Abstract
DNA lesions can block replication forks and lead to the formation of single-stranded gaps. These replication complications are mitigated by DNA damage tolerance mechanisms, which prevent deleterious outcomes such as cell death, genomic instability, and carcinogenesis. The two main tolerance strategies are translesion DNA synthesis (TLS), in which low-fidelity DNA polymerases bypass the blocking lesion, and homology-dependent repair (HDR; postreplication repair), which is based on the homologous sister chromatid. Here we describe a unique high-resolution method for the simultaneous analysis of TLS and HDR across defined DNA lesions in mammalian genomes. The method is based on insertion of plasmids carrying defined site-specific DNA lesions into mammalian chromosomes, using phage integrase-mediated integration. Using this method we show that mammalian cells use HDR to tolerate DNA damage in their genome. Moreover, analysis of the tolerance of the UV light-induced 6–4 photoproduct, the tobacco smoke-induced benzo[a]pyrene-guanine adduct, and an artificial trimethylene insert shows that each of these three lesions is tolerated by both TLS and HDR. We also determined the specificity of nucleotide insertion opposite these lesions during TLS in human genomes. This unique method will be useful in elucidating the mechanism of DNA damage tolerance in mammalian chromosomes and their connection to pathological processes such as carcinogenesis.
Keywords: error-prone DNA repair, homologous recombination repair, recombinational repair
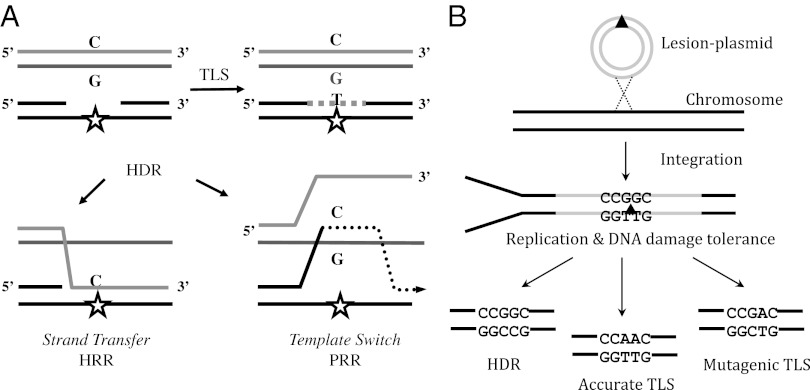
DNA damage is abundant, caused by both external agents such as sunlight and tobacco smoke and intracellular byproducts of metabolism, amounting to about 50,000 lesions per day per cell (1). Despite the presence of effective DNA repair mechanisms that eliminate lesions and restore the original DNA sequence, DNA replication often encounters unrepaired lesions that have escaped repair. These DNA damages may cause arrest of replication forks and the generation of postreplication gaps (2, 3). To complete replication and prevent the formation of double-strand breaks, which are highly deleterious, cells use DNA damage tolerance (DDT) mechanisms. These include translesion DNA synthesis (TLS) and homology-dependent repair (HDR), which enable bypass of the lesions and completion of replication, without removing the lesions from DNA. HDR uses the sequence from the intact sister chromatid to patch the single-stranded template region carrying the lesion. [We term HDR the pathways of DNA damage tolerance that rely on the homologous sister chromatid, also termed postreplication repair (PRR), damage avoidance, template switch, copy choice recombination, and homologous recombination repair.] This is carried out either by physical transfer of the segment complementary to the damaged template [also termed homologous recombination repair (HRR)] or by copying the complementary strand from the sister chromatid (template switch or postreplication repair). TLS employs specialized low-fidelity DNA polymerases to replicate across modified template bases, and it is inherently mutagenic due to the miscoding properties of most DNA lesions (4–6). Both HDR mechanisms are inherently accurate (1) (Fig. 1A). This may explain why it is the major tolerance mechanism in Escherichia coli (7–11). The balance between TLS and HDR in Saccharomyces cerevisiae is less clear, with a possible dependence on assay conditions (12–16). Despite its mutagenic nature TLS has an important role in protecting mammalian cells from DNA damage. This is evident from the skin cancer predisposition and sunlight sensitivity of patients with xeroderma pigmentosum variant who carry germ-line mutations in polη, a major TLS DNA polymerase (17, 18), and the embryonic death of mice in which polζ, another major TLS polymerase, was knocked out (19). As for HDR, it is not yet known whether it functions in mammalian chromosomes to tolerate DNA damage during replication.
Fig. 1.
Principle of the DNA damage tolerance assay for chromosomes of mammalian cells. (A) Schematic presentation of the two cellular strategies for DNA damage tolerance, TLS and HDR. (B) Principle of the unique TLS/HDR method, based on phage integrase-mediated integration into mammalian chromosomes of a plasmid carrying a site-specific lesion. See text for details.
A major obstacle in the study of DNA damage tolerance is the lack of high-resolution methods for simultaneously assaying TLS and HDR during chromosomal replication. Traditionally, chromosomal induced mutagenesis in a reporter gene (e.g., hypoxanthine-guanine phosphoribosyltransferase) (1) is used as a measure of mutagenic TLS; however, this method is blind to accurate TLS, which is the main TLS pathway for some common types of DNA damage, and HDR is much more difficult to measure (20). Plasmid model systems were instrumental in the study of DNA damage tolerance via TLS (e.g., refs. 21–25) and HDR (12, 26); however, they do not obviate the need for studying TLS and HDR at the chromosomal level. To that end we developed a unique method, in which both TLS and HDR can be simultaneously analyzed at a single-nucleotide resolution in chromosomes of mammalian cells in culture. We show that both TLS and HDR share the burden of DNA damage tolerance for each of three different DNA lesions, albeit at different proportions. We also present the spectrum of nucleotides inserted across these lesions during chromosomal TLS.
Results
Unique Assay for High-Resolution Simultaneous Measurement of TLS and HDR in Mammalian Chromosomes.
