Abstract
The recent discovery of the direct oxidation of spermine via spermine oxidase (SMO) as a mechanism through which specific antitumor polyamine analogues exert their cytotoxic effects has fueled interest in the study of the polyamine catabolic pathway. A major byproduct of spermine oxidation is H2O2, a source of toxic reactive oxygen species. Recent targeted small interfering RNA studies have confirmed that SMO-produced reactive oxygen species are directly responsible for oxidative stress capable of inducing apoptosis and potentially mutagenic DNA damage. In the present study, we describe a second catalytically active splice variant protein of the human spermine oxidase gene, designated SMO5, which exhibits substrate specificities and affinities comparable to those of the originally identified human spermine oxidase-1, SMO/PAOh1, and, as such, is an additional source of H2O2. Importantly, overexpression of either of these SMO isoforms in NCI-H157 human non-small cell lung carcinoma cells resulted in significant localization of SMO protein in the nucleus, as determined by confocal microscopy. Furthermore, cell lines overexpressing either SMO/PAOh1 or SMO5 demonstrated increased spermine oxidation in the nucleus, with accompanying alterations in individual nuclear polyamine concentrations. This increased oxidation of spermine in the nucleus therefore increases the production of highly reactive H2O2 in close proximity to DNA, as well as decreases nuclear spermine levels, thus altering the protective roles of spermine in free radical scavenging and DNA shielding, and resulting in an overall increased potential for oxidative DNA damage in these cells. The results of these studies therefore have considerable significance both with respect to targeting polyamine oxidation as an antineoplastic strategy, and in regard to the potential role of spermine oxidase in inflammation-induced carcinogenesis.
Keywords: carcinogenesis, H2O2, oxidation, polyamine, SMO
The naturally occurring polyamines, spermine, spermidine and putrescine, are polycations that are abundant and essential in both prokaryotic and eukaryotic cells. These molecules have been implicated in multiple cellular functions and processes, including proliferation, differentiation, apoptosis, gene expression and cell signaling [1–6]. The strict maintenance of polyamine homeostasis is critical for proper cell function, and is regulated at the levels of biosynthesis, uptake, efflux and catabolism. In cancer cells, the regulatory components of polyamine homeostasis are often disrupted, leading to increased levels of intracellular polyamines, thus promoting increased growth and proliferation, and, potentially, tumor progression [7–11]. This tumor-associated dysregulation of polyamine metabolism has led to the development of several classes of polyamine analogues targeted towards inducing the catabolic enzymes, which in turn deplete intracellular polyamines, and arrest tumor cell growth [7,12–16].
Until recently, it was presumed that polyamine catabolism was solely regulated by the rate-limiting enzyme spermidine/spermine N1-acetyltransferase [17]. The resulting acetylated polyamines, N1-acetylspermidine and N1-acetylspermine, are substrates for either cellular export or further back-conversion via the constitutively- expressed N1-acetylpolyamine oxidase [18]. Recent cloning and characterization of N1-acetylpolyamine oxidase has confirmed its preference for the acetylated polyamines as substrates, and no significant activity is observed when spermine is used as substrate [19,20]. Our cloning of a second polyamine oxidase, human spermine oxidase-1 (SMO/PAOh1), and its confirmation as a spermine oxidase, identified a second mechanism through which the polyamines are catabolized, and has led to increased interest in the exploitation of polyamine catabolism for antitumor therapy [21–23].
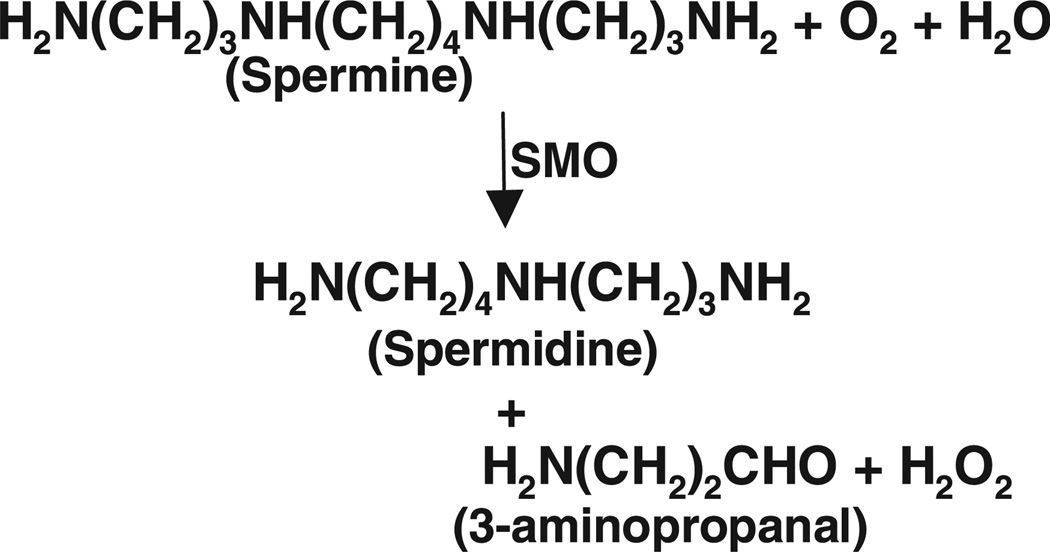
SMO/PAOh1 is an FAD-containing enzyme (EC 1.5.3.–) that directly catabolizes spermine as its preferred substrate, resulting in spermidine, 3-aminopropanal and, importantly, H2O2 (Fig. 1). H2O2 is a readily diffusible source of cellular reactive oxygen species (ROS), and has been linked to the cytotoxicity observed in specific tumor cell types following treatment with antitumor, polyamine catabolism-inducing polyamine analogues [2,12,24–26]. The finding that spermine oxidase activity, and thus H2O2 production, can be induced in a cell and tumor type-specific manner by certain polyamine analogues [22,25,27] adds considerably to the importance of investigating this new pathway member because its regulation may contribute to the facilitation of tumor cell apoptosis [5]. Additionally, studies overexpressing the mouse spermine oxidases have provided evidence that SMO directly induces DNA damage via H2O2-related oxidative stress, and that this damage renders the cell more sensitive to subsequent radiation exposure and apoptosis [28]. Furthermore, induction of spermine oxidase in gastric and lung epithelial cells by Helicobacter pylori and tumor necrosis factor-α, respectively, has been linked to increased ROS production and DNA damage [29,30], thus implicating SMO as a potential molecular link between chronic inflammation and epithelial carcinogenesis. In addition, we have recently demonstrated elevated SMO expression in prostatic epithelial tissue from prostate cancer patients compared to prostate disease-free control patients [31].
Fig. 1.
Catalysis of spermine oxidation by human spermine oxidase. SMO catalyzes the cleavage of spermine, resulting in backconversion to spermidine, and the generation of 3-aminopropanal and H2O2.
