Abstract
Orientation of the cell division axis is essential for the correct development and maintenance of tissue morphology, both for symmetric cell divisions and for the asymmetric distribution of fate determinants during, for example, stem cell divisions. Oriented cell division depends on the positioning of the mitotic spindle relative to an axis of polarity. Recent studies have illuminated an expanding list of spindle orientation regulators, and a molecular model for how cells couple cortical polarity with spindle positioning has begun to emerge. Here, we review both the well-established spindle orientation pathways and recently identified regulators, focusing on how communication between the cell cortex and the spindle is achieved, to provide a contemporary view of how positioning of the mitotic spindle occurs.
Keywords: Spindle, Microtubules, Oriented cell division, Polarity, Mitosis, Centrosome
Introduction
All multicellular animals are tasked with two fundamental developmental challenges: generating cellular diversity and forming three-dimensional tissues, both of which initiate from a single-celled zygote. Cellular diversity is spawned by cell divisions yielding non-identical daughters, and tissue morphogenesis is established through the precise three-dimensional arrangement of cell divisions that form the overall architecture of the organism. Both of these essential challenges are resolved in part through oriented cell division, which regulates embryogenesis, organogenesis and cellular differentiation. Notably, oriented cell divisions, and hence asymmetric cell divisions, remain crucial throughout adulthood as well, functioning as the basis for tissue homeostasis during growth and wound repair. One primary feature of oriented cell division is the proper positioning of the mitotic spindle relative to a defined polarity axis. In principle, spindle orientation is achieved through signaling pathways that provide a molecular link between the cell cortex and spindle microtubules. These pathways are thought to elicit both static connections and dynamic forces on the spindle to achieve the desired orientation prior to cell division. Although our knowledge of the signaling molecules involved in this process and our understanding of how they each function at the molecular level remain limited, collective efforts over the years have shed light on the importance of spindle orientation to animal development and function. Moreover, emerging evidence shows an association between improper spindle orientation and a number of developmental diseases as well as tumor formation. The study of spindle orientation is therefore fundamental to both developmental biology and human disease.
Over a century ago, Oscar Hertwig discovered that sea urchin embryos biased the orientation of the mitotic spindle along their long axis, which led to a model (the ‘Hertwig Rule’) in which cells orient divisions in response to mechanical forces (Hertwig, 1884). The Hertwig model stated that mechanical regulation was the primary determinant of spindle orientation as cells sensed shape changes in response to external forces. Later discoveries, including those in ascidian embryos, in which cell division orientation correlated with differential daughter cell fate and size, suggested a molecular basis for spindle orientation (Conklin, 1905). Conklin reasoned that the ability to alter division orientation to achieve autonomous fate specification during development must rely upon internal molecular regulation rather than external mechanics. The modern molecular era has now vastly expanded our understanding of oriented cell division, with studies showing support for both models.
Genetic identification of the first spindle orientation regulators occurred nearly two decades ago (Cheng et al., 1995; Etemad-Moghadam et al., 1995; Kraut et al., 1996; Zwaal et al., 1996). Since then, the ‘parts list’ of proteins required for proper spindle orientation has grown tremendously, and includes components from several signaling pathways that couple the mitotic spindle to cortical polarity complexes in a variety of cell types from a diverse group of organisms. More recent studies utilizing genetic screening technologies and improved cell culture-based systems have expanded that list further. Moreover, these recent studies have begun to view spindle orientation through a more molecularly focused lens that allows better insight into how these parts fit together during oriented cell divisions.
The ability to bias the orientation of cell division via regulated spindle positioning is conserved from yeast to mammals (Siller and Doe, 2009). For the scope of this Review, we have chosen to focus on selective, well-studied examples from metazoan model organisms. We first highlight several developmental processes that rely on regulated spindle orientation/asymmetric cell divisions and provide an overview of how cell polarity, and hence spindle orientation, is first established in these examples. A particular focus will be placed on the mechanisms by which cortical polarity cues ‘capture’ spindle microtubules, as this process represents an early step in the spindle orientation process. Although microtubule capture itself is likely to be a conserved and generalized aspect of spindle orientation, the underlying molecular pathways appear to be diverse, highlighting the importance of further understanding of the mechanisms involved. We describe several well-established spindle orientation components as well as newly identified factors that regulate communication between the cell cortex and the mitotic spindle, with an aim to provide readers with an updated view of how different cell types regulate the position of the mitotic spindle.
