Abstract
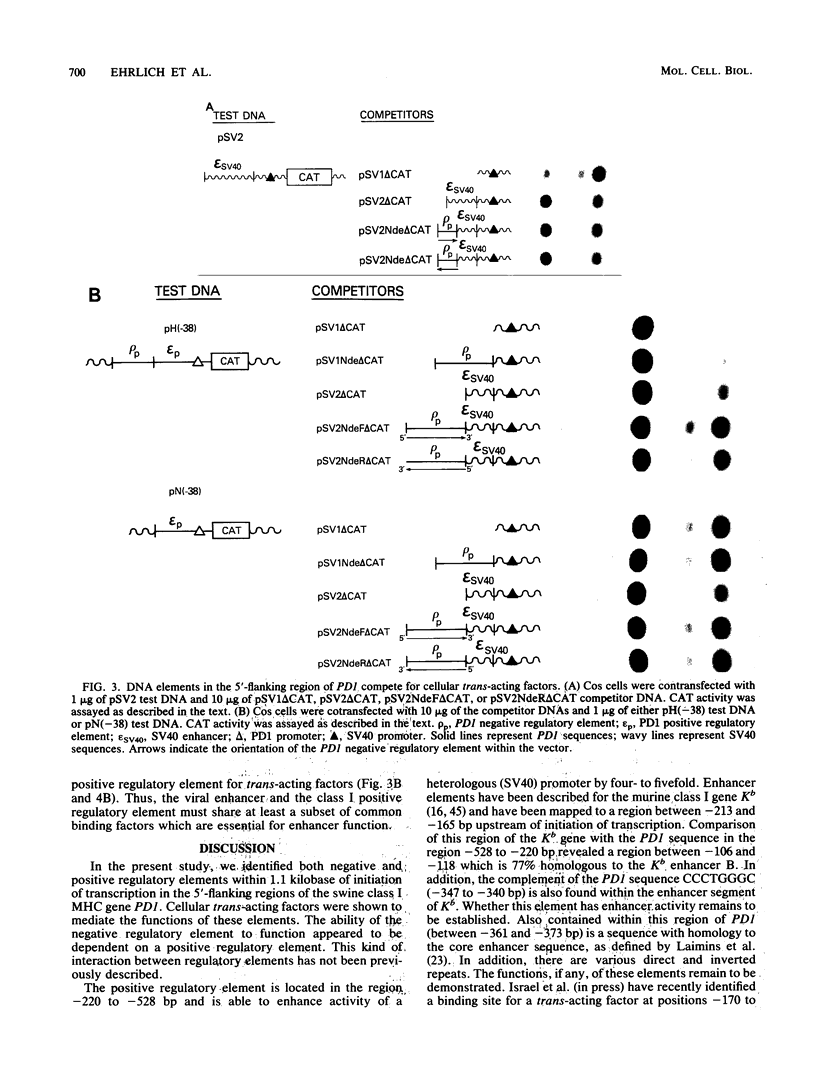
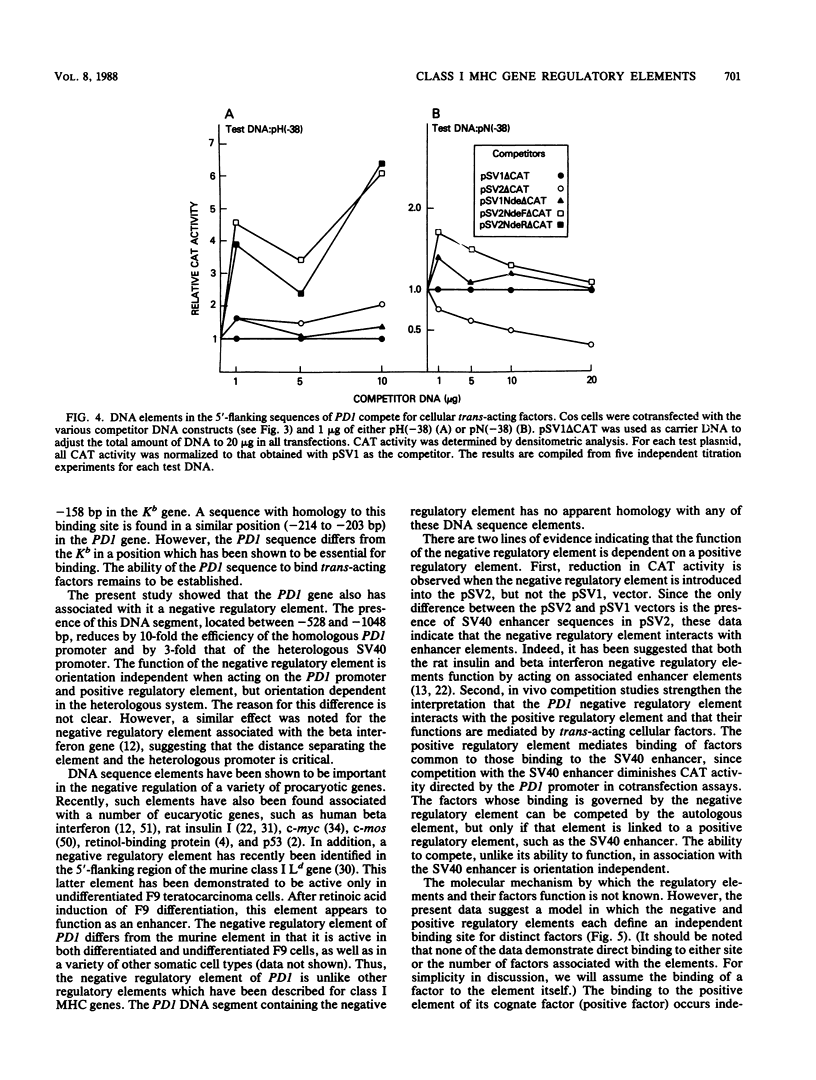
Regulatory DNA sequence elements were functionally identified in the 5'-flanking region of a gene, PD1, which encodes a porcine classical transplantation antigen. Both a positive regulatory element and a novel negative regulatory DNA element were mapped within 1.1 kilobases upstream of exon 1. The negative regulatory element reduced the activity of both the homologous PD1 promoter and a heterologous simian virus 40 promoter. In vivo competition experiments indicated that the functions of the PD1 positive and negative regulatory elements are mediated by distinct cellular trans-acting factors. The PD1 positive regulatory element interacted with cellular factors in common with those binding to the simian virus 40 enhancer. Finally, the negative regulatory element required the presence of a positive regulatory element to function. This interaction between positive and negative regulatory elements represents a novel mechanism for regulating gene expression.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bienz-Tadmor B., Zakut-Houri R., Libresco S., Givol D., Oren M. The 5' region of the p53 gene: evolutionary conservation and evidence for a negative regulatory element. EMBO J. 1985 Dec 1;4(12):3209–3213. doi: 10.1002/j.1460-2075.1985.tb04067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrone O. R., Milstein C. Control of HLA-A,B,C synthesis and expression in interferon-treated cells. EMBO J. 1982;1(3):345–349. doi: 10.1002/j.1460-2075.1982.tb01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Paul P., Font M. P., Cohen O., Sayagh B., Marcadet A., Busson M., Mahouy G., Cann H. M., Dausset J. Analysis of HLA class I genes with restriction endonuclease fragments: implications for polymorphism of the human major histocompatibility complex. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6289–6292. doi: 10.1073/pnas.80.20.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni V., Pirozzi A., Blance C., Cortese R. Negative control of liver-specific gene expression: cloned human retinol-binding protein gene is repressed in HeLa cells. EMBO J. 1987 Mar;6(3):631–636. doi: 10.1002/j.1460-2075.1987.tb04801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T., Lapierre L. A., Fiers W., Strominger J. L., Pober J. S. Recombinant human tumor necrosis factor increases mRNA levels and surface expression of HLA-A,B antigens in vascular endothelial cells and dermal fibroblasts in vitro. Proc Natl Acad Sci U S A. 1986 Jan;83(2):446–450. doi: 10.1073/pnas.83.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J. J., Weiss E. H., Paulson M., Flavell R. A. Duplicated gene pairs and alleles of class I genes in the Qa2 region of the murine major histocompatibility complex: a comparison. EMBO J. 1985 Dec 1;4(12):3203–3207. doi: 10.1002/j.1460-2075.1985.tb04066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Faulk W. P., Sanderson A. R., Temple A. Distribution of MHC antigens in human placental chorionic villi. Transplant Proc. 1977 Jun;9(2):1379–1384. [PubMed] [Google Scholar]