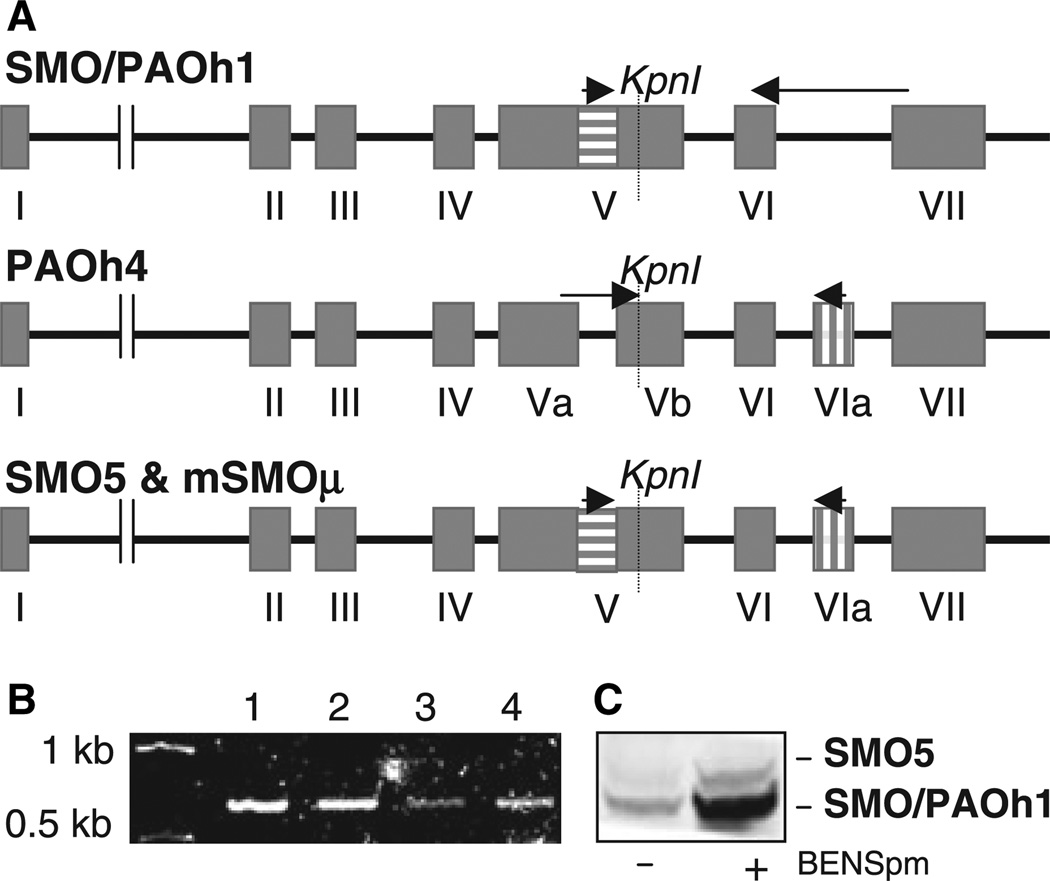
Subsequent to the initial characterization of SMO/PAOh1, three additional splice variants have been identified [32]; however, the purified recombinant proteins of these variants have failed to exhibit significant oxidase activity on the natural polyamines. Based on the exon structures of previously identified human spermine oxidases, we suspected the existence of an additional isoform that possesses a combination of the gene segments present in the two longest variants: the active SMO/PAOh1 and the inactive PAOh4 (Fig. 2A) [32]. However, this hypothetical isoform was not identified when using the standard reverse transcription PCR protocol that produced spermine oxidases 1–4. A mouse spermine oxidase isoform, mSMOµ, has been identified that possesses an exon structure identical to that of our hypothetical spermine oxidase-5, or SMO5. Importantly, this mouse isoform has high spermine oxidase activity, and exhibits a significantly greater degree of localization in the nucleus than the other mouse polyamine oxidases thus far described [33]. We therefore sought to determine whether the corresponding SMO5 isoform exists in human cells.
Fig. 2.
Expression of human spermine oxidase splice variants in lung epithelial cells. (A) Exon structures of spermine oxidase isoforms. SMO5 includes both the internal region of exon V that is absent from the inactive PAOh4 isoform, as well as exon VIa, which is absent from the active isoform, SMO/PAOh1. SMO5 and the active, mouse isoform, mSMOμ, have identical exon structures. Arrows indicate isoform-specific primers utilized for RT- and realtime PCR. (B) Expression of SMO5 mRNA in human lung cells. Total RNA was extracted from: (1) A549; (2) A549 + 10 µm BENSpm; (3) H157; or (4) Beas2B cells. One µg of each RNA sample was used for RT-PCR with primers specific for SMO isoform 5, resulting in 653 bp fragments that were separated and visualized on a 1% agarose gel stained with ethidium bromide. (C) Western blot of endogenous SMO/PAOh1 and SMO5 protein. To visualize relative levels of the two active SMO isoforms, A549 cells were treated with 10 µm BENSpm for 24 h, total protein was extracted, and immunoblotting was performed using a human SMO antibody that recognizes both isoforms.
In the present study, we confirmed the existence of SMO5 in human normal and tumor cell lines. SMO5 cDNA was subcloned and sequenced, and active protein was produced, purified and assayed for kinetic properties and substrate specificities. NCI-H157 human non-small cell lung carcinoma cells were stably transfected, and individual clones selected that overexpress each of the isoforms of interest to confirm cellular localization. In contrast to results observed with the homologous mouse SMO isoforms, each of the three human isoforms examined were present in similar amounts in the nucleus, with SMO/PAOh1 and SMO5 both demonstrating functional oxidase activity in both nuclear and cytoplasmic compartments, as determined by measurement of H2O2 production, as well as HPLC analysis of localized polyamine pools.
Previous reports have described transient overexpression of SMO/PAOh1 in the transformed human kidney cell line HEK-293 [22], and Amendola et al. [28] have described the establishment of stable populations overexpressing several of the mouse SMO isoforms in a mouse neuroblastoma line. The cell lines created in the present study, however, represent the first stable overexpression of any of the human spermine oxidase isoforms in human tumor cells. Furthermore, we report the first direct visualization of the localization of human spermine oxidase in a human tumor cell line. We also identify and classify SMO5 as a third human polyamine catabolic enzyme that possesses the ability to produce reactive oxygen species as a toxic byproduct, while altering intracellular polyamine concentrations. Most importantly, the surprising abundance of both active isoforms, SMO/PAOh1 and SMO5, in the nucleus, emphasizes the potential of these ROS-producing enzymes to either modulate cancer cell response to the antitumor polyamine analogues, or, in cells stimulated by infection and/or inflammation, act as the source of ROS with the potential to produce mutagenic oxidative DNA damage, thus providing a link between inflammation and carcinogenesis.
Results
SMO5 is expressed in human lung cell lines
To determine whether the SMO5 splice variant of the spermine oxidase gene is present in human tissue, we performed RT-PCR using a primer pair specific to SMO5. Specifically, primers were located in the internal region of exon V and in exon VIa, both of which are only found together in the SMO5 splice variant (Fig. 2A). The resulting 653 bp amplification product was detected in all cell lines tested, including the nonsmall cell lung carcinoma lines, NCI-H157 and NCI-A549, as well as the non-tumorigenic lung epithelial cell line, Beas2B (Fig. 2B).
Real-time PCR using the same primers as above for SMO5, as well as primers specific for SMO/PAOh1 (Fig. 2A), suggested that SMO5 mRNA is expressed to a much lower extent than SMO/PAOh1 in both H157 and A549 cell lines (data not shown), possibly accounting for the fact that SMO5 was not identified during our initial cloning of the spermine oxidase isoforms from NCI-H157 cells. We confirmed the relative abundance of the two proteins using western blot analysis of A549 cells, which have previously been shown to express relatively high levels of SMO/PAOh1 [27]. In both untreated cells and in those treated with bis(ethyl)norspermine (BENSpm), a polyamine analogue known to induce spermine oxidase, a 65 kDa band corresponding to SMO5 was apparent at a much lower intensity than the 61.9 kDa SMO/PAOh1 protein band (Fig. 2C).