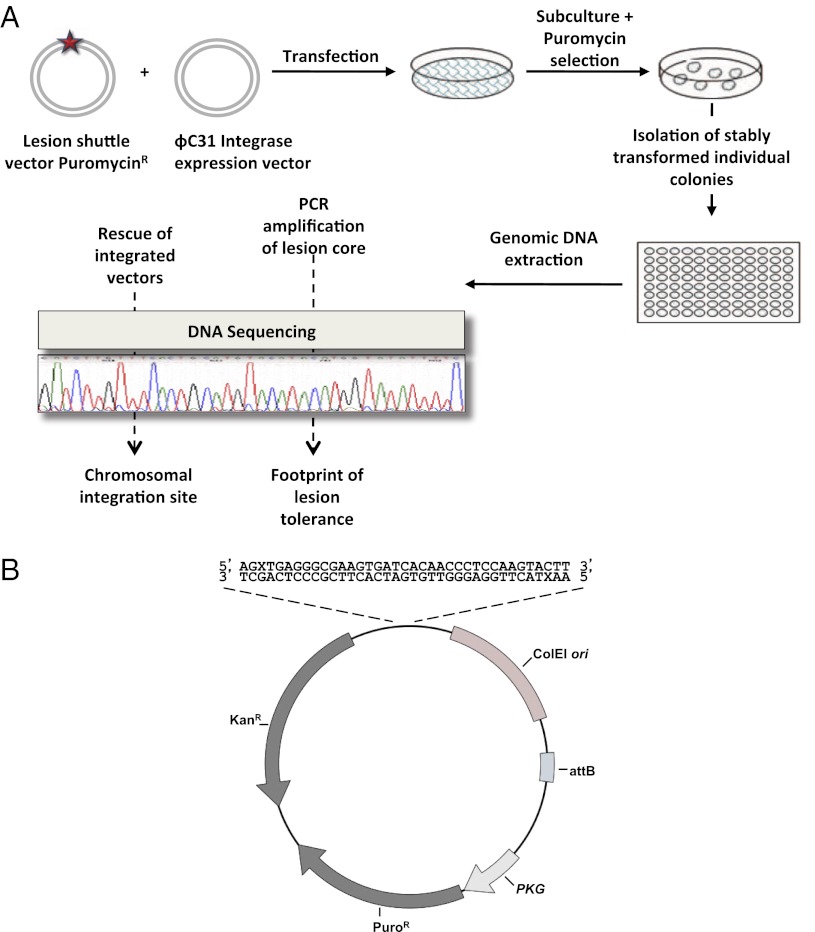
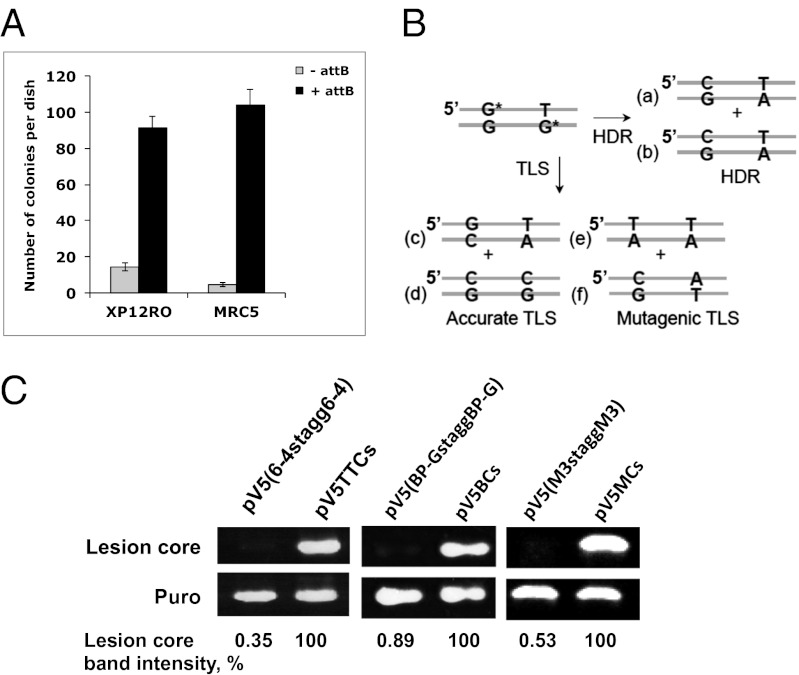
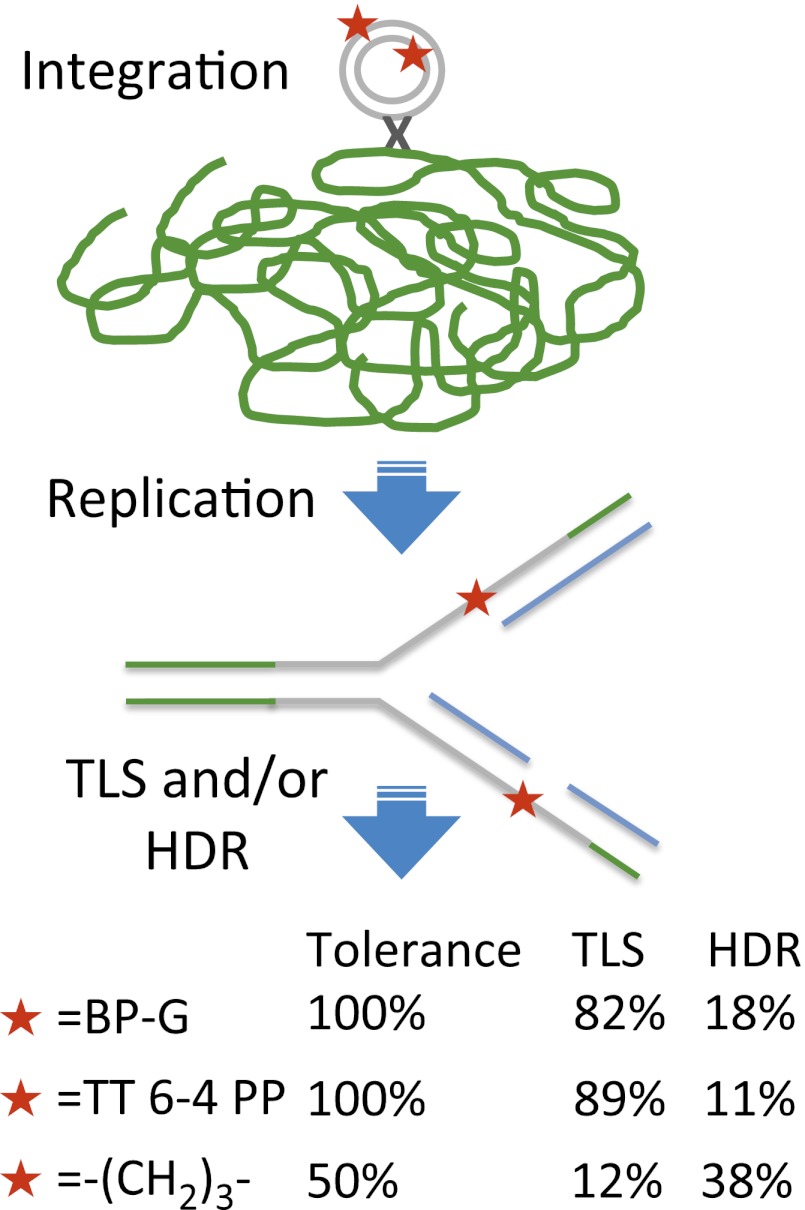
This unique assay is based on a shuttle vector carrying a site-specific lesion, which is stably integrated into chromosomes of mammalian cells using an ectopically expressed phage recombinase (Fig. 1B). Cells are cotransfected with a synthetic DNA vector that contains a selectable marker (puromycinR), a recognition site for the integrase of phage ϕC31, and a defined site-specific lesion, and a second plasmid that expresses ϕC31 integrase (27). Under our assay conditions, replication occurs while the lesion is in the chromosomal locus into which the plasmid had integrated. Therefore, completion of replication and subsequent cell divisions entail that the cell coped with the lesion through the use of DNA damage tolerance mechanisms. To select cells into which the plasmid was stably integrated and then tolerated, puromycin selection is applied. Subsequently, chromosomal DNA is extracted from puromycin-resistant colonies and subjected to DNA sequence analysis in the vicinity of the lesion (Fig. 2A). The colonies arose primarily via integrase-mediated insertions, as indicated by the substantially lower number of colonies obtained in control experiments with plasmids lacking the attB integration sequence, which is the target of the integrase (Fig. 3A and Table 1).
