Abstract
The role of conventional graphene-oxide in biosensing has been limited to that of a quenching substrate or signal transducer due to size inconsistencies and poor supramolecular response. We overcame these issues by using nanoscale GOs (nGO) as artificial receptors. Unlike conventional GO, nGOs are sheets with near uniform lateral dimension of 20 nm. Due to its nanoscale architecture, its supramolecular response was enhanced, with demonstrated improvements in biomacromolecular affinities. This rendered their surface capable of detecting unknown proteins with cognizance not seen with conventional GOs. Different proteins at 100 and 10 nM concentrations revealed consistent patterns that are quantitatively differentiable by linear discriminant analysis. Identification of 48 unknowns in both concentrations demonstrated a >95% success rate. The 10 nM detection represents a 10-fold improvement over analogous arrays. This demonstrates for the first time that the supramolecular chemistry of GO is highly size dependent and opens the possibility of improvement upon existing GO hybrid materials.
INTRODUCTION
Graphene oxide (GO), a chemically exfoliated graphene derivative, has enjoyed a resurgence since its initial discovery.1–3 This is due, in part, to GO’s improved solution processability compared to graphene by incorporation of oxygenated groups in its molecular structure. These chemical groups, including hydroxyl and epoxide groups in the basal plane and carboxyl groups on its exposed edges,4 also enable additional supramolecular interactions that can be exploited for various functional applications including biosensing5 and drug delivery.6 Despite various advances in GO synthesis, however, investigation toward the effect of GO size on supramolecular response has remained challenging due to the predominantly micrometer-sized, yet polydisperse (submicrometer to tens of micrometers) nature of chemically exfoliated GO.7 This, in turn, led to their usage in many applications. While usages of these “conventional GOs” have been popular, it has contributed toward inconsistent sensing responses and obscured important size-based phenomenon.
These issues are particularly evident in bioassays, where GO was commonly used for its unprecedented fluorescence quenching capability.8,9 Most commonly, GO was used in lock-and-key sensing to quench the fluorescence of “key” molecules, such as DNA aptamers,10 antibodies,11 peptides,12 or target-specific fluorophores,13 etc. (see reviews by Loh et al.14 and Liu et al.15 for background). While this approach has served the design of the sensors, it also reduced GO’s role to that of substrate, emulating current strategies, such as enzymelinked immunosorbent assay (ELISA).16,17 It therefore limits the detection to the predictable/expected binding events predetermined by the “key” and sheds little light on the behavior of biomacromolecules toward the GO surface. Furthermore, despite the usage of specific recognition elements, reproducibility issues still persisted due the noise generated by the polydispersity and lower supramolecular responses inherent to micrometer-sized conventional GOs. For the purpose of recognizing biomolecular targets, this size and polydispersity presents a practical challenge, as molecular absorption on GO exhibits significant edge to basal plane preferences.18–20 The molecular affinities of GO should thus differ according to GO size, as the edge length to basal plane ratio scales inversely to its radius (circumference α 1/r). A GO nanocolloid (nGO),21 defined as a nanoscale GO flake with nanometer-sized diameter, should therefore present increased edge-binding effects. Principally, in a nGO, the increased edge to basalplane ratio will present an increased density of carboxylate groups, which are chemical functionalities that exist only on the GO edges.4 The nGO therefore possesses higher charge density,21 which may engender a more consistent and pronounced supramolecular binding and release profile. Conversely, the predominantly micrometer-sized conventional GO lack these attributes. Their supramolecular responses are therefore liable to be attenuated versus the nGOs. By summarizing the differences between nGO and conventional GO, we therefore expect a tighter grouping in nGO’s supramolecular response profile (due to nGO’s size consistency) and an elevated signal level (due to a more pronounced supramolecular binding and release response).
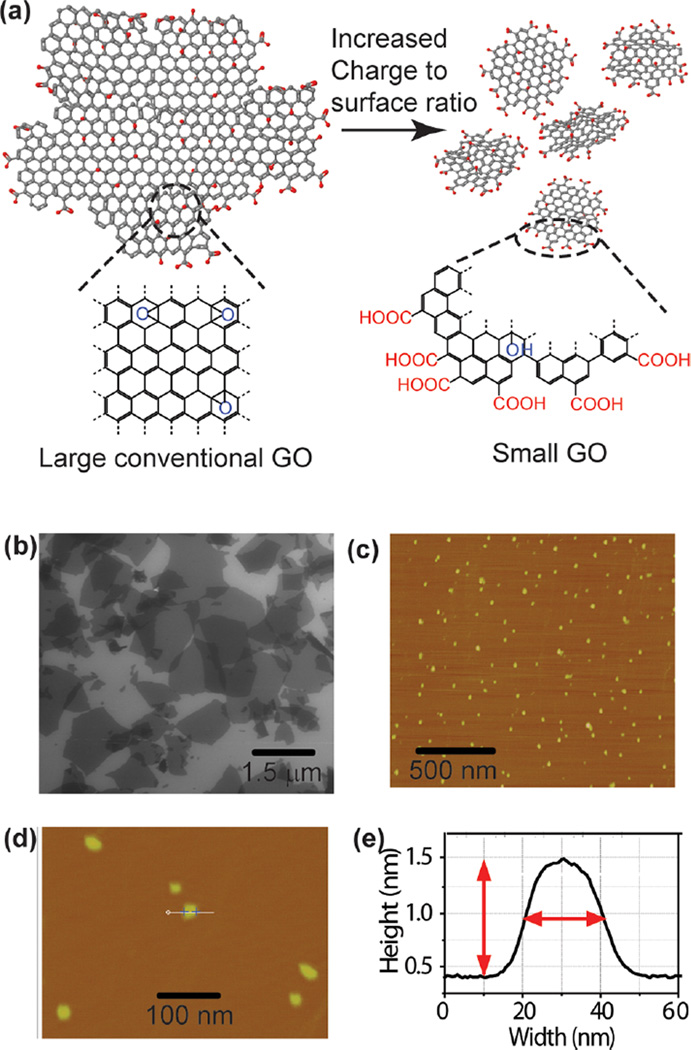
Although postsynthetic size separation approaches have been used to size select and/or decrease the polydispersity of conventional GO,22,23 these approaches have required surface modification agents that can contaminate and alter the colloidal processability of GO sheets and their binding affinities. Conversely, nGOs obtained through the exfoliation of graphite nanofibers produce nanoflakes with very uniform diameters and elevated ζ-potential due to the distinct chemical groups present on GO edges (–COOH, Figure 1a).21 They are also much less likely to aggregate in physiological conditions. For use in “chemical nose/tongue” sensing that requires an array of highaffinity receptors with cross-reactivity toward many analytes, these nGOs are potentially ideal as they maintain the chemical diversity present on the basal plane of conventional GOs (typically micrometer sized, Figure 1b), while enhancing the magnitude of attractive or repulsive electrostatic forces to proteins due to increased edge carboxylate functionalities. This can therefore build upon our previous investigations toward protein–GO bindings, which demonstrated the GO–protein bindings to conserve the enzyme’s secondary structure and is reversible.24 Additionally, as nGO differs with conventional GO in size by orders of magnitude, a broad investigation of supramolecular responses that contrast the two can elucidate size-based chemistry that has not been previously explored.