SMO5 protein purification
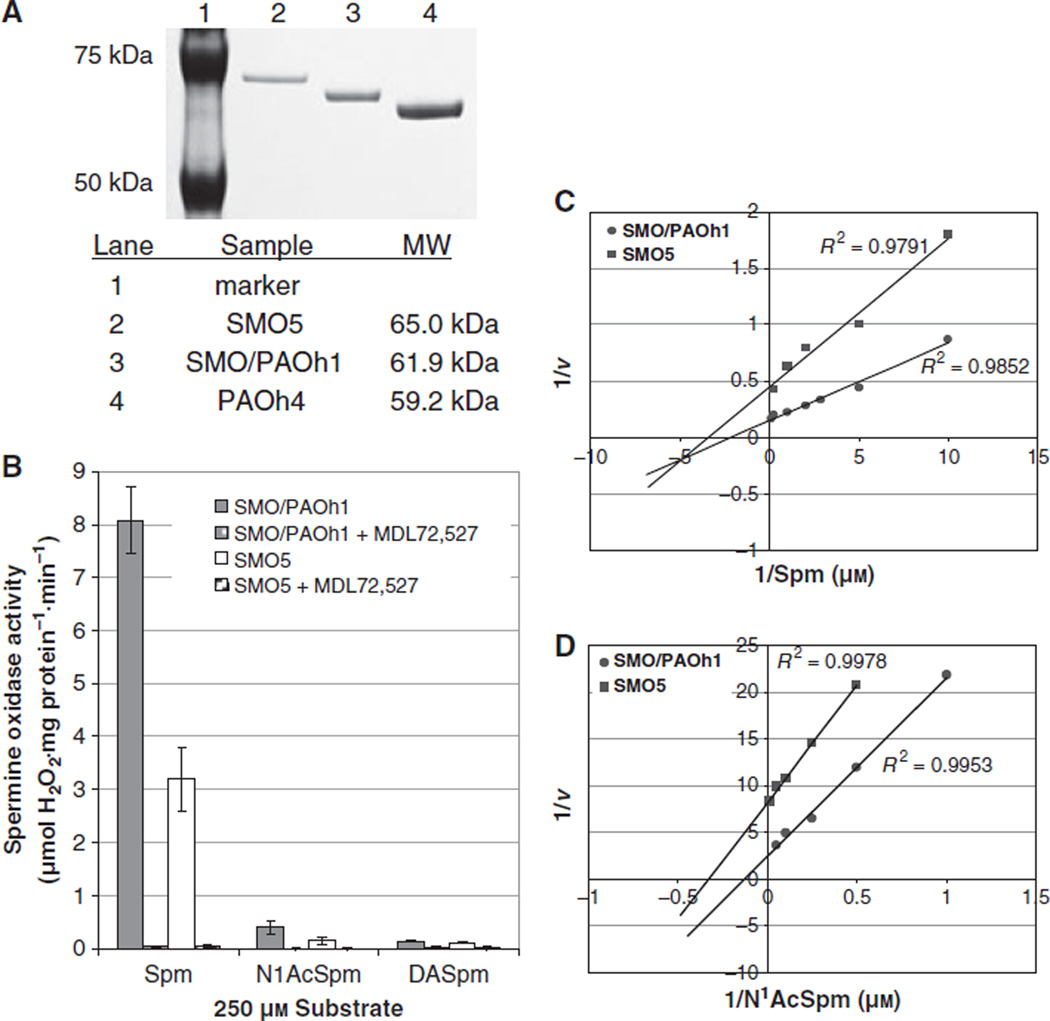
To further study and characterize the SMO5 protein, SMO5 cDNA was subcloned into the pET15b bacterial expression vector. The 1984 bp cDNA (GenBank accession no. EF032141) includes both the internal region of exon V that exists in SMO/PAOh1, as well as exon VIa that is present in PAOh4 (Fig. 2A). Both of these regions are also present in the nuclear-localized mouse spermine oxidase isoform, mSMOμ, which shares 89% nucleotide identity with the human SMO5 cDNA. Isopropyl thio-β-d-galactoside induction of transformed BL21(DE3) Escherichia coli resulted in the production of SMO5 protein, the largest of the SMO isoforms, consisting of 586 amino acids, with a predicted molecular mass of approximately 65 kDa, as observed by SDS/PAGE analysis (Fig. 3A). As previously reported for the PAOh1/SMO protein [23], the majority of SMO5 protein produced in this bacterial expression system localizes to inclusion bodies, and therefore was denatured and refolded prior to analysis. All SMO5 protein production and purification steps were performed in parallel with SMO/PAOh1 and PAOh4 proteins for comparison.
Fig. 3.
Characteristics of recombinant SMO5 protein. (A) Recombinant SMO/PAOh1, PAOh4 or SMO5 protein was expressed and purified from transformed E. coli cells. Dialyzed, refolded proteins were analyzed by SDS/PAGE, visualized by staining with Coomassie blue, and photographed. (B) Substrate specificity and specific activities of purified SMO5 versus SMO/PAOh1. Oxidase activity was assessed for each purified protein using 250 µm of various potential substrates ± an equimolar concentration of the polyamine oxidase inhibitor, MDL72,527. Substrates used were spermine (Spm), N1-acetylspermine (N1AcSpm), or N1,N12-diacetylspermine (DASpm). Data represent the mean ± SE of three separate experiments, each performed in triplicate. (C) Representative Lineweaver–Burk plots of purified SMO/PAOh1 or SMO5 using spermine or N1-acetylspermine as substrates. Increasing concentrations of each substrate were added to each of the purified enzymes and spermine oxidase activity was determined as a function of µmol H2O2·mg protein−1·min−1.
Polyamine oxidase activity and substrate specificity of SMO5
Polyamine oxidase activity assays of the purified recombinant SMO5 and SMO/PAOh1 proteins revealed nearly identical substrate specificities for the two isoforms. Both enzymes clearly exhibit a strong preference for spermine as the primary substrate over all other naturally occurring polyamines, with SMO/PAOh1 having a specific activity approximately 2.5-fold greater than that of SMO5 (Fig. 3B). N1-acetylspermine was the only other polyamine to be oxidized by the proteins, but this activity was less than 10% of that observed when using spermine with the same enzyme. Similar to SMO/PAOh1, SMO5 exhibited virtually no oxidase activity when using N1,N12-diacetylspermine as substrate, and there was a complete absence of oxidation when N1-acetylspermidine, N8-acetylspermidine, spermidine, or the polyamine analogues, BENSpm or N1-ethyl- N11-(cyclopropyl)methyl-4,8,diazaundecane (CPENSpm), were presented as potential substrates. MDL72,527, an inhibitor of the polyamine oxidases, was also effective as an inhibitor of SMO5 activity, as demonstrated by a greater than 99% reduction in the oxidation of spermine.
Purified, recombinant PAOh4 was inactive as an oxidase with all substrates tested (data not shown). The only difference between PAOh4 and SMO5 is the absence of a 53 amino acid central region of exon V, including residues 283–335. According to structure analyses and molecular modeling of homologous proteins, many residues in the 3′ half of this region are highly conserved components of the FAD-binding domain, and are therefore essential for catalysis [28,33–35].
Kinetic properties of SMO5
Polyamine oxidase activity of SMO5 and SMO/PAOh1 was measured using increasing concentrations of either spermine or N1-acetylspermine as substrate (Fig. 3C). SMO5 and SMO/PAOh1 displayed very similar affinities for spermine, as determined by Lineweaver–Burk transformation of the Michaelis–Menton equation, with Km values of 0.5 and 0.6 µm, respectively. Both enzymes possessed a calculated Km of approximately 3.0 µm when N1-acetylspermine was used as substrate. SMO/PAOh1 consistently demonstrated a kcat value approximately 2.4-fold that of SMO5 with either of the substrates examined. Specifically, SMO/PAOh1 exhibited kcat values for spermine and N1-acetylspermine of 7.55 s−1 and 0.28 s−1, respectively, whereas those for SMO5 measured 3.11 s−1 and 0.12 s−1. The most dramatic kinetic differences, however, were seen in the velocities at which each enzyme was capable of oxidizing spermine as opposed to N1-acetylspermine. With both enzymes, the kcat value of spermine is approximately 27-fold that of N1-acetylspermine.