Fig. 2.
Outline of the DNA damage tolerance assay in chromosomes of mammalian cells. (A) Cultured cells were cotransfected with the lesion shuttle vector and the ϕC31 expression plasmid. In parallel, cells were transfected with a similarly constructed control plasmid without a lesion. Forty-eight hours after transfection the cells were subcultured, and after 24 additional hours they were subjected to puromycin selection. Approximately 14 d later resistant colonies were counted, picked, and individually transferred into a 96-well plate for further growth. When they reached suitable confluence, the cells in each individual well were harvested, and their chromosomal DNA was extracted. This DNA was used as a template in PCR reactions aimed to amplify the lesion core area. Amplified samples were then subjected to DNA sequence analysis. (B) Structure of the lesion shuttle vector pLSV5L. PuroR, puromycin resistance gene under the phosphoglycerate kinase (PGK) promoter; attB, phage ϕC31 integrase attachment site; kanR, kanamycin resistance gene; X stands for a lesion. Two lesions in the staggered configuration are shown.
Fig. 3.
(A) Effect of ϕC31-integrase activity on the efficiency of plasmid integration into the chromosomes of mammalian cells. Human XP12RO and MRC5 cells were cotransfected with a mixture of the integrase expression vector pKGϕC31-int and pLSV5 (black bar) or pLSV5nA (gray bar) that lacks the integrase recognition sequence attB. Following transfection the cells were grown under puromycin selection for 12 d, until visible colonies were formed. Culture plates were then fixed and counted. The results shown are an average of three different transfections. (B) Products expected from TLS and HDR of two staggered BP-G lesions during chromosomal DNA damage tolerance. (C) Persistence of DNA lesions in XPA cells. Human XP12RO cells were transfected with the lesion shuttle vectors pLSV5TT6-4stagg6-4, pLSV5BP-GstaggBP-G, pLSV5M3staggM3, or their corresponding control vectors (without lesions). The vectors were extracted from the cells 3 d after transfection and used as templates in PCR reactions aimed to amplify the region containing the lesions (lesion core) and in parallel PCR reactions aimed to amplify a fragment of the puromycin resistance gene (puro) as an internal reference. Quantification of the images of the ethidium bromide-stained gel was done using ImageJ software. The percentage of the lesion-free background in each lesion shuttle vector is shown relative to the corresponding control vector with no lesion (100%), normalized to the intensity of the “puro” bands for each plasmid.
Table 1.
Integration sites of the lesion shuttle vector in human chromosomes
| Chromosome | Location | ϕC31-int mediated?* | Known hotspot?† | No. independent events | Genomic context | Gene | |
| 1 | 1 | q41 | √ | No | 1 | Intron | USH2A |
| 2 | 2 | p11.1 | √ | No | 1 | intergenic | |
| 3 | 2 | q23.2 | √ | Yes | 1 | intergenic | |
| 4 | 3 | p23 | √ | No | 1 | Intergenic | |
| 5 | 4 | p13 | x | No | 1 | Intergenic | |
| 6 | 4 | q35.1 | √ | No | 2 | Intergenic | |
| 7 | 5 | q14.3 | √ | No | 1 | Intron | MGC33214 |
| 8 | 6 | p21.1 | √ | No | 1 | intergenic | |
| 9 | 7 | p14.1 | √ | Yes | 1 | Intergenic | |
| 10 | 8 | p22 | √ | No | 3 | Intron | DLC1 |
| 11 | 9 | p13.2 | √ | No | 1 | Intron | PAX5 |
| 12 | 9 | p21.2 | √ | No | 1 | Intergenic | |
| 13 | 10 | q22.1 | √ | Yes | 1 | Intergenic | |
| 14 | 12 | q22 | x | No | 1 | Intergenic | |
| 15 | 13 | q13.3 | √ | No | 1 | Intron | DCLK1 |
| 16 | 19 | q13.31 | √ | Yes | 11 | Intron | ZNF223 |
| 17 | 20 | p13 | √ | No | 1 | Intron | CDC25B |
| 18 | X | q22.3 | x | No | 1 | Intergenic | |
| Total sites: | 31 | ||||||
Human XPA cells were stably transfected with a lesion shuttle vector containing two BP-G lesions in a staggered configuration, after which the integrated plasmids were rescued along with the chromosomal junction sequences. The chromosomal DNA sequences at the junction sites were determined by PCR followed by DNA sequence analysis. The detailed protocol is described in Materials and Methods, and the sequences in the insertion junctions are in Table S8. The genes in which integration was observed were: USH2A, usher syndrome 2A; MGC33214, transmembrane protein 161B; DLC1, Rho GTPase-activating protein 7; PAX5, paired box protein Pax-5; DCLK1, doublecortin-like kinase 1; ZNF223, zinc finger protein 223; CDC25B, cell division cycle 25 homolog B.
Integrase-mediated insertion events were identified on the basis of the DNA sequence at the insertion site (which are shown in Table S8). √, ϕC31 integrase-mediated insertion; x, random insertion.
Previously identified ϕC31 integrase-mediated integration hotspots are described in ref. 27.