Figure 1.
Conventional GO and nGO. (a) A structural model of conventional GO and nGO showing chemical functionalities suitable usage as artificial protein receptor. Additionally, the increased carboxylate functionalities on nGO edges are also highlighted. (b) SEM micrograph of conventional GO sheets showing size distribution from submicrometer to several micrometers. (c, d) AFM micrograph of nGO showing nearly uniform 20 nm diameters. (e) Representative AFM profile of a nGO flake from (d), showing width of 20 nm and height of 1.1 nm.
An additional favorable attribute of nGO is the highly flexible planar scaffold that can mimic highly responsive natural protein–protein interactions, rendering nGO nondenaturing. Coupling this flexibility with the fact that overall attractive or repulsive interactions between nGO and analyte proteins will be enhanced due the increased charge density of nGO (edge to surface area ratio scales proportionally to 1/r), we expect nGO (Figure 1c–e) to be advantageous as compared to conventional GO synthesized from graphite powders. In the past, these GO size-/shape-dependent investigations have leaned heavily on electronic properties25–28 and gas absorptions,18–20 therefore this investigation will enable a better understanding of whether size effects extend toward complex supramolecular chemistry as well.
In the past, we have reported usage of GO as artificial enzyme receptors and have found the binding process to be protein compatible and able to conserve the structure and function of protein over extended periods (24 h).24 Here, by comparing nGO and conventional GO’s performance as artificial receptors, we thus examine changes in the GO’s capability to recognize different proteins surfaces through binding. In this way, the analyte protein responses from nGO and conventional GO were systematically classified using linear discriminant analysis (LDA) (Figure 2a,b). With this, we demonstrated that nGOs response profiles are very distinct as compared to their conventional counterparts. Particularly, nGO appears to be a selective high-affinity protein receptor with better signal consistency than a conventional GO. It is therefore an ideal artificial receptor for nose/tongue sensors.29,30 A small nGO nose array could therefore in theory discriminate and identify wide varieties of targets and would not be limited by the number of available specific receptors (“keys”). Compared to analogous state-of-the-art gold nanoparticle protein sensor arrays, nGO’s appear to be able to replace a wide variety or state-of-the-art gold-nanoparticle surface/ligands in the nose sensing paradigm (six surfaces, 100 nM sensitivity), all the while obtaining a 10-fold improvement in sensitivity (10 nM). Because this exact methodology has been translated to complex biofluids environments with only slight attenuation in sensitivity,31–37 it may therefore enable usage of GO in wider context.
Figure 2.
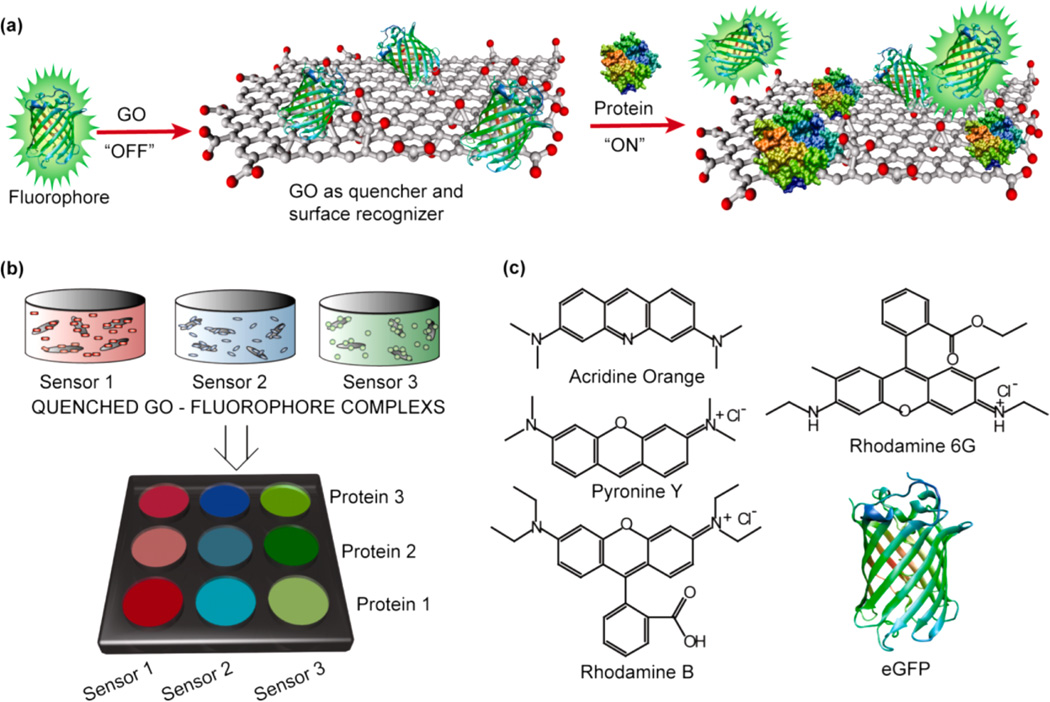
Design and preparation of nGO-based sensor array. (a) Investigation of GO–protein interactions was conducted using the fluorescence displacement transduction. In this mechanism, fluorescent reporters were initially quenched (‘off’) though GO binding. Displacement of quenched fluorophore by analyte proteins restores the fluorescence. Statistical analysis of fluorophore displacement using LDA allows insight into changes in GO–proteins interactions as a function of “nanosizing” the GO flakes (nGO, 20 nm diameter). (b) In all, LDA patterns from a three sensor array show nGO engendered enhanced fluorescent restoration as compared to conventional GO (despite comparable fluorophore binding constants). Using the LDA profiles of various analyte proteins enabled identifications of “blind samples”. In all, nGO was able to reproduce binding/fluorescent results due to enhanced protein interactions. Conventional GO generally failed due to lower fluorescent response and polydispersity. (c) Structure of the fluorescent reporters used in the sensor array.