Overexpression of SMO in NCI-H157 cells
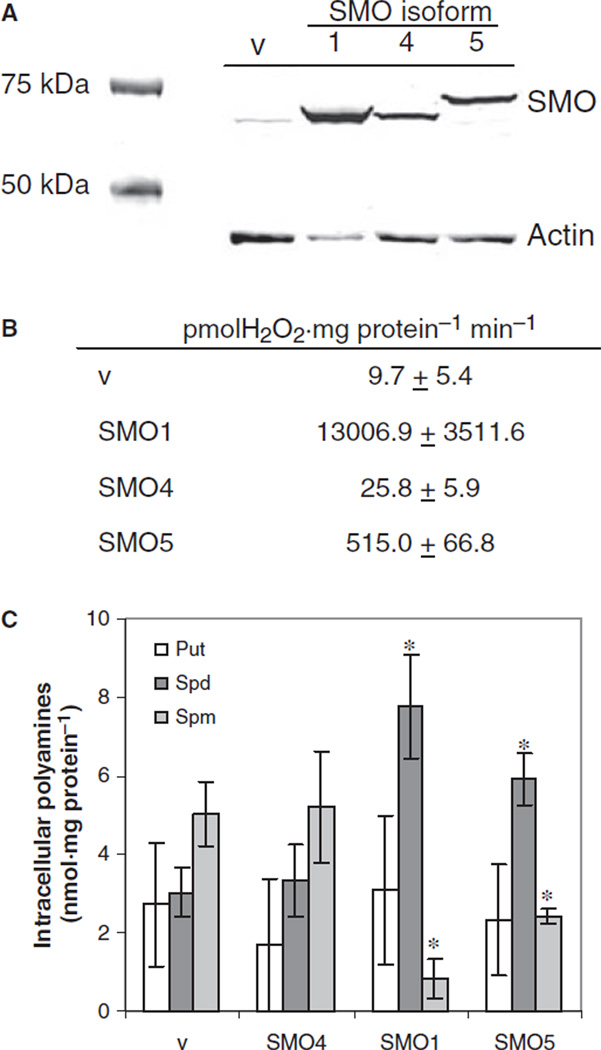
Expression vectors containing coding sequences of SMO/PAOh1, PAOh4 and SMO5 were constructed to determine localization of each protein following stable transfection into H157 human non-small cell lung carcinoma cells. Human SMO antibody [30,31] was used to screen individual stable clones for overexpression of the three SMO isoforms via western blotting. This process was facilitated by the fact that basal expression of SMO in H157 cells is nearly undetectable by western blot analysis. Clones with the highest amounts of each exogenous isoform were selected for further experiments, and designated SMO1, SMO4 and SMO5 (Fig. 4A). Real-time PCR using isoform-specific primer pairs verified specific expression of individual splice variants (data not shown).
Fig. 4.
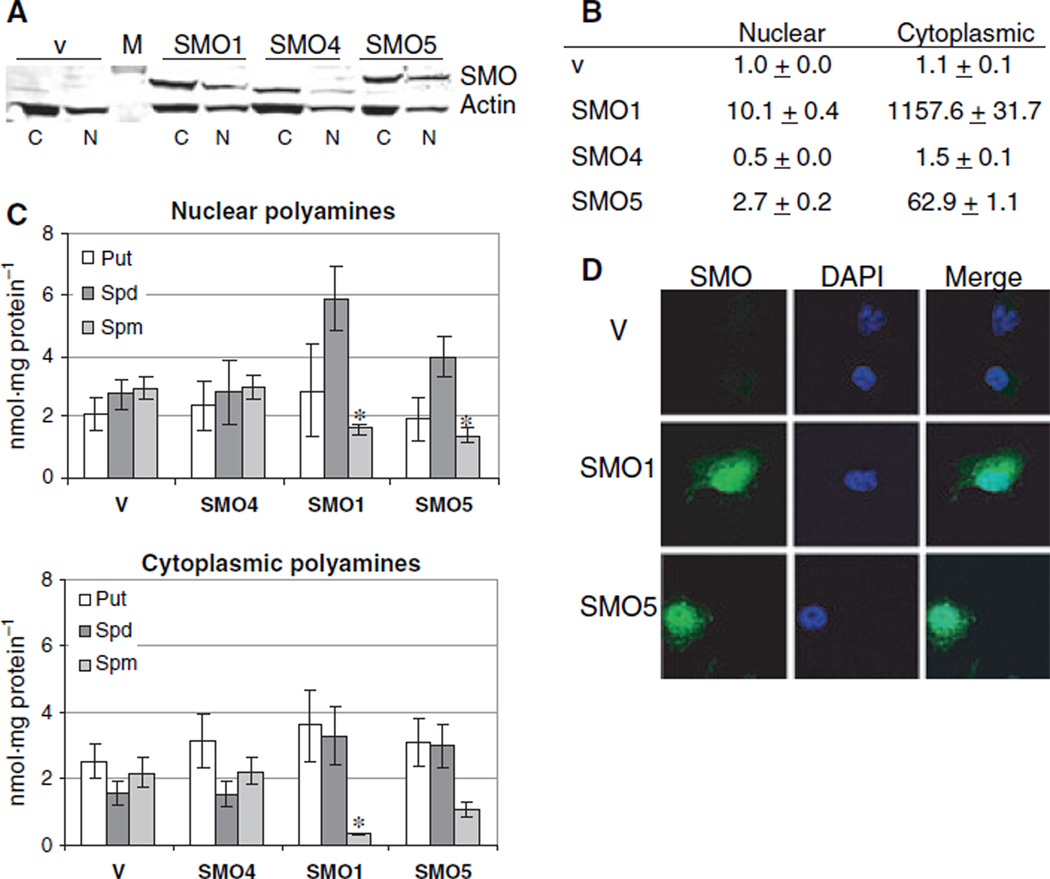
Overexpression of SMO isoforms in NCI-H157 cells. (A) Western blot of total cellular protein from empty vector-transfected control H157 cells (v) and H157 cells stably overexpressing SMO/PAOh1 (SMO1), PAOh4 (SMO4) or SMO5 (SMO5). Total protein (30 µg per lane) was separated on a 10% Novex gel, and immunoblotting was performed using SMO and actin antibodies. Dye-conjugated secondary antibodies were used to detect bound proteins using the Odyssey infrared detection system. The illustrated blot is representative of three separate experiments. (B) For SMO activity assays, 50 µL of cell lysate from each overexpressing cell line was used per reaction, in triplicate, and SMO activity was calculated relative to milligrams of total cellular protein, as determined by the Bradford assay. Graph represents means with standard errors of three separate experiments. (C) Intracellular levels of the polyamines putrescine (Put), spermidine (Spd) and spermine (Spm) were determined by HPLC analysis of dansyl chloride-labeled cell lysates. Data represents the means ± SE of four separate determinations. *Statistically significant differences in individual polyamine levels of SMO overexpressing cells relative to vector control cells (P < 0.02).
SMO activity assays of total cellular protein were used to verify that these cell lines were overexpressing functional spermine oxidase proteins. Not surprisingly, the SMO/PAOh1 clone, SMO1, displayed the highest activity (Fig. 4B). Cells overexpressing SMO5 also displayed significant spermine oxidase activity. As expected, cells overexpressing the inactive splice variant, PAOh4 (clone SMO4), by western blot and real- time PCR, demonstrated spermine oxidase activity no greater than that of the empty vector-transfected control cells.
Polyamine pool analysis of the overexpressing SMO cells further confirmed the increased SMO activity, with decreases in intracellular spermine pools and increased spermidine levels in SMO1 and SMO5 cells. These changes in spermine and spermidine levels in both SMO1 and SMO5 overexpressing cell lines were statistically significant compared to those levels in the vector control cell line (P < 0.02). As expected, cells overexpressing PAOh4 exhibited a polyamine pool profile similar to control cells containing the empty expression vector (Fig. 4C). Importantly, in spite of the much larger amount of SMO activity observed in the SMO/PAOh1 clone relative to the SMO5 clone when assayed in vitro with saturating substrate conditions, there are only slight differences in the intracellular polyamine pools of the two, indicating that the activity of SMO5 is sufficient to catabolize the amount of available spermine in the cell.
Localization of SMO in H157 cells
Western blot analysis and quantification of nuclear and cytoplasmic protein extracts indicated that all three isoforms, SMO/PAOh1, SMO5 and PAOh4, exhibit similar localization patterns when transfected into H157 cells, with significant amounts of spermine oxidase protein present in the nucleus, as well as the cytoplasm (Fig. 5A). This is in contrast to the data regarding the mouse SMO isoforms, of which only the SMO5 homologue has been shown to exist in the nucleus. Because all other mouse spermine oxidases, including the predominant splice variant homologue, show localization exclusively in the cytoplasm [33], our finding that SMO/PAOh1 is present in the nucleus was novel and unexpected. SMO activity assays of SMO/PAOh1 and SMO5 overexpressing nuclear and cytoplasmic extracts corroborate these data, showing spermine oxidase activity and H2O2 generation in both nuclear and cytoplasmic extracts (Fig. 5B). Polyamine pool analyses also verified functional spermine oxidase activity in both fractions, with cells overexpressing SMO/PAOh1 and SMO5 displaying increased nuclear and cytoplasmic spermidine, with significantly diminished spermine levels, compared to vector control or PAOh4-overexpressing cells (Fig. 5C). Furthermore, confocal microscopy of immunofluorescent staining of SMO in cells overexpressing each of the isoforms provided direct visualization and confirmation of protein localization without the possibility of contamination of one fraction with another during protein preparation (Fig. 5D).