The vectors used contained a defined site-specific lesion on both strands, 31 nt apart (Fig. 2B). In this configuration progression of replication requires that tolerance occurred on both strands, either by TLS or by HDR. To discriminate between the two tolerance mechanisms we engineered opposite the lesions mismatched nucleotides, which are infrequently incorporated during TLS. For example, on the basis of plasmid assays, the DNA adduct benzo[a]pyrene-guanine (BP-G), which is formed by tobacco smoke, is bypassed relatively accurately in human cells (∼90% insertion of the correct C), with the main mutagenic event being misinsertion of an A, which together account for 94% of all TLS events (28). Thus, when constructing lesion shuttle vectors with BP-G lesions (Fig. 3B), we engineered opposite the lesions T and/or G, which are infrequently inserted by TLS (2% and 4%, respectively) (28). In this configuration, the sequence obtained after tolerating the lesions and cell propagation for multiple generations is indicative of the tolerance mechanism and enables discrimination between accurate TLS, mutagenic TLS, and HDR (Fig. 3B). For example, accurate TLS of the BP-G in the upper strand (Fig. 3B) will place a C opposite the BP-G, leading after propagation to a G:C base pair (Fig. 3B, product c). Similarly, accurate bypass of the BP-G on the lower strand (Fig. 3B) will place C opposite the BP-G, leading to the formation of product d. Mutagenic TLS on the other hand is expected to place primarily A opposite BP-G, which after propagation will yield a transversion mutation with a T:A base pair being generated instead of G:C—product e in Fig. 3B for BP-G in the upper strand and product f for BP-G in the lower strand. HDR of the BP-G in the upper strand will place the G from the complementary replicated sister chromatid opposite the BP-G, leading after propagation to the formation of a C:G base pair (Fig. 3B, product a), whereas when this occurs on the BP-G on the lower strand, this will produce a T:A base pair (Fig. 3B, product b). Note that the two possible HDR events yield identical products (Fig. 3B, a and b), which are different from the four distinct TLS products (Fig. 3B, c–f).
Our experimental scheme must ensure that the engineered lesions are not eliminated by excision repair before replication. This was resolved by using a human xeroderma pigmentosum A (XPA) cell line, which is totally deficient in nucleotide excision repair (NER). To examine whether the DNA lesions indeed persist in the plasmid DNA and are not removed by the fortuitous nonspecific action of nucleases and polymerases before its integration, we transfected XPA cells with a vector carrying two staggered closely opposed BP-G adducts (pLSV5[BP-GstaggBP-G]), and 3 d after transfection the plasmid contents were extracted and subjected to PCR analysis. The rationale was that if the lesions persisted, the fragment carrying the staggered lesions would not be amplified, whereas if some repair occurred, even on one of the strands, this would give rise to an amplified band. As a control we PCR amplified the corresponding fragment from the control intact plasmid without any lesions, using the same primers. To evaluate whether the amount of the plasmids is similar, we PCR amplified a fragment from the puromycin-resistance gene, which is present on both the lesion-carrying plasmid and the control plasmid. As can be seen in Fig. 3C, the lesion-carrying region could not be amplified, in contrast to the two control PCR reactions, which yielded the expected bands. Similar results were observed also with a plasmid carrying two staggered closely opposed TT 6–4 photoproduct (PP) adducts and an artificial trimethylene lesion, which were used in subsequent experiments (Fig. 3C). Thus, the XPA background ensures that lesions that are substrates for NER persist in the plasmid DNA before its integration, and there was no nonspecific processing of the plasmid that caused removal of the lesions.
Benzo[a]pyrene-Guanine and TT 6–4 PP Are Effectively Tolerated in Human Chromosomes.
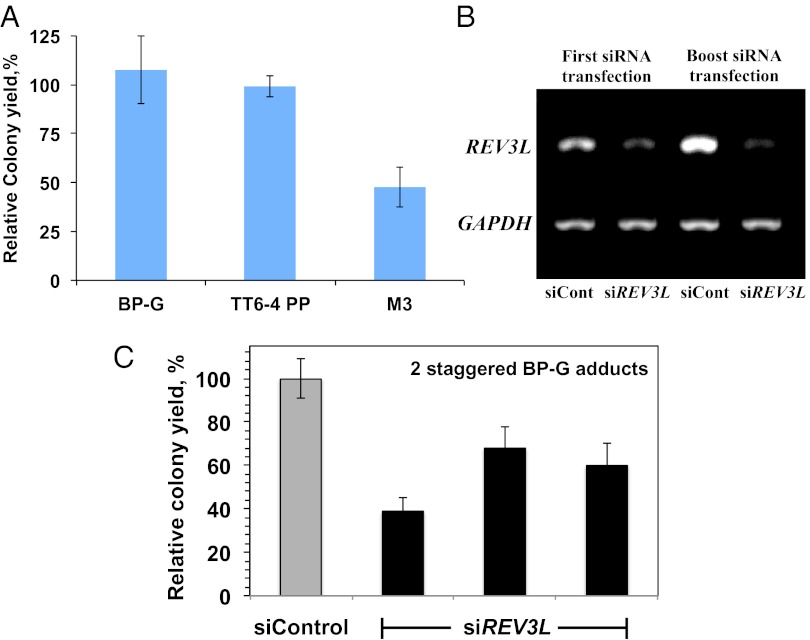
We examined the ability of human cells to tolerate the BP-G adduct by stably transfecting human XPA cells with a lesion-shuttle vector carrying the BP-G lesions and used it to assay tolerance in the same XPA cells. As a control a similarly constructed vector, but with no lesion, was used. The number of stable colonies obtained is affected both by integration and by tolerance efficiencies. However, the relative colony yield (RCY), i.e., the ratio between the number of colonies obtained from the lesion shuttle vector and that obtained from the control shuttle vector, can be taken as an estimate of the efficiency of tolerance of this lesion. Similar numbers of puromycin-resistant colonies were obtained with the BP-G–carrying and the control vectors, suggesting that the BP-G lesion was efficiently tolerated during chromosomal replication (Fig. 4A and Tables S1 and S2).
Fig. 4.
Analysis of the tolerance of DNA lesions integrated into human chromosomes. (A) Relative colony yields of tolerance of two-staggered DNA lesions integrated into the chromosomes of human XPA cells. The detailed data are presented in Tables S1–S6. (B) RT-PCR quantification of REV3L mRNA after treatment with siRNA and a boost treatment after 24 additional hours. (C) Relative colony yields of tolerance of two-staggered BP-G adducts in XPA cells treated with REV3L siRNA or a control siRNA. Results of three different transfections are presented.
We similarly assayed two additional DNA lesions: (i) a TT 6–4 PP adduct, representing one of the two major UV light-induced DNA lesions [unlike BP-G, this is not a bulky lesion, but it distorts DNA (1)], and (ii) an artificial and extreme type of lesion, a trimethylene [(CH2)3] insert into the DNA backbone. We have previously shown, using a gapped plasmid TLS assay, that this hydrocarbon non-DNA insert, termed trimethylene (M3), can be tolerated by TLS both in E. coli (29) and in human cells (28, 30). As can be seen in Fig. 4A and Tables S3 and S4, the relative colony yield for the TT 6–4 PP adduct was 99%, suggesting that the human cells fully tolerated it. In contrast, the colony relative yield of the M3 lesion was 48%, indicating that about half of the cells were unable to cope with this artificial non-DNA insert in their chromosomes (Fig. 4A and Tables S5 and S6).
