2000 Sep;113( Pt 17):3117–23. doi: 10.1242/jcs.113.17.3117. [DOI] [PubMed] [Google Scholar]
- 15.Wyatt L, Wadham C, Crocker LA, Lardelli M, Khew-Goodall Y. The protein tyrosine phosphatase Pez regulates TGFbeta, epithelial-mesenchymal transition, and organ development. J Cell Biol. 2007 Sep 24;178(7):1223–35. doi: 10.1083/jcb.200705035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds AB, Kanner SB, Wang HC, Parsons JT. Stable association of activated pp60src with two tyrosine-phosphorylated cellular proteins. Mol Cell Biol. 1989 Sep;9(9):3951–8. doi: 10.1128/mcb.9.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. Binding of transforming protein, P47gag-crk, to a broad range of phosphotyrosine-containing proteins. Science. 1990 Jun 22;248(4962):1537–9. doi: 10.1126/science.1694307. [DOI] [PubMed] [Google Scholar]
- 18.Dorssers LC, van Agthoven T, Dekker A, van Agthoven TL, Kok EM. Induction of antiestrogen resistance in human breast cancer cells by random insertional mutagenesis using defective retroviruses: identification of bcar-1, a common integration site. Mol Endocrinol. 1993 Jul;7(7):870–8. doi: 10.1210/mend.7.7.8413311. [DOI] [PubMed] [Google Scholar]
- 19.Cabodi S, Tinnirello A, Di Stefano P, Bisaro B, Ambrosino E, Castellano I, et al. p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res. 2006 May 1;66(9):4672–80. doi: 10.1158/0008-5472.CAN-05-2909. [DOI] [PubMed] [Google Scholar]
- 20.Burnham MR, Harte MT, Richardson A, Parsons JT, Bouton AH. The identification of p130cas-binding proteins and their role in cellular transformation. Oncogene. 1996 Jun 6;12(11):2467–72. [PubMed] [Google Scholar]
- 21.Honda H, Oda H, Nakamoto T, Honda Z, Sakai R, Suzuki T, et al. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas. Nat Genet. 1998 Aug;19(4):361–5. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- 22.Defilippi P, Di Stefano P, Cabodi S. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol. 2006 May;16(5):257–63. doi: 10.1016/j.tcb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997 Mar 4;94(5):1680–5. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchetot C, Chagnon M, Dube N, Halle M, Tremblay ML. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005 Jan;35(1):44–53. doi: 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005 Jan;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Guo C, Chen Y, Shulha HP, Schnetz MP, LaFramboise T, et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods. 2008 Feb;5(2):163–5. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montero JC, Seoane S, Ocana A, Pandiella A. Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors. Clin Cancer Res. 2011 Sep 1;17(17):5546–52. doi: 10.1158/1078-0432.CCR-10-2616. [DOI] [PubMed] [Google Scholar]
- 28.Yarom N, Jonker DJ. The role of the epidermal growth factor receptor in the mechanism and treatment of colorectal cancer. Discov Med. 2011 Feb;11(57):95–105. [PubMed] [Google Scholar]
- 29.Hirata R, Chamberlain J, Dong R, Russell DW. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat Biotechnol. 2002 Jul;20(7):735–8. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 30.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32(1):e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011 Feb;12(2):104–17. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 33.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005 Dec;5(12):921–9. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 34.Bohnacker T, Marone R, Collmann E, Calvez R, Hirsch E, Wymann MP. PI3K{gamma} Adaptor Subunits Define Coupling to Degranulation and Cell Motility by Distinct PtdIns(3,4,5)P3 Pools in Mast Cells. Sci Signal. 2009 Jun 9;2(74):ra27. doi: 10.1126/scisignal.2000259. [DOI] [PubMed] [Google Scholar]
- 35.Riggins RB, DeBerry RM, Toosarvandani MD, Bouton AH. Src-dependent association of Cas and p85 phosphatidylinositol 3′-kinase in v-crk-transformed cells. Mol Cancer Res. 2003 Apr;1(6):428–37. [PubMed] [Google Scholar]
- 36.Li E, Stupack DG, Brown SL, Klemke R, Schlaepfer DD, Nemerow GR. Association of p130CAS with phosphatidylinositol-3-OH kinase mediates adenovirus cell entry. J Biol Chem. 2000 May 12;275(19):14729–35. doi: 10.1074/jbc.275.19.14729. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg GS, Alexander DB, Pellicena P, Zhang ZY, Tsuda H, Miller WT. Src phosphorylates Cas on tyrosine 253 to promote migration of transformed cells. J Biol Chem. 2003 Nov 21;278(47):46533–40. doi: 10.1074/jbc.M307526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patwardhan P, Shen Y, Goldberg GS, Miller WT. Individual Cas phosphorylation sites are dispensable for processive phosphorylation by Src and anchorage-independent cell growth. J Biol Chem. 2006 Jul 28;281(30):20689–97. doi: 10.1074/jbc.M602311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akagi T, Murata K, Shishido T, Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway by utilizing focal adhesion kinase and H-Ras. Mol Cell Biol. 2002 Oct;22(20):7015–23. doi: 10.1128/MCB.22.20.7015-7023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang F, Kumar S, Zhang ZY. Proteomic approaches to studying protein tyrosine phosphatases. Mol Biosyst. 2007 May;3(5):308–16. doi: 10.1039/b700704n. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Guo A, Yu J, Possemato A, Chen Y, Zheng W, et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. PNAS. 2007 Mar 6;104(10):4060–4. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, Zhang X, Guda K, Lawrence E, Sun Q, Watanabe T, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci U S A. 2010 Feb 9;107(6):2592–7. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang P, Zhao Y, Zhu X, Sedwick D, Zhang X, Wang Z. Cross-talk between Phospho-STAT3 and PLC{gamma}1 Plays a Critical Role in Colorectal Tumorigenesis. Mol Cancer Res. 2011 Oct;9(10):1418–28. doi: 10.1158/1541-7786.MCR-11-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.