- Frels W. I., Bluestone J. A., Hodes R. J., Capecchi M. R., Singer D. S. Expression of a microinjected porcine class I major histocompatibility complex gene in transgenic mice. Science. 1985 May 3;228(4699):577–580. doi: 10.1126/science.3885396. [DOI] [PubMed] [Google Scholar]
- Friedman R. L., Stark G. R. alpha-Interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature. 1985 Apr 18;314(6012):637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Zinn K., Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985 Jun;41(2):509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Pellegrino M. A., Ng A. K., Ferrone S. Role of antigen density in immune lysis of interferon-treated human lymphoid cells: analysis with monoclonal antibodies to the HLA-A,B antigenic molecular complex and to Ia-like antigens. Scand J Immunol. 1981 Nov;14(5):529–535. [PubMed] [Google Scholar]
- Israel A., Kimura A., Fournier A., Fellous M., Kourilsky P. Interferon response sequence potentiates activity of an enhancer in the promoter region of a mouse H-2 gene. Nature. 1986 Aug 21;322(6081):743–746. doi: 10.1038/322743a0. [DOI] [PubMed] [Google Scholar]
- Israël A., Kimura A., Kieran M., Yano O., Kanellopoulos J., Le Bail O., Kourilsky P. A common positive trans-acting factor binds to enhancer sequences in the promoters of mouse H-2 and beta 2-microglobulin genes. Proc Natl Acad Sci U S A. 1987 May;84(9):2653–2657. doi: 10.1073/pnas.84.9.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Israël A., Le Bail O., Kourilsky P. Detailed analysis of the mouse H-2Kb promoter: enhancer-like sequences and their role in the regulation of class I gene expression. Cell. 1986 Jan 31;44(2):261–272. doi: 10.1016/0092-8674(86)90760-9. [DOI] [PubMed] [Google Scholar]
- Klein J. The major histocompatibility complex of the mouse. Science. 1979 Feb 9;203(4380):516–521. doi: 10.1126/science.104386. [DOI] [PubMed] [Google Scholar]
- Korber B., Hood L., Stroynowski I. Regulation of murine class I genes by interferons is controlled by regions located both 5' and 3' to the transcription initiation site. Proc Natl Acad Sci U S A. 1987 May;84(10):3380–3384. doi: 10.1073/pnas.84.10.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L., Holmgren-König M., Khoury G. Transcriptional "silencer" element in rat repetitive sequences associated with the rat insulin 1 gene locus. Proc Natl Acad Sci U S A. 1986 May;83(10):3151–3155. doi: 10.1073/pnas.83.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson L. A., Fisher C. A., Whelan J. P. Striking paucity of HLA-A, B, C and beta 2-microglobulin on human neuroblastoma cell lines. J Immunol. 1983 May;130(5):2471–2478. [PubMed] [Google Scholar]