Because previous studies have indicated that polζ is involved in TLS across BP-G lesions (28, 31), we repeated the TLS/HDR experiments in XPA cells in which the expression of REV3L, the catalytic subunit of polζ, was knocked down by siRNA (Fig. 4B). As can be seen in Fig. 4C, the relative colony yield and thus the efficiency of DDT decreased about twofold under these conditions compared with cells treated with a nontargeting control siRNA, suggesting that polζ is involved in TLS across BP-G in this system. The real effect might be bigger, because the cells still express some polζ (Fig. 4B). DNA sequence analysis showed no significant difference in the balance of TLS/HDR between the two situations (Table S7). Thus, survival is reduced when the expression of polζ is knocked down, and HDR does not seem to serve as an efficient backup under these conditions, as indicated by the lack of change in DNA sequence signature. A possible explanation is that polζ is involved also in HDR, as recently reported (32, 33).
DNA Damage Tolerance Events Are Broadly Distributed Along the Human Genome.
To identify the chromosomal sites in which the lesion shuttle vector had integrated, we rescued from colonies the shuttle vector along with flanking chromosomal sequences and subjected them to DNA sequence analysis across the plasmid–chromosome junctions. As can be seen in Table 1 and Table S8, of the 31 isolates that were sequenced, 90% (28/31) were integrase mediated, as indicated by the DNA sequence flanking the integration sites (Table S8). The 31 isolates mapped to 18 different loci that were distributed over 15 different chromosomes. Four sites were previously reported to be insertion hotspots of ϕC31-mediated integration (27), and 11 were new sites. A major locus responsible for 35% (11/31) of the isolates is a known ϕC31-integration hotspot in chromosome 19 q13.31, where an intron of the zinc finger protein 223 (ZNF223) gene is located (Table 1). Most other integration sites (75%; 15/20) were scored once, whereas two additional integration sites were scored two and three times. The integration loci were located in both intergenic regions (61%; 11/18) and gene introns (39%; 7/18). Taken together, a broad distribution of chromosomal sites was obtained.
Both TLS and HDR Operate to Tolerate DNA Lesions in Human Chromosomes.
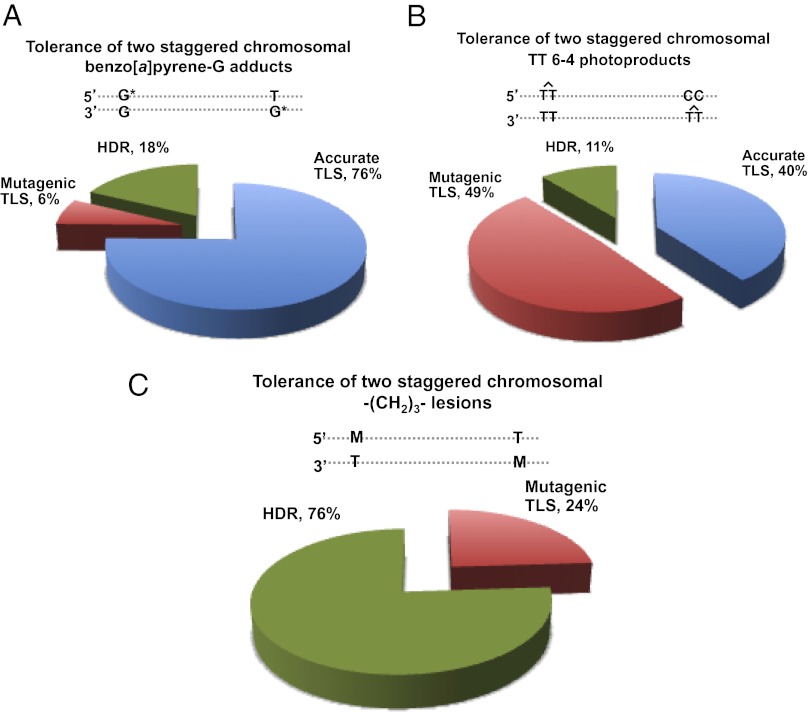
Analysis of the DNA sequence was carried out to determine the type of event that enabled the cells to replicate their DNA across the BP-G lesions (Fig. 5A and Table S1). DNA sequences obtained from 90 colonies revealed that 82% (74/90) carried the signature of TLS. The main event was accurate TLS, which accounted for 76% (68/90) of all tolerance events, and 92% (68/74) of TLS events. Mutagenic TLS accounted for only 6% (6/90) and showed the expected preference for insertion of an A opposite the lesion. Interestingly, 18% (16/90) of the sequences carried the signature of HDR. Thus, both TLS and HDR operate to bypass the BP-G lesions; however, TLS dominates over HDR under our conditions.
Fig. 5.
Categories of DNA damage tolerance events of DNA lesions integrated into human chromosomes. (A–C) The data shown are for pairs of BP-G adducts (A), TT 6–4 photoproducts (B), and M3 lesions (C), in a staggered configuration. The detailed data are presented in Tables S1–S6, respectively.
DNA sequence analysis of the tolerance events of the TT 6–4 adducts showed that 89% (40/45) were via TLS (Fig. 5B and Table S3). In contrast to BP-G, TLS across the TT 6–4 PP was highly mutagenic, accounting for 55% of all TLS events. Thus, both TLS and HDR tolerate the TT 6–4 PP lesion, although TLS appears to have a bigger role under our conditions, performing a highly mutagenic bypass.
Analysis of the DNA sequence changes that had occurred during tolerance of the M3 lesions revealed that the majority of the colonies carried the sequences originally present opposite the M3 lesions, suggesting tolerance by HDR (76%; 29/39 events), whereas only 24% (9/39) of the sequences carried the signature of TLS (Fig. 5C and Table S5).
To examine whether the assay can be used with a single lesion, we constructed a vector with a single TT 6–4 PP lesion and assayed its tolerance after integration in the genome of human XPA cells. As can be seen in Table S9, 53 events were due to TLS, and 53 events were due to replication and HDR combined (because the two have an identical signature, as discussed above). Deducting 50% of the events for replication leaves no observed HDR event. Thus, in this case TLS accounts for >98%, whereas HDR accounts for <2% (less than 1/53), compared with 89% and 11%, respectively, in the staggered configuration. This difference is within the experimental error of the assay system. As for the mutagenicity, in the single-lesion configuration mutagenic TLS accounted for 75% of the TLS events compared with 55% in the staggered configuration, a difference that is statistically not significant (χ2-test, P = 0.203).