In the process of examining the size-dependent supra-molecular differences between nGO and conventional GO using the nose-sensing platform, consistency and reproducibility of responses were examined by way of blind sample testing. In this series of experiments, analytes were blinded and randomized by a third party, and the resultant fluorescent responses were compared to “training matrix” responses obtained a priori using the known specimens. In this manner, we report that at both 100 and 10 nM protein concentrations, nGO’s demonstrated near perfect reproducibility (24/24 at 100 nM and 22/24 at 10 nM), while conventionally synthesized GO’s were considerably subpar (17/24 at 100 nM and general inability to distinguish proteins at 10 nM). In addition to improved protein affinities due to its size and enhanced edge effects, this also demonstrates nGO’s superiority with regard to reproducible supramolecular response.
RESULTS
Sensor Array Design
The nose arrays were constructed by complexing fluorescent reporter molecules with nGOs and conventional GOs separately. In each respective array, the GO was used as both a recognition element and a fluorescence quencher.9 A fluorescence signal is triggered when reporter ligands are displaced from the GO surface by analyte proteins through binding competition38 (Figure 2a). Rare instances of fluorescent decrease can occur if fluorophores aggregate on analyte proteins and self-quench. In total, five different fluorophores were used in the initial array (acridine orange, rhodamine B, pyronine Y (PY), rhodamine 6G (R6G), Histagged emerald green fluorescent protein (eGFP)), with three generating most effective responses (PY, rhodamine 6G, and eGFP) (Figure 2c). Here, PY is cationic, R6G is zwitterionic, and they were bound to GOs through a combination of aromatic π–π stacking and electrostatic interactions. eGFP was His-tagged to create a hydrophobic domain for binding with graphitic patches on the GO surface. Supramolecular sensing responses were recorded with eight different proteins with noticeable pairs in molecular weight (MW) and isoelectric point (pI) for added challenge (Table S1). For example, hemoglobin (Hem) and bovine serum albimin (BSA) share a MW of ~65 kDa, and histone (His) and lysozyme (Lys) both share a pI of 11. LDA binding profiles were generated using both conventional GO and nGO arrays at 100 and 10 nM protein concentrations, and identifications of 24 unknowns were performed at each concentration (48 total).
Materials Characterization
Physical characterization of the nGOs and conventional GOs was conducted prior to usage (Figure 1b,c). The conventional GOs were obtained through a modified Hummer’s method by exfoliation of graphite powder.39 Uniform nGOs of ~20 nm in diameter were synthesized using a graphite nanofibers precursor.21 Both conventional GOs and nGOs were dispersed in 5 mM phosphate buffer at physiological pH 7.4. For nGOs, dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern, London, U.K.) revealed a ζ potential of −54.4 ± 5.6 mV. Complementary characterizations of nGOs using atomic force microscopy (AFM) and transmission electron microscopy (TEM) revealed diameters of 19.2 ± 1.7 (fwhm) and 23.6 ± 3.6 nm, respectively (Figures 1d,e and S2b,c). For conventional GO, scanning electron microscopy (SEM) revealed high polydispersity with a distribution of 2.01 ± 2.20 µm (Figures 1b and S4).
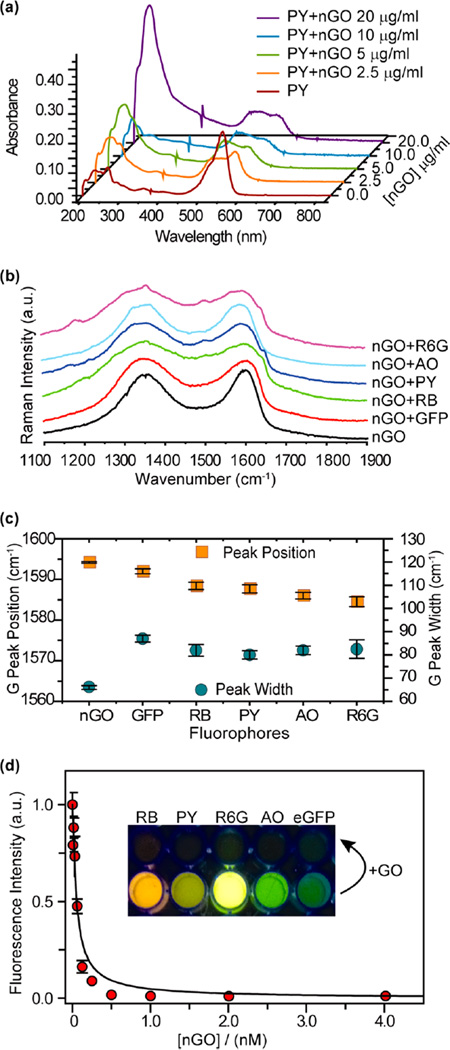
With the physical properties of the GOs understood, we next characterized the GO:fluorescent reporter complexes. Demonstration of GO:reporter molecule complexation was established through UV–vis absorbance measurements and Raman spectroscopy. We began with an absorbance titration assay using nGOs and fluorophores. Because absorption effects are instantaneous according to the Franck–Condon principle, absorbance spectroscopy can be used to characterize changes to fluorophores upon complexation.40 We observed that with addition of nGOs, the absorbance peak (λmax) of the small fluorophore molecules changed through development of secondary peaks, providing evidence of complexation. For the case of PY (Figure 3a), the characteristic peak at 546 nm was significantly broadened (fwhm from 46 to 116 nm), with a new peak emerging at 484 nm. The broadening was likely due to the cumulative absorbance of complexed and free molecules. Similar changes were observed with R6G, AO, and RB but not eGFP (Figure S5). The lack of change in eGFP is due to the morphology of eGFP. As the fluorescent residues of eGFP (Ser65-dehydroTyr66-Gly67)41 are encased inside the β-barrel, they are decoupled from direct electronic interactions with the nGOs. Although direct interactions were not observed between eGFP and nGOs, quenching could still occur through fluorescence resonant energy transfer.8,9 Similar absorbance titrations were also conducted using the conventional GOs. For all cases, results similar to those of nGOs were obtained. From the conventional GO:PY titration, for example, λmax shifted from 546 to 484 nm with corresponding broadening (Figure S6).
Figure 3.
Spectroscopic characterization of nGO complexes. (a) UV–vis spectra of PY before and after complexation with nGO. (b) Raman D and G bands from various nGO complexes used in this report. (c) Summary of G band peak shift (frequency) and broadening (width) due to nGO complexation with various fluorophores. (d) Fluorescence titration of eGFP with nGO. Inset shows the fluorophore solutions before and after the addition of nGO.