Fig. 5.
Functional SMO/PAOh1 and SMO5 are found in both the cytoplasm and nucleus of overexpressing NCI-H157 cells. (A) Western blot analysis of nuclear and cytoplasmic SMO protein from H157 cells overexpressing SMO/PAOh1 (SMO1), PAOh4 (SMO4), SMO5 (SMO5) or empty vector (v). Nuclear (N) or cytoplasmic (C) proteins (30 µg per lane) were separated by SDS/PAGE, transferred to poly(vinylidene difluoride), and incubated with an antibody specific for human SMO. Blot is representative of three independent experiments. M, molecular weight marker. (B) Fold-change in SMO activity of nuclear and cytoplasmic proteins. Nuclear or cytoplasmic protein (25 µL per reaction) was assayed for oxidase activity on spermine, in triplicate. Values were calculated relative to protein amount, as determined by BioRad DC quantification, and represent means ± SD calculated within a single, representative experiment that was repeated three times. (C) Localized polyamine pool analysis. Aliquots of nuclear and cytoplasmic extracts used above for SMO activity assays were dansylated and analyzed for localized polyamine content via HPLC. Concentrations of putrescine (Put), spermidine (Spd), and spermine (Spm) represent duplicate injections in three separate experiments, with standard errors. Cells lines SMO1 and SMO5 both demonstrate active spermine oxidase in both fractions, as depicted by the observed decreases in spermine with accumulation of spermidine. Again, asterisks indicate statistically significant differences in spermine levels of SMO overexpressing cells relative to vector control cells (P < 0.05). Although spermidine levels do appear to be elevated in the cell lines overexpressing active SMO isoforms, these changes were not found to be statistically significant. (D) Both cytoplasm and nuclei stain for SMO/PAOh1 and SMO5 in overexpressing H157 cells. Stably transfected H157 cells were fixed on chamber slides, permeabilized, and incubated with the SMO antibody (1 : 500), followed incubation with Alexa Fluor488 anti-rabbit secondary serum, and DAPI staining. Confocal microscopy was performed with a × 60 objective. Column 1, SMO; column 2, DAPI staining of nuclei, column 3, columns 1 and 2 merged.
Discussion
Several studies have confirmed that the oxidation of polyamines by SMO plays an important role in the antitumor effects of multiple antitumor polyamine analogues that act as inducers of SMO expression [2,12,21,22,25,27]. More recently, a series of studies have implicated SMO activity as a source for potentially mutagenic ROS production and as a direct link between infection, inflammation and carcinogenesis [29–31]. Consequently, a better understanding of the physical cellular localization of SMO has gained in importance. A highly active mouse spermine oxidase isoform, mSMOµ, was recently reported by Cervelli et al. [33] to be localized in both the nucleus and cytoplasm, and its nuclear localization was considered to be unique among the mouse SMO isoforms [28,33]. Although we had previously cloned a number of human spermine oxidase splice variants, as well as truncated proteins, no homologue of the mouse mSMOµ, which is essentially a combination of the human isoforms 1 and 4, had been identified. The discovery of its existence in the mouse suggested the likelihood of a human homologue, and led us to pursue the identification of human SMO5. Furthermore, the implications of spermine oxidase activity in the nucleus prompted us to further investigate the localization of the human SMO isoforms.
SMO5 mRNA was detected by RT-PCR in all human lung cell lines examined. The addition of exon VIa to SMO/PAOh1 to make SMO5 had little effect on the substrate affinities of the purified proteins. However, the kcat values of SMO5 were some-what lower for both spermine and N1-acetylspermine than were those of SMO/PAOh1, thus accounting for the observed difference in specific activities. Because the only difference in the gene structures of the two enzymes is the presence of exon VIa, it appears that its presence may actually hamper the efficiency with which spermine is oxidized by human SMO. Molecular modeling of homologous mouse isoforms [28] place this specifically mammalian, highly conserved, 31 amino acid region on the surface loop of the enzyme in close proximity to the FAD-binding domain, where it may potentially interfere with the reaction. In spite of this difference, by possessing a specific activity of approximately 3 µmol H2O2·mg protein−1·min−1, purified SMO5 protein still very efficiently oxidizes spermine. Furthermore, it should be noted that these proteins have been denatured and refolded; thus, the possibility of altered activities cannot be excluded.
A second region that varies among the three enzymes studied, located within exon V, is present in the active SMO/PAOh1 and SMO5 enzymes, but absent from the inactive PAOh4 protein. This entire 53 amino acid region is highly conserved in mammals, and can be subdivided into a 5′-end that is suggested to be involved in nuclear localization of the mouse spermine oxidase [28], and a 3′ region, which structural modeling of homologous proteins has demonstrated to play an essential role in the oxidase activity of the enzyme through interacting with the FAD cofactor [34,35]. A mouse SMOμ mutant protein was purified that lacks the 31 residues of the 5′ end of this region, while retaining all of its catalytic activity [28]. Intriguingly, the human version of this entire 53 amino acid region is flanked by splice sites, resulting in the production of several catalytically inactive SMO proteins [32], including the PAOh4 protein utilized in the present study. Although many isoforms have been identified for the mouse SMO protein, none display this same pattern of splicing. Analysis of nucleotide sequences surrounding this region reveal a base change (G to A) at the 3′ end of the removed human sequence, resulting in the creation of a 3′ splice consensus sequence. This base change is present only in the human SMO sequence, as even the chimpanzee (Pan troglodytes) sequence (XM 001163724.1), which is approximately 99% homologous to human SMO, retains the guanine nucleotide that has also been reported for Canis familiaris (XM 542910.2), Bos taurus (XM 577020.2), Rattus norvegicus (XM 001079707.1) and Mus musculus (AF495853.1). The essential bases of the 5′ splice consensus sequence are present in human, chimp, rat and mouse sequences, whereas those of the dog and cow show variations in one of the two critical bases.
Despite the findings that, in the cell line studied, endogenous SMO5 appears to be less abundant than SMO/PAOh1, and the maximum velocity of the purified protein is somewhat lower than that of SMO/PAOh1, it is imperative to recognize that SMO5 is an active, efficient oxidase in the polyamine catabolic pathway. This is especially demonstrated by the altered polyamine pool concentrations detected in cells overexpressing SMO5 compared to those transfected with SMO/PAOh1. In spite of the lower expression level of exogenous spermine oxidase displayed by the SMO5 cells as detected by western blot, as well as the lower specific activity of SMO5 and potential for splicing to the inactive form, these cells still exhibit a decrease in spermine and increase in spermidine pools that is only slightly less than that observed in cells overexpressing SMO/PAOh1. This difference is even smaller when considering the 25-fold difference in activity between the two overexpressing cell lines when lysates are assayed in vitro with saturating spermine concentrations. It appears that the oxidase activity of SMO5 is sufficient to efficiently oxidize almost all spermine available in the cell, whereas the extremely high activity of SMO/PAOh1 is limited by intracellular substrate amounts.
In the present study, we demonstrate that both active human isoforms, SMO5 and the predominant splice variant, SMO/PAOh1, exist in the nucleus. By contrast, Cervelli et al. [33] previously reported the existence of only mSMOμ, the mouse SMO5 homologue, in the nucleus, whereas the other mouse SMO splice variants localized exclusively in the cytoplasm. The differences in localization observed may be a result of the indirect detection of the mouse SMO isoforms through interaction with a V5 tag antibody [28,33], as opposed to our detection using an antibody to human spermine oxidase [30,31]. The immunofluorescent detection of SMO in the vector control H157 cells presented here provides direct evidence of nuclear localization of endogenous spermine oxidase in these cells. As noted above, two regions of the human SMO gene are present or absent in different combinations in the three isoforms studied here. Bianchi et al. [28] have suggested that, in the mouse SMO protein, which does not contain evidence of a nuclear localization sequence [33], both the 5′ end of the internal region of exon V, and exon VIa must be present for nuclear localization of their tagged construct. Although highly conserved, with an amino acid identity of 70% to the mouse sequence, exon VIa is apparently not essential for localization of the human protein because SMO/PAOh1 shows significant translocation to, and activity in, the nucleus without it. Furthermore, no apparent differences in localization are observed between SMO/PAOh1 and SMO5, the latter of which contains the extra exon.