Discussion
Despite the importance of DNA damage tolerance in mammalian cells as a protective mechanism against genome instability and cancer, the scarcity of high-resolution assays for measuring TLS and HDR, especially in chromosomes, has hampered progress in the field. The TLS/HDR assay presented in this study enables one to simultaneously analyze TLS and HDR in mammalian chromosomes at a single-nucleotide resolution. At this point the assay needs to be performed in excision-repair-deficient cells, to prevent elimination of the lesion from DNA before replication; however, it is expected to provide useful mechanistic insight even with this limitation. We used lesion shuttle vectors with two staggered closely opposed lesions. The staggered lesion configuration, first used by Lawrence in the yeast S. cerevisiae (12) and later adopted to the chicken DT40 cell line (23), has the advantage that tolerance must occur on both strands and therefore eliminates potential biases between an intact and a lesion-carrying strand. A similar approach was recently taken by Pages et al. (11), who inserted a plasmid carrying a defined lesion into a specific site in the E. coli genome. Consistent with previous reports they found that the overwhelming majority of tolerance events involved HDR (damage avoidance). Unlike in the E. coli system, in our system the plasmid is integrated into various genomic sites, enabling the future analysis of the effect of genome location on DNA damage tolerance, e.g., the balance between TLS and HDR.
DNA damage tolerance mechanisms provide a major defense against base damages that interfere with DNA replication and may lead to the collapse of replication forks and the formation of double-strand breaks (1, 2, 34). Although there was significant evidence for the operation of TLS in tolerating replication-blocking lesions, there are limited hints on the activity of HDR in mammalian chromosomes (20, 35). Using a model assay system based on a gap-lesion plasmid and a homologous dsDNA, we have previously shown that gap-filling HDR functions in mammalian cells and operates via a strand transfer mechanism (26). However, the results presented here indicate that both TLS and HDR operate in mammalian chromosomes to tolerate DNA lesions. Chromosomal TLS across BP-G was mostly accurate (92%), highlighting the need for high-resolution assays such as the one described here, because assays measuring mutagen-induced chromosomal mutations are blind to accurate TLS and therefore cannot score most TLS events across BP-G. The main mutagenic event was a GC→ΤΑ transversion caused by misinsertion of an A opposite the BP-G, consistent with previous results obtained with plasmid model systems (28, 36–38) and with chromosomal mutagenesis assays monitoring BP-induced mutations in chromosomal reporter genes (39, 40).
TLS across TT 6–4 PP was highly mutagenic, with a typical GTT→TTT hotspot. This is consistent with the results that we have previously obtained with nonreplicating gapped plasmids in mammalian cells (28, 41) and with an earlier report using a replicative plasmid (42). A recent study reported that TLS across a TT 6–4 PP in a replicating plasmid is largely accurate (43). This difference does not stem from a replicative vs. a gapped nonreplicating plasmid as those authors suggested, because an earlier study using a replicative plasmid found that TLS is highly mutagenic (42), like in our study. The reason for the difference is not known yet, but it should be pointed out that the TLS that we have monitored across chromosomal TT 6–4 PP was highly mutagenic.
Interestingly, both TLS and HDR were involved in the tolerance of each of the three lesions that were studied, although at different proportions. It is not clear what determines the labor division between TLS and HDR. Possible factors involved may include chromosomal location, the type of DNA damage, and the cell cycle, following the finding that like in S. cerevisiae cells (16, 44), the TLS in mammalian cells occurs both in the S and the G2 phases (45). Interestingly, mammalian cells are very effective in tolerating BP-G, despite its bulkiness, and TT 6–4 PP, despite the significant deformation that this lesion causes. The situation is different for the artificial M3 lesions. In this case, tolerance was not fully efficient, and most of it was via HDR.
Although the identity of the DNA polymerase involved in the synthesis step of HDR is not clear, there were reports on the involvement of DNA polymerases η in an in vitro system (46) and of Rev1 and DNA polymerase ζ in cultured cells (32). The action of these or other polymerases in HDR is not expected to affect the sequence signatures that we have observed in our system, because they are expected to be distributed all over the synthesis patch, in contrast to the targeted and semitargeted base signatures that have been observed. In addition, the error frequency of DNA polymerase η is ∼1% (47), and that of the yeast DNA polymerase ζ about 0.1% (48), much lower than the frequencies that we have observed. Why is HDR, which is inherently error-free, not always preferred over the inherently error-prone TLS? TLS is a very localized “repair” operation, the worst outcome being a point mutation. Moreover, an elaborate regulation (22, 49–51) operates to minimize the mutagenic load caused by TLS. In contrast, HDR, despite its inherent accurate mechanism, is more complex and involves both the damaged strand and the replicated sister chromatid. Given the abundance of repetitive sequences in mammalian genomes, such a DNA transaction may cause a higher risk of rearrangements. Such events were indeed observed in cells lacking polζ (52). Thus, in mammals the selective benefit of avoiding potentially deleterious rearrangements may have led to the selection of regulatory mechanisms that sometimes favor TLS over HDR. The availability of the unique TLS/HDR assay will facilitate further studies of the mechanisms of DNA damage tolerance in mammalian cells and their role in preventing genome instability and various pathologies such as cancer.
Materials and Methods
Plasmid Construction.
Plasmid pLSV5 was constructed in four steps. First, the puromycin resistance gene under the phosphoglycerate kinase (PGK) promoter was cloned into the kanamycin-carrying plasmid pSKSL (53) to generate the plasmid pSKSL-puro. The puromycin resistance gene was obtained by PCR from pRetrosuper, using the following primers: 5′-GGTAGGGAATTCGCTTTTCCAAGGCAGTC-3′ and 5′-GATGCATGGGATCCTGCGCTCCTTTCGG-3′. The 1,705-bp PCR fragment was digested with BglII and BamHI and cloned into pSKSL between those sites. In the second step, an attB recognition sequence was added to pSKSL-puro. The attB recognition sequence was obtained from the plasmid pTA-attB (27) by PCR amplification, using the primers 5′-GAGCGGCCGCCAGTGTGATGGAGATCTGCA-3′ (containing a BstXI site) and 5′-CTAGTAAC GGC CGCCACACCGCTGGAATTC-3′ (containing a BglII site). The PCR product (374 bp) was cut with BstXI and BglII and ligated to BstXI-BglII cut-modified pSKSL-puro. In the third step, the resulting vector (pSKSL-p-attB) was digested with BstXI and AhdI, its termini converted to blunt ends using DNA polymerase I Klenow fragment, and religated to eliminate the BstXI site. The last step involved the creation of a new BstXI site and the addition of a 925-bp spacer between the BsTXI and BspQI sites. The spacer was obtained by PCR amplification of pRetrosuper, using the primers 5′-TTTTTTCCATGGCCACACCGCTGGAGCAAAAACAGGAAGGCAAA-3′ and 5′-TTTTTTCCATGGTGGACTAATCGCCTCTACGC-3′. The PCR product was digested with NcoI and ligated to an AflII-digested vector, yielding the 5,332-bp plasmid pLSV5.