Raman spectroscopy was used to further characterize GO complexes. This was done to examine changes to GO’s electronic structure as a consequence to complexation with the electron donor–acceptor molecules.42 For GO, the two characteristic bands observed in Raman spectroscopy were the G (~1590 cm−1) and D (~1350 cm−1) bands. With electronic doping, the G peak position and bandwidth shift, whereas D band remains almost unchanged.43 In our experiment, nGO, saturated nGO:reporter complexes, and their conventional GO counterparts were deposited on to Si/SiO2 chips. Raman spectra were acquired using a Nanophoton Raman 11 microscope with excitation using a 532 nm laser (Figure 3b). The G band peak position and bandwidth were measured by peak fitting using a Lorentzian function. For GO nanocolloid and conventional GO complexes, the peak position was blue-shifted, and the bandwidth increased, providing further evidence of complexation (Figure 3c). Most significantly the change in G band peak position is minimal for eGFP, indicating little electronic communication between nGOs and eGFP molecules, a similar effect as observed in our UV study. Again, similar results were observed for convetional GO complexes (Figure S7). Taken together, these UV–vis and Raman results bring strong proof of binding between the fluorophores and GOs in solution.
Having characterized GO:reporter complexation, fluorescent titrations were conducted to quantify fluorophore loading (Figures 3d, S8, and S9). The Stern–Volmer binding/quenching constants (KS–V) were obtained using nonlinear regression (Table 1). For both GO systems, it can be seen that eGFP was the most tightly bound, with higher loadings observed in conventional GO. Because eGFP binds GO through hydrophobic interactions between its His-tagged domain and the graphitic patches on GO, it is logical that larger conventional GOs would bind in greater quantities, as KS–V indicated (Table 1). Similarly, as the fluorophores reporters are composed mostly with aromatic rings, their binding mechanism is predominantly due to aromatic/π–π stacking.44–46 With this in mind, the zwitterionic fluorophores (R6G, RB) observed lower loadings on nGO likely due to increased anionic–anionic repulsion as a consequence of nGO’s increased charge density, thus screening the π–π stacking in close proximity. As PY and AO do not contain negative charges, binding was similar between nGO and conventional GO.
Table 1.
Summary of Stern–Volmer (KS–V) Binding Constants from Fluorescent Titrations and Nonlinear Regressiona
| KS–V (109 M−1) | KS–V (µg−1) | |||
|---|---|---|---|---|
| reporter | nGO | conventional GO | nGO | conventional GO |
| eGFP | 20 ± 2.2 | N/A | 2.9 ± 0.32 | 13 ± 1.2 |
| pyronin Y | 4.3 ± 0.47 | N/A | 0.63 ± 0.070 | 0.71 ± 0.053 |
| rhodamine 6G | 5.7 ± 0.45 | N/A | 0.84 ± 0.067 | 1.42 ± 0.19 |
| acridine orange | 9.4 ± 0.47 | N/A | 1.4 ± 0.070 | 1.1 ± 0.080 |
| rhodamine B | 0.37 ± 0.046 | N/A | 0.055 ± 0.0067 | 0.12 ± 0.013 |
Due to the polydispersity of conventional GO, its molecular weight was not estimated. KS–V in molar concentration units was thusly calculated only for nGO complexes. Mass-based KS–V was calculated for both nGO and conventional GO.
With the GO:reporter affinities determined, proper titration points for the sensor array were selected. This value influences the dynamic range and response probability in each sensor. We first considered the small molecule reporters that exhibited lower loadings than eGFP. To increase the probability of reporter displacement in the event of analyte protein binding, we selected a titration point with higher reporter loadings, with the rationale that higher reporter coverage on the GO will provide less space to accommodate both reporter and analyte protein, thereby elevating the probability of displacement. After initial experiments, 50% quenching points on the titration curves were used (~2× reporter loading per GO unit versus 100% quenching point). At this titration point, fluorescence response with analyte addition was overwhelmingly positive, indicating increased displacement response. There were also rare instances of negative fluorescence response due to free reporters aggregating on analyte proteins and self-quench (vide infra).47 In contrast to small molecule fluorophores, displacement of eGFP at 100% quenching point consistently produced strong fluorescence regeneration; thus suggesting sufficient loading (as evidenced by elevated KS–V). The fully bound titration point was hence used for eGFP. Identical titration points were used for both nGO and conventional GO arrays.
Protein Recognition: nGO versus Conventional GO, a Size-Dependent Effect
Once the loading points were selected, we compare nGO and conventional GO arrays’ ability to differentiate analyte proteins. Eight proteins of varying size and charge were tested, with several protein pairs having similar molecular weights and/or pI values (Table S1). We conducted the initial sensing tests at 100 nM analyte protein concentration, which is detection limit of the gold nanoparticle systems.47 Protein identification and classification were first performed using the nGO and conventional GO arrays using all five reporters [R6G, PY, eGFP, acridine orange (AO), and rhodamine B (RB)]. The responses were compiled into a training matrix, and distinct combinations of fluorescent responses were observed for each analyte protein (Figure 4a,c, Table S2). In all, five replicates were obtained for each analyte protein in each sensor, producing 200 data points (5 sensors × 8 proteins × 5 replicates) for each array. Comparing fluorescent signal intensity in Figure 4a,c, it can be observed that nGOs appear to have higher reporter displacement rates compared to conventional GOs. Considering the magnitude of reporter displacement as a consequence of binding competition between the reporter ligands and protein analytes for the GO surface, it can be surmised that proteins appear to possess higher affinity for nGO surfaces than conventional GOs. While the classification and “spread” of individual data points within each classification class can be dependent on GO polydispersity issues, the increased fluorescent signal observed in the nGO receptors is likely attributable to a GO size-dependent effect. There is, therefore, a significant size-induced difference in GO–protein binding behavior. These enhanced responses are possibly a consequence of the increased charge and defect density on nGO surfaces,21 which enabled a protein mimetic surface with improved supramolecular binding mechanisms (electrostatic, hydrophobic, hydrogen bonding). It also suggests that edges may play critical roles in the protein binding response of GO systems.
Figure 4.