The present studies also demonstrated the nuclear presence of the inactive isoform, PAOh4, when overexpressed in H157 cells. It is possible that SMO proteins that are metabolically inactive as oxidases are serving other functions in the nucleus and in the cell in general. A homologous protein, lysine-specific demethylase 1 (LSD1) [36], was recently described which also functions in the nucleus. Although it possesses little oxidase activity on the polyamines, it does play a very important role as a histone lysine demethylase component of transcriptional repression complexes [36]. LSD1 is an FAD-dependent enzyme that acts on mono- and dimethylated lysine 4 of histone H3 through an oxidase reaction almost identical to the activity of SMO [36,37]. Furthermore, the active site of LSD1 is highly homologous to that of SMO/PAOh1 and SMO5 [36]. Therefore, the possibility exists that the spermine oxidase splice variants that do not oxidize spermine may oxidize other, as yet undefined, substrates.
Most importantly, our experiments demonstrating that spermine oxidase activity exists in the nucleus, in the forms of both SMO/PAOh1 and SMO5, emphasizes the functional significance of these SMO enzymes in relation to the production of H2O2 and subsequent ROS. Generally, spermine oxidase activity is very low in cells. However, in cancer cells, where polyamine levels are frequently increased, spermine oxidase activity can be induced by antitumor polyamine analogues, in a tumor-specific manner, leading to ROS generation, lethal DNA damage and apoptotic cell death, as previously reported [2,25,27].
At least equally important to analogue-induced spermine oxidation as a means for chemotherapeutic intervention are our recent discoveries that ROS produced by SMO causes potentially mutagenic DNA damage in cells stimulated by inflammatory cytokines or the infectious agent, H. pylori, the causative agent of peptic ulcers and gastric cancer [29,30,38]. These studies have significant implications in that they strongly implicate spermine oxidase as one direct link between inflammation and carcinogenesis. As such, the proximal nature of SMO-generated H2O2 to DNA may prove to be critical [39].
Either pharmacologically via polyamine analogue treatment, or as a result of inflammation and/or infection, the induction of SMO-produced H2O2 in the nucleus has the obvious potential to increase apoptotic effects, due to the proximity of the ROS source to the target DNA, where the possibility of detoxification of the ROS prior to damage is reduced. Additionally, the oxidation of spermine to spermidine diminishes nuclear spermine pools. Because spermine has essential roles in the the protection of DNA, including free radical scavenging and DNA shielding, this reduction would further contribute to the likelihood of detrimental DNA damage [1]. Our discovery that two active spermine oxidase isoforms, including the predominant splice variant, produce highly reactive H2O2 proximal to DNA may be essential to understanding the mechanism of action of the antitumor polyamine analogues, as well as may further contribute to our understanding of the role of spermine oxidase as a link between inflammation and carcinogenesis.
Experimental procedures
Cell lines and culture conditions
Cell lines used were NCI-H157 human non-small cell lung carcinoma, A549 human lung adenocarcinoma and Beas2B transformed, non-tumorigenic, human lung epithelium (ATCC, Manassas, VA, USA). H157 and A549 cells were maintained in RPMI 1640 media (Mediatech, Inc., Herndon, VA, USA) containing 9% iron-supplemented fetal bovine serum (Hyclone, Logan, UT, USA) and 1% penicillin and streptomycin. Beas2B cells were maintained in LHC-9 serum-free medium (Invitrogen, Carlsbad, CA, USA) with 1% penicillin and streptomycin. All cells were kept at 37 °C, 5% CO2. A549 cells were also treated for 24 h with 10 µm BENSpm, an antitumor polyamine analogue that induces expression of SMO/PAOh1 mRNA in these cells.
SMO splice variant-specific mRNA expression in human lung cell lines
The existence of SMO5 mRNA in various human lung cell lines was verified using RT-PCR with primers specific for the exon structure of SMO5 to amplify a 653 bp fragment (Fig. 1). Primer sequences used were: 5′-GATCCCGGCGGACCATGTGATTGTG-3′ and 5′-TTTACGGCGCCCCTGTTAGCATCC-3′. Total cellular RNA was isolated from cells using Trizol reagent according the manufacturer’s instructions, and reverse transcriptase PCR was performed using SuperScriptII One-Step RT-PCR with Platinum Taq (Invitrogen).
Expression levels were further quantified using SYBR green-mediated real-time PCR with primer pairs specific for SMO5, SMO/PAOh1, PAOh4 or GAPDH. Trizol-extracted RNA was treated with DNAse I, and cDNA was produced using M-MLV reverse transcriptase with an oligo d(T) primer (Invitrogen). QuantiTect SYBR green Taq polymerase was purchased from Qiagen (Valencia, CA, USA), and real-time PCR was performed on a BioRad iCycler My IQ single color real-time PCR detection system (Hercules, CA, USA). Primer sequences used for SMO5 were the same as those described above for RT-PCR. SMO/PAOh1 real-time primers were: 5′-GATCCCGGCGGACCATGTGATT GTG-3′ and 5′-CCTGCATGGGCGCTGTCTTTG-3′. PAOh4 primers were: 5′-GCCCCGGGGTGTGCTAAA GAG-3′ and 5′-TTTACGGCGCCCCTGTTAGCATCC-3′. GAPDH primers were: 5′-GAAGGTGAAGGTCGGA GTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. All primers were manufactured by Invitrogen.
Human SMO antibody production and western blot analysis
Rabbit, polyclonal, anti-human SMO serum was produced by standard methods using the peptide sequence Ac-CIHWDQASARPRGPEIEPR-amide, and antisera were collected and affinity purified [30]. The peptide corresponds to amino acids 263–281 of SMO/PAOh1, located in the 5′ region of exon 5, thus recognizing splice variants 1, 4 and 5.
For detection of endogenous SMO5 and PAOh1 /SMO proteins, A549 cells were treated with BENSpm as above and total cellular protein extracted. Expression of SMO was detected by western blotting using 30 µg of total protein per lane on 10% Bis-Tris Novex gels (Invitrogen) as previously reported [30]. Briefly, gels were run at 200 V in Mops buffer, transferred onto activated Immunblot poly(vinylidene difluoride) membrane (BioRad) for 1 h at 30 V, and blocked for 1 h in Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE, USA). Rabbit SMO and mouse actin (Sigma, St Louis, MO, USA) primary antibodies were then added together at dilutions of 1 : 1000 and 1 : 1500, respectively, with 0.1% Tween 20 in blocking buffer for 1 h at room temperature. Following washes with NaCl/Pi-Tween, blots were incubated with rabbit IgG4 IRDye800 (Rockland Immunochemicals, Gilbertsville, PA, USA) and mouse IgG4 Alexa Fluor680 (Molecular Probes, Carlsbad, CA, USA) dye-conjugated secondary antibodies (1 : 4000 each, 0.1% Tween 20, in blocking buffer, protected from light, for 45 min), which allowed detection of each protein using an Odyssey infrared detection system and software (LI-COR Biosciences).
Construction and purification of recombinant SMO5/pET15b
PAOh4 cDNA was cloned into the pET15b bacterial expression vector (Novagen, Madison, WI, USA) using the method previously described for SMO/PAOh1 [23]. The resulting pET15b constructs, containing either SMO/PAOh1 or PAOh4, were then combined to produce SMO5/pET15b. Specifically, the XbaI to KpnI fragment of SMO/PAOh1 in pET15b was isolated and ligated to the purified KpnI to XbaI fragment of PAOh4 in pET15b. The resultant SMO5/pET15b cDNA therefore contains exons of SMO/PAOh1 that are 5′ of the KpnI site, as well as all exons of PAOh4 that are 3′ of the KpnI site (Fig. 1A). The SMO5/pET15b cDNA sequence was verified using an ABI Prism automated sequencer (Applied Biosystems, Foster City, CA, USA). Restriction and modification enzymes were purchased from New England Biolabs (Beverly, MA, USA).