The synthesis of short oligonucleotides containing a site-specific BP-G adduct, a TT 6–4 PP, or an M3 lesion,was previously described (28). The lesion shuttle vector pV5(BP-GstaggBP-G), which contains two site-specific BP-G lesions in a staggered configuration, was prepared using modified oligonucleotides containing a BP-G adduct. It was constructed in several steps as follows (Fig. S1 and Table S10): First, a 12-mer oligo (marked B and 1BS in Fig. S1A and Table S10, respectively) carrying the BP-G was extended to a 61-mer oligonucleotide by ligating a 12-mer oligo (marked A and 1AS in Fig. S1A and Table S10, respectively) to its 5′ end and a 37-mer oligo (marked C and 1CS in Fig. S1A and Table S10, respectively) to its 3′ end, using a 45-mer oligo (marked D and 1DS in Fig. S1A and Table S10, respectively) as a scaffold (Fig. S1A). The resulting 61-mer elongated oligonucleotide 1ES (Fig. S1B and Table S10) was separated from the scaffold and excess oligonucleotides on a 12% denaturing polyacrylamide gel containing 8 M urea. The purified 61-mer oligo with site-specific BP-G was then annealed to a complementary 68-mer oligo (1EL, Table S10) with single BP-G modification that was generated in a similar manner. The resulting double-stranded duplex that contained a 5′ overhang complementary to a cleaved BspQI site and a 3′ overhang complementary to a cleaved BstXI site (Fig. S1B) was ligated to a gel-purified 4,321-bp fragment of pLSV5, obtained by BstXI and BspQI double digestion. The closed circular product pLSV(BP-GstaggBP-G) was purified from other ligation products by electrophoresis, using a 0.8% agarose gel. A gel slice containing the band with the pLSV5(BP-GstaggBP-G) DNA was then cut out and the DNA electroeluted, using an Elutrap apparatus (Schliecher and Schuell). The control vector pVBCs with no lesions was constructed using unmodified 61-mer and 68-mer oligonucleotides (Table S10, sequences 1FS and 1FL, respectively). The vectors containing the TT 6–4 PP and the M3 lesions and their corresponding control vectors were built in a similar manner. The oligonucleotides used for building these vectors are listed in Tables S11 and S12. Plasmid pV5(TT6-4staggTT6-4) was built using elongated oligonucleotides 2EL and 2ES, and its corresponding control, pV5TTCs, was built using oligonucleotides 2FL and 2FS (Table S11). Plasmid pV5(M3-2staggM3) and its corresponding control pV5MCs were built using the oligo pairs 3E2L-3ES, and 3FL-3FS, respectively (Table S12). Oligonucleotides without a lesion were synthesized by Sigma-Aldrich. All oligonucleotides were purified on a 12% or 15% denaturing polyacrylamide gel containing 8 M urea before use.
Cell Culture and Genomic Lesion Tolerance Assay Procedure.
SV40-transformed XPA (XP12RO) human fibroblast cells were cultured in MEM Eagle (Sigma) supplemented with 2 mM l-glutamine (GIBCO/BRL), 100 units/mL of penicillin, 100 μg/mL of streptomycin (Biological Industries), and 15% FBS (HyClone).
The genomic lesion tolerance assay was performed as follows. Cultured cells were grown to 80% confluence in 6-well plates. The cells were then cotransfected with a mixture of a lesion shuttle vector (pV5L, 60 ng) or a control vector (pV5LCo, 60 ng) and the ϕC31-integrase expression vector (pKGphc31o, 2 µg), using 9 µL/well HiPerFect (Qiagen). Forty-eight hours after transfection, the cells were split into two 10-cm dishes and after reattachment were subjected to 0.55 µg/mL puromycin (Sigma) for selection. Approximately 14 d later, resistant colonies were counted, picked, and individually transferred into a 96-well plate for further growth. The number of resistant colonies obtained relative to the number of colonies obtained with the control vector without a lesion is used as a rough estimate of the overall efficiency of DNA lesion tolerance. When it reached suitable confluence (∼5 d later), the genomic DNA of the cells in each well was extracted essentially as described (54). This genomic DNA was then used as a template in a PCR aimed to amplify the lesion core area. The PCR was performed in 96-well format, in a final volume of 10 µL per well. Each PCR included 1 µL genomic DNA (50–200 ng), 0.09 µL Phusion termostable DNA polymerase (2 units/µL; NEB), primers (10 pmol each), dNTPs (0.2 mM final), and Phusion High Fidelity PCR reaction buffer. The reaction consisted of 38 cycles, and the annealing temperature was 60 °C. The primers were 5′TTGGCAGAACATATCCATCG 3′ (forward) and 5′CTGACCTTTTGGTCACATGGC 3′ (reverse). The resulting PCR product was 258 bp long for the BP-G and 6–4TT constructs and 257 bp long for the M3 constructs. The PCR products were sequenced using the primers 5′ ATTAATGAATCGGCCAACG 3′ or 5′CTGACCTTTTGGTCACATGGC 3′.
Plasmid Rescue.
For plasmid rescue, genomic DNA of individual colonies or pools was extracted using the DNeasy Kit (Qiagen). This genomic DNA was then digested with a combination of restriction enzymes XbaI and XmaJI that did not cut within the lesion shuttle vector sequence. In a typical reaction, 10 µg of DNA was digested with 30 units of each enzyme for 16 h. DNA was then extracted with phenol/chloroform, precipitated in ethanol, and ligated under dilute conditions (5 ng/µL) with bacteriophage T4 DNA ligase. The ligated DNA was then used to transform electrocompetent DH10B bacteria cells that were subsequently plated on agar plates containing kanamycin. Plasmid DNA was prepared from resulting colonies and sequenced using the following primers: attL, 5′ CAGTGAGGCACCTATCTCAGC 3′; attR, 5′ CGCTATGTGTTCTGGGAAATC 3′.