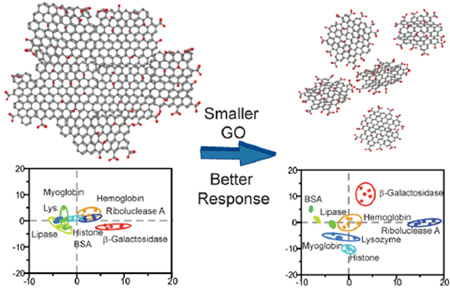
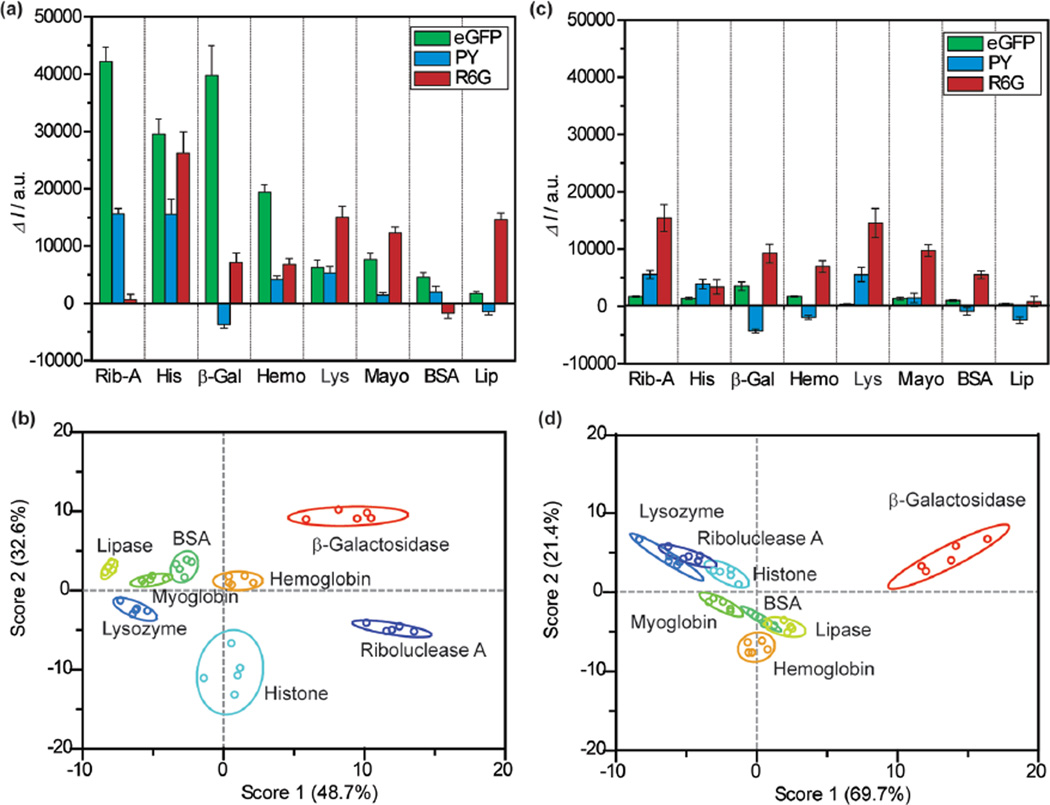
Sensing of proteins at 100 nM concentration. (a) Fluorescence response (ΔI) of the nGO array in presence of 100 nM analyte proteins (Rib-A, His, β-Gal, Hem, Lys, Mayo, BSA, and Lip). Bar height represents average of five replicates. (b) Canonical score plot for the nGO array as calculated by LDA using data from (a). The canonical plot showed that all proteins are accurately classified (100%). (c) Fluorescent response from the conventional GO array at 100 nM analyte protein concentrations. (d) Canonical score plot for the conventional GO array showing compressed responses as compared to nGO.
To delineate analyte responses systematically we used LDA,48,49 a statistical technique for data classification, dimensionality reduction, and machine learning. Accuracy was measured using the Jacknife classification, a method that removes classification bias.48 At 100 nM using the initial nGO array, we observed an LDA/Jacknife accuracy of 100% that is considerably higher than the performance of individual sensors. Complete 100% classification could also be achieved using only three sensors in the array (eGFP, R6G, and PY). Consequently, a training matrix considering only these three elements was constructed. This finalized training matrix with 120 data points from 40 test cases (3 sensors × 8 proteins × 5 replicates) produced canonical factors of 48.7, 32.6, and 18.7% in LDA (Figure 4b). The magnitude of the canonical factors is of particular interest, as a significant contribution from each sensor dimension indicates multiple axes of differentiation.
Using LDA, 100 nM sensing results from the conventional GO array were also analyzed. Due to its aforementioned lower fluorescent response (Figure 4c), it yielded appreciatively worse LDA separation (Figure 4d). Most notably, the 95% confidence ellipse produced by the large GO array contained visible overlaps between a numbers of proteins (Figure 4d). In the case of 100 nM protein detection, although the Jacknife analysis indicated 100% separation using five sensors (AO, RB, eGFP, R6G, and PY) and 98% separation with three sensors (eGFP, R6G, and PY), the individual canonical plots were significantly compressed and overlapping (69.7, 21.4, and 8.9%), thus giving it less significance for identification of unknown specimens.
To better understand the LDA classifications, their canonical factors were correlated with raw fluorescence numbers (Figure S10a). For the nGO array, each canonical factor appeared predominantly driven by individual sensor response. Specifically, factor 1 was driven in large part by the eGFP response (0.952 Pearson coefficient), factor 2 by PY (−0.839), and factor 3 by R6G (0.941). The fact that each sensor dimension was dictated by a canonical factor was particularly noteworthy, as this indicates sufficient affinity and reporter displacement in each sensor system, thus validating the high affinity between nGO and proteins. For the conventional GO sensors, we observed a more subdued response, with less distinction between individual sensors due to the lower fluorescent signals from decreased reporter displacements. In particular, although factor 1 here showed strong correlation to PY (0.9768), factor 2 showed correlations to both R6G (0.8079) and eGFP (0.7132), leaving little leeway for factor 3, which showed weak separation (canonical factor 8.9%) and weak correlation to sensor responses (Figure S10b). To better understand the basis for the nGOs sensor responses, we also correlated fluorescent results with analyte protein MW and pIs. In particular, we observed that nGO:PY response appears to correlate well with protein charges (pI), suggesting that this response may be driven essentially by electrostatics (Figure S10 c). The results thus demonstrated the improved ability of nGOs to differentiate analyte proteins with accuracy and repetition as compared to conventional GO.
To examine the reproducibility of responses in the nGOs and conventional GOs arrays, we utilized the aforementioned LDA results as training sets (“training matrix”) for identification of unknown specimens. The efficacies of the nGO and conventional GO arrays were contrasted by a series of 24 unknown samples (consisting of the aforementioned eight protein species) each prepared, randomized, and blinded by separate researchers. Identifications were made by Mahalanobis distance-square proximities to known group centers from the training matrix. For the case of nGOs, the unknown specimens were identified with 100% accuracy (24/24 samples, Table S3). The conventional GO array performed less effectively, with only 71% of unknowns accurately identified (17/24 samples, Table S4). Presumably, this is a consequence of nGO’s higher protein affinity, enhanced response, and conventional GO’s polydispersity (sample to sample variations). The inability of conventional GOs to identify the unknowns also suggests that its training matrix classifications accuracy will decrease with increased sampling.