Oriented/asymmetric cell divisions during metazoan development
It should be noted that oriented/asymmetric cell division, as discussed throughout this Review, is defined by two mutually coupled events: (1) establishment of a cortical polarity axis by the unequal distribution of polarity proteins and cell fate determinants; and (2) alignment of the mitotic spindle with respect to this polarity axis. In certain cases, the polarity axis coincides with a tissue or body axis of the organism. We will focus our discussion primarily on how these two processes are linked at the molecular level using four examples: the Caenorhabditis elegans zygote, Drosophila neuroblasts, Drosophila sensory organ precursors and mammalian epidermal cells. As gene and protein names often vary between species, please see Table 1 for the naming of orthologs.
Table 1.
Identified regulators of spindle orientation
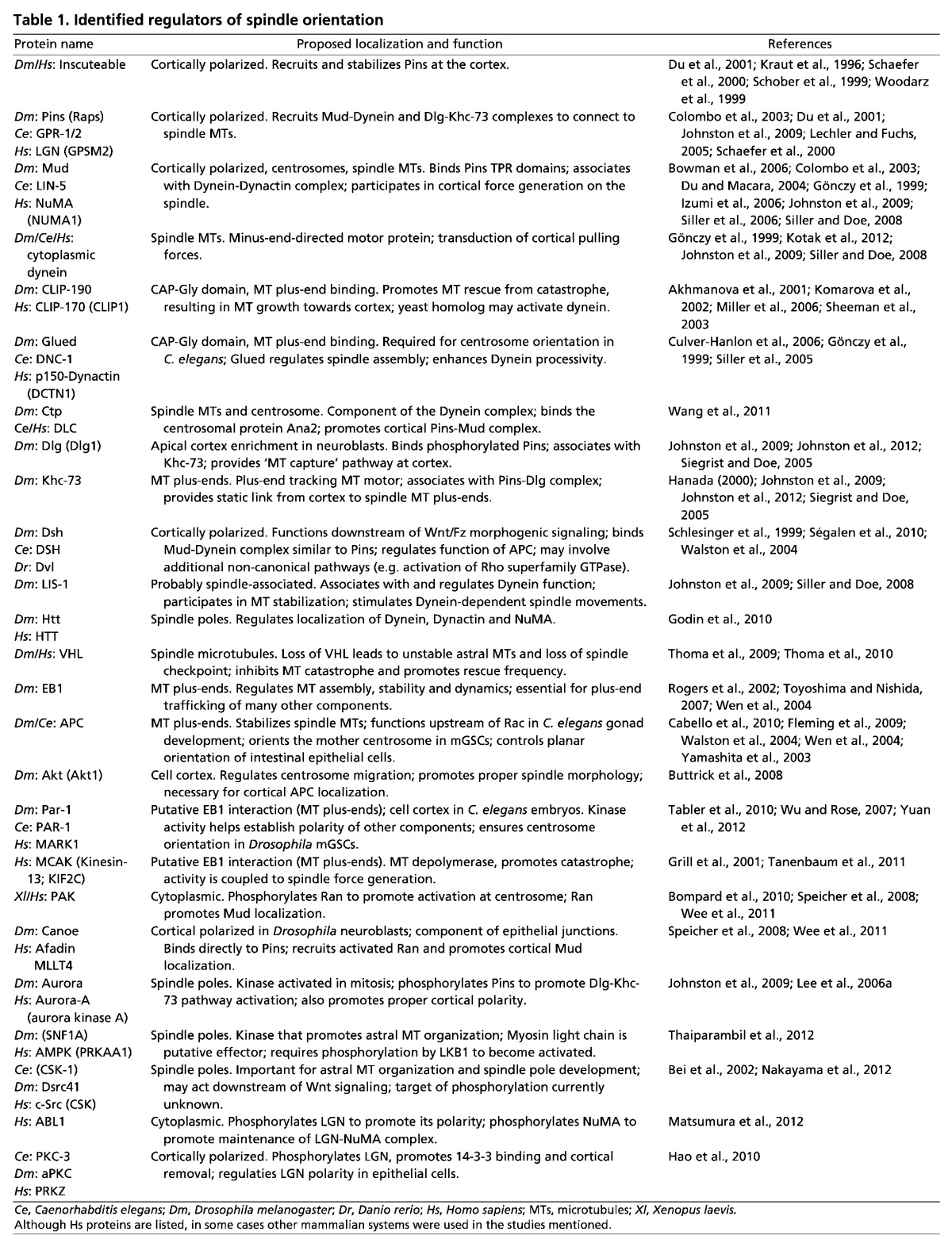
The first series of divisions in the C. elegans zygote produces siblings that are asymmetric in both size and fate (Fig. 1A). Early genetic studies revealed a set of evolutionarily conserved partitioning-defective (PAR) proteins that are necessary for establishing an anterior-posterior (A-P) cortical polarity axis prior to the first zygotic division (Kemphues et al., 1988). Subsequent studies demonstrated that the PAR complex also regulates spindle orientation and the unequal force generation exerted on the spindle poles (Etemad-Moghadam et al., 1995; Grill et al., 2001; Kemphues et al., 1988). Defects in PAR complex genes result in mispositioning of the mitotic spindle, loss of daughter cell asymmetry and, ultimately, non-viable animals. As shown in Fig. 1A, two successive divisions with A-P spindle orientations produce a four-cell embryo containing a blastomere known as the EMS blastomere. Spindle orientation along the A-P axis and subsequent asymmetric cell division of the EMS cell then generates an E daughter cell, which will give rise to endodermal lineages, and a MS daughter cell, which will form mesodermal lineages. Spindle orientation in the one-cell stage and the P2 blastomere cell requires the activity of GPR-1/2 and LIN-5, which constitute an evolutionarily conserved non-canonical G-protein signaling network (Colombo et al., 2003; Gönczy, 2008; Werts et al., 2011). Mutations in Wnt signaling pathway components result in misalignment of the EMS spindle and mis-specification of germ cell layers (Schlesinger et al., 1999; Walston et al., 2004) (Fig. 1A). By manipulating contact sites at the four-cell stage, Goldstein showed that cell-cell contacts establish a site that captures centrosomes via emanating microtubules to orient cell divisions (Goldstein, 1995). Actin-rich contact sites between the EMS and P2 cells determined spindle orientation and influenced partitioning of fate information necessary for gut specification (Goldstein, 1995; Waddle et al., 1994). Collectively, these studies suggest that spindle orientation is an essential determinant of cell fate specification during C. elegans development.