Supplementary Material
Acknowledgments
We thank Michele Calos (Department of Genetics, Stanford University School of Medicine) for her generous gift of plasmid carrying the attB site and the ϕC31 integrase gene. Z.L. is the Incumbent of the Maxwell Ellis Professorial Chair in Biomedical Research. This work was supported by grants from the Flight Attendant Medical Research Institute, the Israel Science Foundation (Grant 1136/08), the Leona M. and Harry B. Helmsley Charitable Trust (to Z.L.), and the US National Institutes of Health/National Cancer Institute (Grant CA099194 to N.E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 6258 (volume 110, number 16).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216894110/-/DCSupplemental.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd Ed. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.Friedberg EC. Suffering in silence: The tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6(12):943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair. 2006;5(12):1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9(4):729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104(40):15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupp WD, Wilde CE, 3rd, Reno DL, Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971;61(1):25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 8.Smith KC. Recombinational DNA repair: The ignored repair systems. Bioessays. 2004;26(12):1322–1326. doi: 10.1002/bies.20109. [DOI] [PubMed] [Google Scholar]
- 9.Izhar L, et al. Analysis of strand transfer and template switching mechanisms in Escherichia coli: Predominance of strand transfer. J Mol Biol. 2008;381:803–809. doi: 10.1016/j.jmb.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol Cell. 2002;10(4):917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 11.Pagès V, Mazón G, Naiman K, Philippin G, Fuchs RP. Monitoring bypass of single replication-blocking lesions by damage avoidance in the Escherichia coli chromosome. Nucleic Acids Res. 2012;40(18):9036–9043. doi: 10.1093/nar/gks675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci USA. 2005;102(44):15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456(7224):915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 14.Blastyák A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28(1):167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hishida T, Kubota Y, Carr AM, Iwasaki H. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature. 2009;457(7229):612–615. doi: 10.1038/nature07580. [DOI] [PubMed] [Google Scholar]
- 16.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465(7300):951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399(6737):700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285(5425):263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 19.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18(1):174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Xiao W, McCormick JJ, Maher VM. Identification of a protein essential for a major pathway used by human cells to avoid UV- induced DNA damage. Proc Natl Acad Sci USA. 2002;99(7):4459–4464. doi: 10.1073/pnas.062047799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriya M, et al. Targeted mutations induced by a single acetylaminofluorene DNA adduct in mammalian cells and bacteria. Proc Natl Acad Sci USA. 1988;85(5):1586–1589. doi: 10.1073/pnas.85.5.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avkin S, et al. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol Cell. 2006;22(3):407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 23.Szüts D, Marcus AP, Himoto M, Iwai S, Sale JE. REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36(21):6767–6780. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci USA. 2009;106(28):11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon JH, Bhatia G, Prakash S, Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases kappa and zeta in human cells. Proc Natl Acad Sci USA. 2010;107(32):14116–14121. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adar S, Izhar L, Hendel A, Geacintov N, Livneh Z. Repair of gaps opposite lesions by homologous recombination in mammalian cells. Nucleic Acids Res. 2009;37(17):5737–5748. doi: 10.1093/nar/gkp632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalberg TW, et al. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357(1):28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 28.Shachar S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28(4):383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maor-Shoshani A, Ben-Ari V, Livneh Z. Lesion bypass DNA polymerases replicate across non-DNA segments. Proc Natl Acad Sci USA. 2003;100(25):14760–14765. doi: 10.1073/pnas.2433503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adar S, Livneh Z. Translesion DNA synthesis across non-DNA segments in cultured human cells. DNA Repair. 2006;5(4):479–490. doi: 10.1016/j.dnarep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, et al. hREV3 is essential for error-prone translesion synthesis past UV or benzo[a]pyrene diol epoxide-induced DNA lesions in human fibroblasts. Mutat Res. 2002;510(1–2):71–80. doi: 10.1016/s0027-5107(02)00253-1. [DOI] [PubMed] [Google Scholar]
- 32.Sharma S, et al. REV1 and polymerase ζ facilitate homologous recombination repair. Nucleic Acids Res. 2012;40(2):682–691. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane DP, Shusterman M, Rong Y, McVey M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 2012;8(4):e1002659. doi: 10.1371/journal.pgen.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansen JG, Tsaalbi-Shtylik A, de Wind N. Functional interactions between DNA damage signaling and mutagenic translesion synthesis at post-replicative gaps. Cell Cycle. 2009;8(18):2857–2858. doi: 10.4161/cc.8.18.9409. [DOI] [PubMed] [Google Scholar]
- 35.Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30(3):684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang JL, Maher VM, McCormick JJ. Kinds of mutations formed when a shuttle vector containing adducts of benzo[a]pyrene-7,8-diol-9,10-epoxide replicates in COS7 cells. Mol Cell Biol. 1987;7(3):1267–1270. doi: 10.1128/mcb.7.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandes A, et al. Mutagenic potential of stereoisomeric bay region (+)- and (-)-cis-anti-benzo[a]pyrene diol epoxide-N2-2′-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37(28):10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 38.Avkin S, et al. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: The role of DNA polymerase kappa. J Biol Chem. 2004;279(51):53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Mazur-Melnyk M, de Boer JG, Glickman BW. A comparison of mutational specificity of mutations induced by S9-activated B[a]P and benzo[a]pyrene-7,8-diol-9,10-epoxide at the endogenous aprt gene in CHO cells. Mutat Res. 1999;423(1–2):23–32. doi: 10.1016/s0027-5107(98)00221-8. [DOI] [PubMed] [Google Scholar]
- 40.Lambert B, et al. Mutations induced in the hypoxanthine phosphoribosyl transferase gene by three urban air pollutants: Acetaldehyde, benzo[a]pyrene diolepoxide, and ethylene oxide. Environ Health Perspect. 1994;102(Suppl 4):135–138. doi: 10.1289/ehp.94102s4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z. Reduced fidelity and increased efficiency of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair. 2008;7:1636–1646. doi: 10.1016/j.dnarep.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gentil A, et al. Mutagenicity of a unique thymine-thymine dimer or thymine-thymine pyrimidine pyrimidone (6-4) photoproduct in mammalian cells. Nucleic Acids Res. 1996;24(10):1837–1840. doi: 10.1093/nar/24.10.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon JH, Prakash L, Prakash S. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase zeta in mouse and human cells. Genes Dev. 2010;24(2):123–128. doi: 10.1101/gad.1872810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141(2):255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 45.Diamant N, et al. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012;40(1):170–180. doi: 10.1093/nar/gkr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McIlwraith MJ, et al. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20(5):783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Matsuda T, et al. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312(2):335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 48.Zhong X, et al. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34(17):4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 50.Bienko M, et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell. 2010;37(3):396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 51.Livneh Z. Keeping mammalian mutation load in check: Regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle. 2006;5(17):1918–1922. doi: 10.4161/cc.5.17.3193. [DOI] [PubMed] [Google Scholar]
- 52.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res. 2006;66(1):134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 53.Tomer G, Livneh Z. Analysis of unassisted translesion replication by the DNA polymerase III holoenzyme. Biochemistry. 1999;38(18):5948–5958. doi: 10.1021/bi982599+. [DOI] [PubMed] [Google Scholar]
- 54.Ramírez-Solis R, et al. Genomic DNA microextraction: A method to screen numerous samples. Anal Biochem. 1992;201(2):331–335. doi: 10.1016/0003-2697(92)90347-a. [DOI] [PubMed] [Google Scholar]