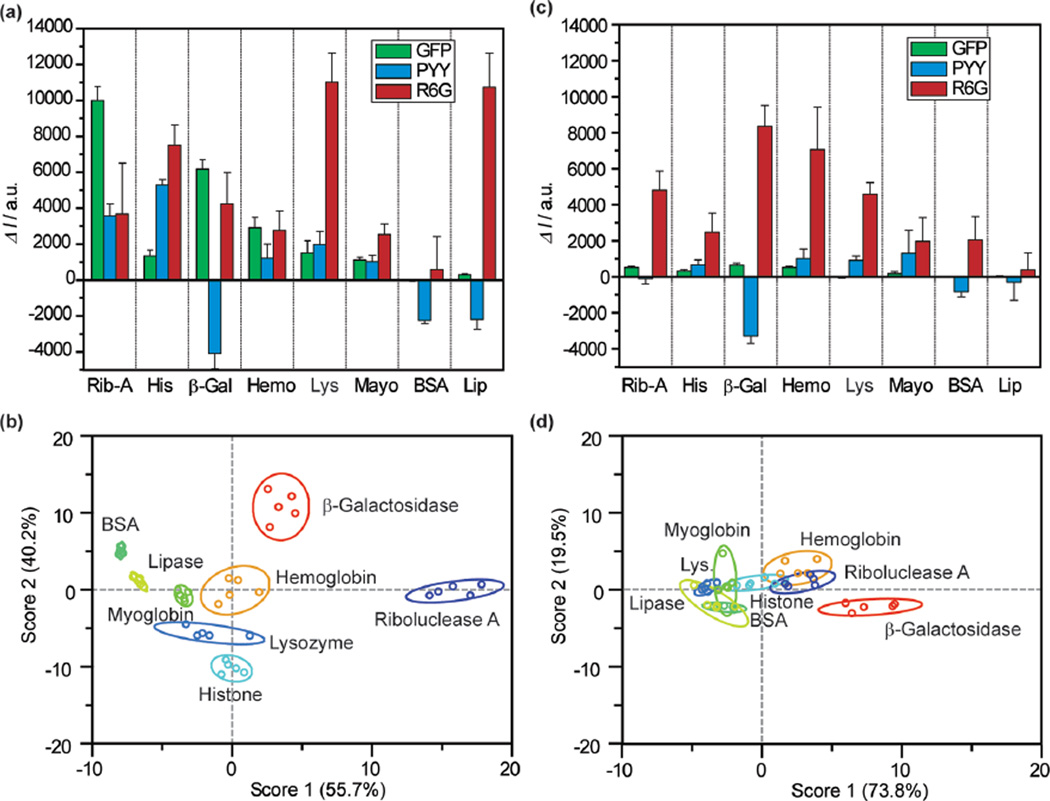
After compiling the GO responses at 100 nM protein concentration, both arrays were tested again at a 10 nM concentration using the same methodology (Figure 5a, Table S5). For the nGO array, three canonical factors were generated (55.7, 40.2, and 4.1%). Similar to the 100 nM measurements, eGFP was responsible for factor 1, PY with factor 2, and R6G for factor 3 (Figure S11). The canonical factor distributions, however, were changed. At 10 nM, the nGO array appeared to rely more heavily on eGFP and PY for protein classification, as the factor 3 contribution was reduced from 18.7% at 100 nM to 4.1%. As a consequence, although the first two factors produced highly separated LDA responses (as indicated by the lack of overlap in Figure 5b), the attenuated third factor created near proximities between hemoglobin and myoglobin and between lysozyme with histone. Despite this reduced contribution from factor 3, the array performed effectively at 10 nM analyte protein concentration, producing 95% Jacknife classification accuracy within the training matrix (38/40 examples). Testing of unknowns at 10 nM produced successful results, with 92% accurately identified (22/24 samples, Table S6). The monodispersity of nGO likely assisted in reproducing identical responses despite reduced separations in factor 3. Comparably, the training matrix produced by the conventional GO was only capable of 75% classification accuracy, with all canonical plots collapsed toward the origin due to minimal fluorescence response (Figure 5c,d). Due to the low classification accuracy, the conventional GO training matrix was therefore unsuitable for unknown identification.
Figure 5.
Sensing of proteins at 10 nM concentration. (a) Fluorescence response (ΔI) patterns of the nGO array in presence of 10 nM different analyte proteins. (b) Canonical score plot for the nGO array as calculated by LDA, with 95% confidence ellipses. The canonical plot shows that 95% of all protein cases were accurately classified. (c) Fluorescence response (ΔI) patterns of the conventional GO array at 10 nM concentration. (d) Corresponding canonical score plot for the conventional GO array, demonstrating incompetent classification.
DISCUSSION
The ability to control and manipulate supramolecular affinities has long been a recognized challenge in the field of GO biosensing. Much of this issue can be attributed to apathy toward GO polydispersity and to a lack of understanding toward the effect of GO size on its supramolecular chemistry. Previously, the utilization of “lock and key” mechanisms have improved responses by shifting burden of identification from GO to the “key” molecules, thereby bypassing these effects, but consistency in response magnitudes is still challenging due to differing “key” loadings on conventional polydisperse GO. Here, we have demonstrated many of these challenges can be solved by usage of nGO synthesized through exfoliation of carbon nanofibers with uniform diameters.21 Furthermore, we have shown that the fluorescence responses and binding patterns of nGO demonstrate markedly improved molecular cognizance as compared to their conventional larger spatial scale counterparts. The improvement in surface cognizance and sensor accuracy is particularly obvious as demonstrated by the usage of nGO as artificial protein receptors in chemical nose sensor arrays. In contrast to the conventional GO array that exhibits low fluorescent signals and marginal classification results at 100 nM protein concentration, the nGO array demonstrated enhanced response and was successful at classifying and identifying analyte proteins at 10 nM quantities. This represents a 10-fold improvement over current nonamplified sensors.47 This superior detection limit arises from the increased fluorescent signal generated in the nGO array. While polydispersity issues contribute to the reproducibility challenges in the conventional GO array, the improved fluorescent response observed in the nGO array is likely a GO size-dependent phenomenon that cannot be explained on the grounds of GO polydispersity. This enhanced fluorescence response is likely an indication of higher protein affinity, which is derived due to nGO’s elevated charge density and enhanced electrostatic response. The nGOs in this report clearly exhibit a wide range of affinities for different biological molecules and are behaviorally distinct from their conventional counter parts. It therefore demonstrated that GO supramolecular bindings can be greatly affected by their size.