BL(21)DE3 chemically competent E. coli cells were transformed with the SMO/PAOh1, PAOh4 or SMO5 expression construct DNA, and transformants were selected on LB agar plates in the presence of 50 µg·mL−1 ampicillin. Liquid LB cultures were grown and recombinant protein production was induced by the addition of 1 mm isopropyl thio-β-d-galactoside for 3 h at 37 °C. Each protein was denatured, purified by affinity chromatography with Ni-NTA resin (Qiagen), and refolded in the presence of 0.2 µm FAD using a Slide-a-lyzer dialysis cassette (Pierce, Rockford, IL, USA) as previously described [23]. Approximate protein size was verified on a precast 10% Bis-Tris polyacrylamide Novex gel (Invitrogen) stained with Coomassie brilliant blue and photographed using a Kodak EDAS 290 scientific imaging system (New Haven, CT, USA).
Spermine oxidase activity and substrate specificity
Purified recombinant SMO5, along with SMO/PAOh1, was assayed for oxidase activity using a chemiluminescent detection method as previously described [23]. Various polyamines, as well as synthetic polyamine analogues, were assayed as potential substrates at a concentration of 250 µm. Spermine, spermidine, N1,N12-diacetylspermine, N8-acetylspermidine and N1-acetylspermidine were purchased from Sigma, and N1-acetylspermine was purchased from Fluka (Buchs, Switzerland). The polyamine analogues, CPENSpm and BENSpm, were synthesized as previously described [40]. The polyamine oxidase inhibitor, MDL72,527 [41], was also utilized in these experiments, at equimolar concentration as substrate. Protein concentration was determined using the method of Bradford with reagents from BioRad.
Kinetic analysis of polyamine oxidase activity
Oxidase activity was assessed using fixed concentrations of purified enzymes with concentrations of spermine or N1-acetylspermine in the range 0.1–250 µm. Values were plotted and kinetic parameters were determined using the Lineweaver–Burk transformation of the Michaelis–Menton equation.
Construction of spermine oxidase mammalian expression plasmids
For expression studies in mammalian cell lines, SMO5, SMO/PAOh1, or PAOh4 cDNAs were each cloned into the phCMV3 vector (Gene Therapy Systems, San Diego, CA, USA) using PCR with the primer pair 5′-CAATCCTC GAGTATGCAAAGTTGTGAATCCAG-3′ and 5′-TAA TAAGCTTTGGTCCCCTGCTGGAAGAGGTC-3′ and each cDNA in pET15b as the template. PCR products were restriction digested with XhoI and HindIII (underlined primer sequences), and ligated into the same restriction sites of phCMV3. Sequences were verified by sequencing.
Overexpression of human spermine oxidase isoforms in NCI-H157 cells
NCI-H157 human non-small cell lung carcinoma cells were seeded in six-well plates and transfected for 4.5 h with each of the SMO expression constructs from above using 1.3 µg of DNA with 3.7 µL Lipofectin per well (Invitrogen). Stable colonies were isolated following selection with 0.4 mg·mL−1 G418. Colonies were screened for increased expression of SMO by western blotting using 30 µg of total protein per lane as described above. Overexpressing clones for each isoform were further analyzed for spermine oxidase isoform-specific mRNA expression using real-time PCR as described above, as well as for spermine oxidase activity [23] and polyamine pool levels [42], as previously described.
Cellular localization of SMO isoforms
The western blotting method and antibodies described above were also used to detect the presence of the SMO protein in nuclear versus cytoplasmic protein fractions, which were isolated using Pierce NE-PER nuclear and cytoplasmic extraction reagents, and quantified using Bio- Rad DC reagents. Thirty micrograms of lysate was loaded per lane and immunoblotting was performed as described above. These same lysates were further analyzed for spermine oxidase activity, using 25 µL of nuclear or cytoplasmic lysate per reaction in the luminol-based assay previously described [23]. Additionally, 50 µL of nuclear or cytoplasmic lysate were dansylated and analyzed by HPLC for localized quantitation of polyamine levels [42]. Statistical significance of changes in individual polyamine pools was analyzed using Student’s t-test.
To further validate localization of the chosen isoforms, we used immunofluorescent staining of the transfected H157 cells. Cells were plated on eight-well chamber slides, allowed to attach overnight at 37 °C, 5% CO2 and fixed in 10% formalin for 4 h. Slides were placed in 1% Tween 20 for 1 min, then steamed for 40 min in high temperature target retrieval solution (Dako, Carpinteria, CA, USA). SMO antibody was added at 1 : 500 in antibody dilution buffer (Dako) and incubated for 45 min at room temperature. Following NaCl/Pi-Tween washes, anti-rabbit Alexa Fluor488 dye-conjugated secondary serum (Molecular Probes) was added at a dilution of 1 : 100 in Dulbecco’s NaCl/Pi and incubated for 30 min. After additional NaCl/Pi-Tween washes, nuclei were counterstained for 1 min with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma), and cells were observed and analyzed using a Nikon C1si confocal microscope system (Melville, NY, USA) with ×60 oil immersion magnification.
Acknowledgements
This work was funded by NIH Grants CA51085 and CA98454, the Samuel Waxman Cancer Research Foundation, and the Patrick C. Walsh Prostate Cancer Research Fund, for which R. A. C. is the Schwartz Scholar.