- Malissen M., Malissen B., Jordan B. R. Exon/intron organization and complete nucleotide sequence of an HLA gene. Proc Natl Acad Sci U S A. 1982 Feb;79(3):893–897. doi: 10.1073/pnas.79.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marche P. N., Tykocinski M. L., Max E. E., Kindt T. J. Structure of a functional rabbit class I MHC gene: similarity to human class I genes. Immunogenetics. 1985;21(1):71–82. doi: 10.1007/BF00372243. [DOI] [PubMed] [Google Scholar]
- Meruelo D., Kornreich R., Rossomando A., Pampeno C., Mellor A. L., Weiss E. H., Flavell R. A., Pellicer A. Murine leukemia virus sequences are encoded in the murine major histocompatibility complex. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1804–1808. doi: 10.1073/pnas.81.6.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J., Appella E., Ozato K. Negative regulation of the major histocompatibility class I gene in undifferentiated embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9537–9541. doi: 10.1073/pnas.83.24.9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir U., Walker M. D., Rutter W. J. Regulation of rat insulin 1 gene expression: evidence for negative regulation in nonpancreatic cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3180–3184. doi: 10.1073/pnas.83.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. T., Bach F. H., Ploegh H. L., Strominger J. L., Kavathas P., DeMars R. Use of HLA loss mutants to analyse the structure of the human major histocompatibility complex. Nature. 1982 Apr 1;296(5856):454–456. doi: 10.1038/296454a0. [DOI] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers E. F., Yang J. Q., Marcu K. B. A negative transcriptional control element located upstream of the murine c-myc gene. EMBO J. 1986 May;5(5):899–904. doi: 10.1002/j.1460-2075.1986.tb04301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satz M. L., Singer D. S. Effect of mouse interferon on the expression of a porcine major histocompatibility gene introduced into mouse L cells. J Immunol. 1984 Jan;132(1):496–501. [PubMed] [Google Scholar]
- Satz M. L., Wang L. C., Singer D. S., Rudikoff S. Structure and expression of two porcine genomic clones encoding class I MHC antigens. J Immunol. 1985 Sep;135(3):2167–2175. [PubMed] [Google Scholar]
- Schrier P. I., Bernards R., Vaessen R. T., Houweling A., van der Eb A. J. Expression of class I major histocompatibility antigens switched off by highly oncogenic adenovirus 12 in transformed rat cells. 1983 Oct 27-Nov 2Nature. 305(5937):771–775. doi: 10.1038/305771a0. [DOI] [PubMed] [Google Scholar]