The broader prospects of this report underscore a disconnect between the use of polydispersed GO (with diverse and larger spatial size distribution) and expectations of consistent assay results. As the size of conventional GOs can vary from submicrometer to tens of micrometers within the same batch, we show that within each batch, there are likely significant variations in supramolecular affinities, raising the possibility that the results previously obtained with GO and lock-and-key sensing may have been driven by tail-distribution phenomenon (e.g., extremely small or large GO) and are inherently difficult to control. This report, therefore, suggests the possibility to optimize polydisperse GO assays through careful size tunings and highlights a fundamental aspect of GO supramolecular chemistry that has not been previously examined. Currently we have exploited these size effects by demonstrating protein recognition using nGO in buffers. This methodology ensures a clean comparison between the nGO (tens of nanometers) and conventional GO (submicrometer to micrometers).
Previously, gradual evolutions of this approach through surface modifications have enabled sensing in complex biofluids.31–37 However, the improved sensitivities of “nose” sensing through differential receptors are ultimately limited by the number of differential receptors that one desires to pack in an array. The use of a facile array, as demonstrated here, may therefore be more attractive, as it demonstrates preliminary screening capabilities that may complement existing higher-cost ELISA assays. The greater relevance here, however, lies in the prospect of exploiting size-enabled supramolecular properties of GO for future material development. For example, as size scaling can emphasize certain supramolecular properties of GO, size selection can implicitly tune mechanical properties of GO gels, such as elasticity and viscosity, for usages in materials processing. It is also possible that the size scaling of GO can influence the wetting and electrical properties of their films, as smaller GOs possess greater charge density. This paper therefore suggests that there is a largely ignored aspect of GO chemistry that bears great promise for future applications of this two-dimensional material.
METHODS
Materials
nGO was prepared from 20 nm carbon fibers (Catalytic Materials LLC) using KMnO4 oxidation.21 Conventional GO was prepared from graphite powder (Bay carbon, SP-1) using a modified Hummer’s method.39 Fluorescent reporter molecules (acridine orange, rhodamine B, pyronine Y, rhodamine 6G) were purchased from Sigma-Aldrich. His-tagged emerald green fluorescent proteins were expressed according to reported procedure.50 BSA, β-galactosidase (β-Gal, from E. coli), hemoglobin (Hem, from equine heart), histone (His), lipase (Lip), lysozyme (Lys), myoglobin (Mayo) and ribonuclease A (Rib-A) were purchased from Sigma-Aldrich and used as received. 5 mM phosphate buffer (PB, pH 7.4) was used throughout.
Fluorescence Titration
Fluorescent titrations were conducted in 96-well plates by titrating increasing GO concentrations against constant fluorophore concentrations at 200 µL fluid volumes. Appropriate fluorophore concentrations were first determined by evaluating fluorescence responses in absence of the GO quencher. In all cases, concentrations were adjusted to achieve fluorescence ~>25 000 RFU. With fluorophore concentrations fixed, serially diluted GOs were added to adjacent wells, while maintaining overall fluid volumes at 200 µL/well. The mixtures were allowed to equilibrate for 30 min at room temperature before measurement. All experiments were performed in triplicates. Fluorescence measurements were made using a Biotek Synergy 4 plate reader. The fluorescent titration curves were fitted in GraphPAD Prism 5 software using the Stern–Volmer quenching equation. The Stern–Volmer quenching constant therefore encompasses the quantity of fluorophores bound by individual GOs, which can be useful for describing the overall binding (SI).
Analyte Protein Response
Nose arrays were prepared according to procedures described in the main text. Fluorescent titration curves were shown in Figures S8 and S9. Reporter–GO complexes were prepared by incubating fluorescent reporter and GOs at room temperature for 30 min. In a 96-well plate, 190 µL of the resultant complex was pipetted into each well, and 10 µL of analyte proteins was then added to produce a total volume of 200 µL/well, with final concentrations of 100 or 10 nM. For control wells, 10 µL PB was added instead of proteins. Analyte proteins and GO complexes were equilibrated for another 30 min before a final reading. In all cases, fluorescence changes reported were in reference to the control samples. Fluorescent responses at 100 nM analyte protein concentration were recorded for all five GO–reporter complex (GO + eGFP, PY, R6G, AO, or RB). Data were first analyzed with all five complexes and then with three complexes only (eGFP, PY, R6G), per discussion in main text. For the 10 nM analyte protein sensing, only eGFP, PY, and R6G responses were recorded.
Unknown Identification
Unknown samples were prepared, randomized, and identified by three separate researchers. Fluorescence responses of each sample were recorded in triplicate and then averaged using the same procedures as above. Identifications were made by evaluating Mahalanobis distance-square proximities to known group centers from the training matrix.
Supplementary Material
ACKNOWLEDGMENTS
Absorbance measurements were performed at Northwestern University Keck Biophysics facility. Fluorescence measurements were performed at Northwestern University High Throughput Analysis facility. TEM and AFM measurements were performed at the Northwestern NUANCE facility. Dr. D. Latte, Dr. M. Mustata, and Mr. Y. K. Huang for assistance with Raman, AFM, and SEM characterization. S.C. is thankful for a DHS graduate fellowship. This work was funded by the National Cancer Institute Center for Cancer Nanotechnology Excellence (CCNE) initiative at Northwestern University award no. U54CA119341. J.H. acknowledges support from National Science Foundation (DMR CAREER 0955612) and the Alfred P. Sloan Research Foundation. V.M.R. acknowledges support from GM077173.