Fig. 1.
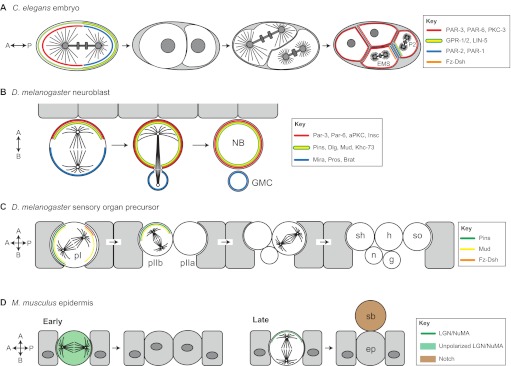
Spindle positioning regulates oriented/asymmetric cell division during metazoan development. (A) Spindle orientation regulates asymmetric cell divisions in early C. elegans development. The first zygotic division produces daughter cells that are asymmetric in both size and fate specification. Prior to division, the cell cortex is polarized along an anterior-posterior (A-P) axis by the activity of PAR (blue) and PKC-3 (red). Spindle orientation is coupled to this polarity axis primarily through the action of the GPR-1/2 and LIN-5 complex (yellow/green), which enriches along the posterior cortex. In subsequent divisions, the Fz-Dsh complex (orange) regulates spindle orientation in the EMS cell. Similar to the one-cell stage, PAR complexes show reciprocal polarity in EMS and P2 cells (Arata et al., 2010), whereas GPR-1/2 influences spindle positioning through asymmetric localization in the P2 cell specifically (Werts, Roh-Johnson and Goldstein, 2011). (B) Drosophila neuroblasts polarize along an apical-basal (A-B) axis through the activity of the Par-aPKC complex (red). Spindle orientation is regulated by the apical Pins complex, which recruits the regulatory proteins Mud, Dlg and Khc-73 (yellow/green). Neuroblast asymmetric divisions result in a larger self-renewed neuroblast (NB) and a smaller ganglion mother cell (GMC), which is specified for neuronal differentiation by the inheritance of the Mira-Pros-Brat complex (blue). (C) Drosophila sensory organ precursor cells (SOPs) in the epithelium of developing wing imaginal discs give rise to the adult mechanosensory bristles. Each SOP progenitor gives rise to five distinct cells that constitute the entire bristle structure: g, glial; h, hair; n, neuron; sh, shaft; so, socket. Spindle orientation in the initial (pI) cell division is regulated by the coordinated action of Pins (green), which positions the spindle within the epithelial plane, and Fz-Dsh (orange), which regulates orientation along the A-P axis. Pins-mediated rotation of the spindle in the pIIb cell then establishes the A-B division orientation necessary for correct specification and positioning of the neuronal and glial cells. (D) In the mouse skin, basal epidermal cells (ep) divide within the epithelial plane early in development, resulting in expansion of the tissue (left). At later developmental stages (right), division orientation switches to an A-B mode via Pins-mediated repositioning of the mitotic spindle. This mode of division positions one daughter, the suprabasal (sb) cell, below the epithelium. The sb cell differentially inherits Notch, which specifies differentiation resulting in stratification of the skin.