Abbreviations
- BENSpm
bis(ethyl)norspermine
- CPENSpm
N1-ethyl-N11-(cyclopropyl)methyl-4,8,diazaundecane
- DAPI
4′,6′-diamidino-2-phenylindole
- LSD1
lysine-specific demethylase 1
- ROS
reactive oxygen species
- SMO/PAOh1
human spermine oxidase-1
- SMO5
spermine oxidase-5
References
- 1.Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA., Jr The natural polyamine spermine functions directly as a free radical scavenger. Proc Natl Acad Sci U S A. 1998;95:11140–11145. doi: 10.1073/pnas.95.19.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ha HC, Woster PM, Yager JD, Casero RA., Jr The role of polyamine catabolism in polyamine analogue-induced programmed cell death. Proc Natl Acad Sci U S A. 1997;94:11557–11562. doi: 10.1073/pnas.94.21.11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pegg AE, McCann PP. Polyamine metabolism and function. Am J Physiol. 1982;243:C212–C221. doi: 10.1152/ajpcell.1982.243.5.C212. [DOI] [PubMed] [Google Scholar]
- 4.Seiler N, Raul F. Polyamines and apoptosis. J Cell Mol Med. 2005;9:623–642. doi: 10.1111/j.1582-4934.2005.tb00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas T, Thomas TJ. Polyamine metabolism and cancer. J Cell Mol Med. 2003;7:113–126. doi: 10.1111/j.1582-4934.2003.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson NE, Mank AR, Prestigiacomo LJ, Bergeron RJ, Casero RA., Jr Growth inhibition of hormone-responsive and -resistant human breast cancer cells in culture by N1, N12-bis(ethyl)spermine. Cancer Res. 1993;53:2071–2075. [PubMed] [Google Scholar]
- 8.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Pledgie A, Casero RA, Jr, Davidson NE. Molecular mechanisms of polyamine analogs in cancer cells. Anticancer Drugs. 2005;16:229–241. doi: 10.1097/00001813-200503000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Kingsnorth AN, Wallace HM, Bundred NJ, Dixon JM. Polyamines in breast cancer. Br J Surg. 1984;71:352–356. doi: 10.1002/bjs.1800710513. [DOI] [PubMed] [Google Scholar]
- 11.Wallace HM, Duthie J, Evans DM, Lamond S, Nicoll KM, Heys SD. Alterations in polyamine catabolic enzymes in human breast cancer tissue. Clin Cancer Res. 2000;6:3657–3661. [PubMed] [Google Scholar]
- 12.Casero RA, Jr, Wang Y, Stewart TM, Devereux W, Hacker A, Wang Y, Smith R, Woster PM. The role of polyamine catabolism in anti-tumour drug response. Biochem Soc Trans. 2003;31:361–365. doi: 10.1042/bst0310361. [DOI] [PubMed] [Google Scholar]
- 13.Casero RA, Jr, Woster PM. Terminally alkylated polyamine analogues as chemotherapeutic agents. J Med Chem. 2001;44:1–26. doi: 10.1021/jm000084m. [DOI] [PubMed] [Google Scholar]
- 14.Casero RA, Jr, Celano P, Ervin SJ, Porter CW, Bergeron RJ, Libby PR. Differential induction of spermidine / spermine N1-acetyltransferase in human lung cancer cells by the bis(ethyl)polyamine analogues. Cancer Res. 1989;49:3829–3833. [PubMed] [Google Scholar]
- 15.Porter CW, Ganis B, Libby PR, Bergeron RJ. Correlations between polyamine analogue-induced increases in spermidine / spermine N1-acetyltransferase activity, polyamine pool depletion, and growth inhibition in human melanoma cell lines. Cancer Res. 1991;51:3715–3720. [PubMed] [Google Scholar]
- 16.Chang BK, Liang Y, Miller DW, Bergeron RJ, Porter CW, Wang G. Effects of diethyl spermine analogues in human bladder cancer cell lines in culture. J Urol. 1993;150:1293–1297. doi: 10.1016/s0022-5347(17)35763-4. [DOI] [PubMed] [Google Scholar]
- 17.Casero RA, Jr, Pegg AE. Spermidine / spermine N1-acetyltransferase – the turning point in polyamine metabolism. FASEB J. 1993;7:653–661. [PubMed] [Google Scholar]
- 18.Seiler N. Polyamine oxidase, properties and functions. Prog Brain Res. 1995;106:333–344. doi: 10.1016/s0079-6123(08)61229-7. [DOI] [PubMed] [Google Scholar]
- 19.Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. Genomic identification and biochemical characterization of the mammalian polyamine oxidase involved in polyamine back-conversion. Biochem J. 2003;370:19–28. doi: 10.1042/BJ20021779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu T, Yankovskaya V, McIntire WS. Cloning, sequencing, and heterologous expression of the murine peroxisomal flavoprotein, N1-acetylated polyamine oxidase. J Biol Chem. 2003;278:20514–20525. doi: 10.1074/jbc.M302149200. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA., Jr Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61:5370–5373. [PubMed] [Google Scholar]
- 22.Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem J. 2002;367:665–675. doi: 10.1042/BJ20020720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA., Jr Properties of purified recombinant human polyamine oxidase, PAOh1/SMO. Biochem Biophys Res Commun. 2003;304:605–611. doi: 10.1016/s0006-291x(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 24.McCloskey DE, Yang J, Woster PM, Davidson NE, Casero RA., Jr Polyamine analogue induction of programmed cell death in human lung tumor cells. Clin Cancer Res. 1996;2:441–446. [PubMed] [Google Scholar]
- 25.Pledgie A, Huang Y, Hacker A, Zhang Z, Woster PM, Davidson NE, Casero RA., Jr Spermine oxidase SMO(PAOh1), Not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J Biol Chem. 2005;280:39843–39851. doi: 10.1074/jbc.M508177200. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Kramer DL, Diegelman P, Vujcic S, Porter CW. Apoptotic signaling in polyamine analogue-treated SK-MEL-28 human melanoma cells. Cancer Res. 2001;61:6437–6444. [PubMed] [Google Scholar]
- 27.Devereux W, Wang Y, Stewart TM, Hacker A, Smith R, Frydman B, Valasinas AL, Reddy VK, Marton LJ, Ward TD, et al. Induction of the PAOh1/SMO polyamine oxidase by polyamine analogues in human lung carcinoma cells. Cancer Chemother Pharmacol. 2003;52:383–390. doi: 10.1007/s00280-003-0662-4. [DOI] [PubMed] [Google Scholar]
- 28.Bianchi M, Amendola R, Federico R, Polticelli F, Mariottini P. Two short protein domains are responsible for the nuclear localization of the mouse spermine oxidase mu isoform. Febs J. 2005;272:3052–3059. doi: 10.1111/j.1742-4658.2005.04718.x. [DOI] [PubMed] [Google Scholar]
- 29.Xu H, Chaturvedi R, Cheng Y, Bussiere FI, Asim M, Yao MD, Potosky D, Meltzer SJ, Rhee JG, Kim SS, et al. Spermine oxidation induced by Helicobacter pylori results in apoptosis and DNA damage: implications for gastric carcinogenesis. Cancer Res. 2004;64:8521–8525. doi: 10.1158/0008-5472.CAN-04-3511. [DOI] [PubMed] [Google Scholar]
- 30.Babbar N, Casero RA., Jr Tumor necrosis factor-alpha increases reactive oxygen species by inducing spermine oxidase in human lung epithelial cells: a potential mechanism for inflammation-induced carcinogenesis. Cancer Res. 2006;66:11125–11130. doi: 10.1158/0008-5472.CAN-06-3174. [DOI] [PubMed] [Google Scholar]
- 31.Goodwin AC, Jadallah S, Toubaji A, Lecksell K, Hicks JL, Kowalski J, Bova GS, deMarzo AM, Netto GJ, Casero RA. Increased spermine oxidase expression in human prostate cancer and prostatic intraepithelial neoplasia tissues. Prostate. 2008:68. doi: 10.1002/pros.20735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray-Stewart T, Wang Y, Devereux W, Casero RA., Jr Cloning and characterization of multiple human polyamine oxidase splice variants that code for isoenzymes with different biochemical characteristics. Biochem J. 2002;368:673–677. doi: 10.1042/BJ20021587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cervelli M, Bellini A, Bianchi M, Marcocci L, Nocera S, Polticelli F, Federico R, Amendola R, Mariottini P. Mouse spermine oxidase gene splice variants. Nuclear subcellular localization of a novel active isoform. Eur J Biochem. 2004;271:760–770. doi: 10.1111/j.1432-1033.2004.03979.x. [DOI] [PubMed] [Google Scholar]
- 34.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 35.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat Struct Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Chaturvedi R, Cheng Y, Asim M, Bussiere FI, Xu H, Gobert AP, Hacker A, Casero RA, Jr, Wilson KT. Induction of polyamine oxidase 1 by Helicobacter pylori causes macrophage apoptosis by hydrogen peroxide release and mitochondrial membrane depolarization. J Biol Chem. 2004;279:40161–40173. doi: 10.1074/jbc.M401370200. [DOI] [PubMed] [Google Scholar]
- 39.Babbar N, Murray-Stewart T, Casero RA., Jr Inflammation and polyamine catabolism: the good, the bad and the ugly. Biochem Soc Trans. 2007;35:300–304. doi: 10.1042/BST0350300. [DOI] [PubMed] [Google Scholar]
- 40.Bergeron RJ, Neims AH, McManis JS, Hawthorne TR, Vinson JR, Bortell R, Ingeno MJ. Synthetic polyamine analogues as antineoplastics. J Med Chem. 1988;31:1183–1190. doi: 10.1021/jm00401a019. [DOI] [PubMed] [Google Scholar]
- 41.Bey P, Bolkenius FN, Seiler N, Casara P. N-2,3-Butadienyl-1,4-butanediamine derivatives: potent irreversible inactivators of mammalian polyamine oxidase. J Med Chem. 1985;28:1–2. doi: 10.1021/jm00379a001. [DOI] [PubMed] [Google Scholar]
- 42.Kabra PM, Lee HK, Lubich WP, Marton LJ. Solid-phase extraction and determination of dansyl derivatives of unconjugated and acetylated polyamines by reversed-phase liquid chromatography: improved separation systems for polyamines in cerebrospinal fluid, urine and tissue. J Chromatogr. 1986;380:19–32. doi: 10.1016/s0378-4347(00)83621-x. [DOI] [PubMed] [Google Scholar]