Footnotes
ASSOCIATED CONTENT
S Supporting Information
Synthesis of nano and conventional GO, UV–vis, Raman and fluorescence titration, training matrices, identification studies, and control experiments of sensing using GO:fluorophore reporters. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Brodie BC. Philos. Trans. R. Soc. London. 1859;149:249. [Google Scholar]
- 2.Hummers WS, Offeman RE. J. Am. Chem. Soc. 1958;80:1339. [Google Scholar]
- 3.Li D, Kaner RB. Science. 2008;320:1170. doi: 10.1126/science.1158180. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Cote LJ, Huang J. Acc. Chem. Res. 2012;45:1356. doi: 10.1021/ar300047s. [DOI] [PubMed] [Google Scholar]
- 5.Wright AT, Anslyn EV. Chem. Soc. Rev. 2006;35:14. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Robinson JT, Sun X, Dai H. J. Am. Chem. Soc. 2008;130:10876. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S, Ruoff RS. Nat. Nano. 2009;4:217. doi: 10.1038/nnano.2009.58. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Cote LJ, Kim F, Huang J. J. Am. Chem. Soc. 2009;132:260. doi: 10.1021/ja906730d. [DOI] [PubMed] [Google Scholar]
- 9.Swathi RS, Sebastian KL. J. Chem. Phys. 2008;129:054703. doi: 10.1063/1.2956498. [DOI] [PubMed] [Google Scholar]
- 10.Lu C-H, Yang H-H, Zhu C-L, Chen X, Chen G-N. Angew. Chem. Int. Ed. 2009;48:4785. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 11.Jung JH, Cheon DS, Liu F, Lee KB, Seo TS. Angew. Chem. Int. Ed. 2010;49:5708. doi: 10.1002/anie.201001428. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M, Yin B-C, Wang X-F, Ye B-C. Chem. Commun. (Cambridge, U. K.) 2011;47:2399. doi: 10.1039/c0cc04887a. [DOI] [PubMed] [Google Scholar]
- 13.Balapanuru J, Yang J-X, Xiao S, Bao Q, Jahan M, Polavarapu L, Wei J, Xu Q-H, Loh KP. Angew. Chem. Int. Ed. 2010;49:6549. doi: 10.1002/anie.201001004. [DOI] [PubMed] [Google Scholar]
- 14.Loh KP, Bao Q, Eda G, Chhowalla M. Nat. Chem. 2010;2:1015. doi: 10.1038/nchem.907. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Dong X, Chen P. Chem. Soc. Rev. 2012;41:2283. doi: 10.1039/c1cs15270j. [DOI] [PubMed] [Google Scholar]
- 16.Van Weemen BK, Schuurs AHWM. FEBS Lett. 1971;15:232. doi: 10.1016/0014-5793(71)80319-8. [DOI] [PubMed] [Google Scholar]
- 17.Engvall E, Perlmann P. Immunochemistry. 1971;8:871. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 18.Ohba T, Kanoh H. J. Phys. Chem. Lett. 2012:511. doi: 10.1021/jz2016704. [DOI] [PubMed] [Google Scholar]
- 19.Radovic LR. J. Am. Chem. Soc. 2009;131:17166. doi: 10.1021/ja904731q. [DOI] [PubMed] [Google Scholar]
- 20.Vladimir Kh D. Carbon. 2002;40:659. [Google Scholar]
- 21.Luo J, Cote LJ, Tung VC, Tan ATL, Goins PE, Wu J, Huang J. J. Am. Chem. Soc. 2010;132:17667. doi: 10.1021/ja1078943. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Liu Z, Welsher K, Robinson J, Goodwin A, Zaric S, Dai H. Nano Res. 2008;1:203. doi: 10.1007/s12274-008-8021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green AA, Hersam MC. J. Phys. Chem. Lett. 2009;1:544. doi: 10.1021/jz900235f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De M, Chou SS, Dravid VP. J. Am. Chem. Soc. 2011;133:17524. doi: 10.1021/ja208427j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai J, Ruffieux P, Jaafar R, Bieri M, Braun T, Blankenburg S, Muoth M, Seitsonen AP, Saleh M, Feng X, Mullen K, Fasel R. Nature. 2010;466:470. doi: 10.1038/nature09211. [DOI] [PubMed] [Google Scholar]
- 26.Jiao L, Zhang L, Wang X, Diankov G, Dai H. Nature. 2009;458:877. doi: 10.1038/nature07919. [DOI] [PubMed] [Google Scholar]
- 27.Kosynkin DV, Higginbotham AL, Sinitskii A, Lomeda JR, Dimiev A, Price BK, Tour JM. Nature. 2009;458:872. doi: 10.1038/nature07872. [DOI] [PubMed] [Google Scholar]
- 28.Son Y-W, Cohen ML, Louie SG. Nature. 2006;444:347. doi: 10.1038/nature05180. [DOI] [PubMed] [Google Scholar]
- 29.John SK. Trends Neurosci. 1991;14:79. doi: 10.1016/0166-2236(91)90025-p. [DOI] [PubMed] [Google Scholar]
- 30.DeVries SH, Baylor DA. Cell. 1993;72:139. doi: 10.1016/s0092-8674(05)80033-9. [DOI] [PubMed] [Google Scholar]
- 31.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10912. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat. Chem. 2009;1:461. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2010;132:5285. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranda OR, Creran B, Rotello VM. Curr. Opin. Chem. Biol. 2010;14:728. doi: 10.1016/j.cbpa.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda OR, Li XN, Garcia-Gonzalez L, Zhu ZJ, Yan B, Bunz UHF, Rotello VM. J. Am. Chem. Soc. 2011;133:9650. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunz UHF, Rotello VM. Angew. Chem. Int. Ed. 2010;49:3268. doi: 10.1002/anie.200906928. [DOI] [PubMed] [Google Scholar]
- 37.Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Haick H. Nat Nano. 2009;4:669. doi: 10.1038/nnano.2009.235. [DOI] [PubMed] [Google Scholar]
- 38.Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782. [Google Scholar]
- 39.Kim F, Luo J, Cruz-Silva R, Cote LJ, Sohn K, Huang J. Adv. Funct. Mater. 2010;20:2867. [Google Scholar]
- 40.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York: Springer; 2006. [Google Scholar]
- 41.Tsien RY. Annu. Rev. Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 42.Dong XC, Fu DL, Fang WJ, Shi YM, Chen P, Li LJ. Small. 2009;5:1422. doi: 10.1002/smll.200801711. [DOI] [PubMed] [Google Scholar]
- 43.Das A, Pisana S, Chakraborty B, Piscanec S, Saha SK, Waghmare UV, Novoselov KS, Krishnamurthy HR, Geim AK, Ferrari AC, Sood AK. Nat. Nano. 2008;3:210. doi: 10.1038/nnano.2008.67. [DOI] [PubMed] [Google Scholar]
- 44.Wang QH, Hersam MC. Nat. Chem. 2009;1:206. doi: 10.1038/nchem.212. [DOI] [PubMed] [Google Scholar]
- 45.Khokhar FS, van Gastel R, Poelsema B. Phys. Rev. B. 2010;82:205409. [Google Scholar]
- 46.Chen RJ, Zhang Y, Wang D, Dai H. J. Am. Chem. Soc. 2001;123:3838. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- 47.You C-C, Miranda OR, Gider B, Ghosh PS, Kim I-B, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat Nano. 2007;2:318. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 48.Manly BFJ. Multivariate Statistical Methods: A Primer. Boca Raton, FL: Chapman & Hall/CRC Press; 2005. [Google Scholar]
- 49.McLachlan GJ. Discriminant Analysis And Statistical Pattern Recognition. Hoboken, NJ: John Wiley & Sons; 2004. [Google Scholar]
- 50.De M, Rana S, Rotello VM. Macromol. Biosci. 2009;9:174. doi: 10.1002/mabi.200800289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.