- Singer D. S., Camerini-Otero R. D., Satz M. L., Osborne B., Sachs D., Rudikoff S. Characterization of a porcine genomic clone encoding a major histocompatibility antigen: expression in mouse L cells. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1403–1407. doi: 10.1073/pnas.79.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Moore K. W., Frelinger J. G., Sher B. T., Shen F. W., Boyse E. A., Hood L. A pseudogene homologous to mouse transplantation antigens: transplantation antigens are encoded by eight exons that correlate with protein domains. Cell. 1981 Sep;25(3):683–692. doi: 10.1016/0092-8674(81)90175-6. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Winoto A., Minard K., Hood L. Clusters of genes encoding mouse transplantation antigens. Cell. 1982 Mar;28(3):489–498. doi: 10.1016/0092-8674(82)90203-3. [DOI] [PubMed] [Google Scholar]
- Strachan T., Sodoyer R., Damotte M., Jordan B. R. Complete nucleotide sequence of a functional class I HLA gene, HLA-A3: implications for the evolution of HLA genes. EMBO J. 1984 Apr;3(4):887–894. doi: 10.1002/j.1460-2075.1984.tb01901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita K., Miyazaki J., Appella E., Ozato K. Interferons increase transcription of a major histocompatibility class I gene via a 5' interferon consensus sequence. Mol Cell Biol. 1987 Jul;7(7):2625–2630. doi: 10.1128/mcb.7.7.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Kress M., Khoury G., Jay G. A transcriptional enhancer and an interferon-responsive sequence in major histocompatibility complex class I genes. Mol Cell Biol. 1986 Oct;6(10):3550–3554. doi: 10.1128/mcb.6.10.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. H., Golden L., Fahrner K., Mellor A. L., Devlin J. J., Bullman H., Tiddens H., Bud H., Flavell R. A. Organization and evolution of the class I gene family in the major histocompatibility complex of the C57BL/10 mouse. Nature. 1984 Aug 23;310(5979):650–655. doi: 10.1038/310650a0. [DOI] [PubMed] [Google Scholar]
- Weiss E., Golden L., Zakut R., Mellor A., Fahrner K., Kvist S., Flavell R. A. The DNA sequence of the H-2kb gene: evidence for gene conversion as a mechanism for the generation of polymorphism in histocompatibilty antigens. EMBO J. 1983;2(3):453–462. doi: 10.1002/j.1460-2075.1983.tb01444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. G., McGeady M. L., Baroudy B. M., Blair D. G., Vande Woude G. F. Mouse c-mos oncogene activation is prevented by upstream sequences. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7817–7821. doi: 10.1073/pnas.81.24.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn K., Maniatis T. Detection of factors that interact with the human beta-interferon regulatory region in vivo by DNAase I footprinting. Cell. 1986 May 23;45(4):611–618. doi: 10.1016/0092-8674(86)90293-x. [DOI] [PubMed] [Google Scholar]