Asymmetric division of Drosophila neuroblasts, the stem cells of the developing fly brain, regulates development of the fly central nervous system (Fig. 1B). Neuroblasts polarize along an apical-basal (A-B) axis and divide in a stem cell-like manner to produce a self-renewed neuroblast and a ganglion mother cell (GMC) that produces differentiated neurons and glia (Doe, 2008). Thus, a relatively small number of neuroblasts can supply the vast number of differentiated neuronal cells that constitute the adult brain. Genetic studies over the past decade have identified three core protein complexes (Fig. 1B) that ensure proper asymmetric neuroblast division: (1) the apical ‘polarity complex’ consisting of the evolutionarily conserved proteins Par-3 (also known as Bazooka), Par-6 and atypical protein kinase C (aPKC) (Petronczki and Knoblich, 2001; Wodarz et al., 1999); (2) the apical ‘spindle orientation complex’ consisting of Inscuteable (Insc), Partner of Inscuteable (Pins; also known as Rapsynoid) and Mushroom body defect (Mud) (Schaefer et al., 2000; Schober et al., 1999; Siller et al., 2006; Yu et al., 2000); and (3) the basal ‘differentiation complex’ consisting of the adapter protein Miranda (Mira) and cell fate markers such as Prospero (Pros), Brain tumor (Brat) and Numb (Betschinger et al., 2006; Lee et al., 2006b). Proper A-B spindle positioning ensures apical inheritance of aPKC, which promotes self-renewal, and basal inheritance of the Miranda complex, which induces neuronal differentiation (Fig. 1B). Defects in these core components uncouple spindle orientation from the polarity axis, disrupting division asymmetry and often resulting in overproliferation of neural stem cells at the expense of differentiated progeny. This dysregulated division pattern can result in neuroblast-based tumors, loss of neuronal production, and lethality (Cabernard and Doe, 2009; Lee et al., 2006b) (see Box 1). Thus, spindle orientation with respect to intrinsic polarity cues ensures proper stem cell homeostasis during development.
Box 1. Oriented/asymmetric cell division and disease
Defects in spindle orientation are associated with malignant neuroblast-based tumors caused by uncontrolled self-renewal divisions in Drosophila (Gonzalez, 2007; Lee et al., 2006b). Several prominent tumor suppressor proteins, including APC, Dlg, VHL (Thoma et al., 2010; Thoma et al., 2009) and LKB1, regulate spindle orientation (Table 1), suggesting that spindle misorientation might contribute to tumor development (Pease and Tirnauer, 2011).
Neurogenesis requires control of cell division orientation in neuroprogenitors at specific developmental stages (Lancaster and Knoblich, 2012) to balance proliferative and neurogenic outcomes (Konno et al., 2008; Peyre et al., 2011). Mutations in the spindle orientation regulators LIS1 (PAFAH1B1) and HTT manifest in Type I lissencephaly and Huntington’s disease, respectively. Interestingly, both LIS1 and HTT regulate the function of cytoplasmic dynein, a potentially universal regulator of spindle orientation (Table 1; see main text). Whether a causative molecular link exists between spindle orientation and neurodevelopmental disorders will be an important future research endeavor.
Components of the intraflagellar transport (IFT) machinery in cilia have been linked to misoriented cell divisions possibly underlying polycystic kidney disease (PKD) (Delaval et al., 2011; Fischer et al., 2006; Hildebrandt and Otto, 2005). Several IFT proteins localize to centrosomes, and mutations in IFT88 result in spindle misorientation (Delaval et al., 2011), leading to improper spatial arrangement of nephron epithelia and cyst development. Interestingly, IFT88 participates in astral microtubule formation possibly through a dynein-dependent transport complex (Delaval et al., 2011). However, mutations in other IFT components appear to drive cystogenesis through mechanisms independent of spindle misorientation (Jonassen et al., 2012).
Intestinal crypt stem cells regulate spindle orientation to produce differentiated progeny. Heterozygous mutations in the tumor suppressor protein APC, which is mutated in ∼85% of colorectal cancer cases (Markowitz and Bertagnolli, 2009), results in spindle misorientation and altered cell shape (Quyn et al., 2010). However, this dependence of intestinal stem cells on division orientation/asymmetric division has been challenged recently and remains controversial (Schepers et al., 2011).
Drosophila sensory organ precursors (SOPs) are ectodermal progenitor cells that give rise to mechanosensory organs of the fly peripheral nervous system (Fig. 1C). Each SOP in the imaginal disc of the developing wing undergoes a series of divisions, the orientation of which is crucial for both asymmetric sibling fate specification and production of the proper structural integrity and alignment of individual sensory wing hairs (Fig. 1C). Cortical spindle orientation cues in SOPs localize through an evolutionarily conserved mechanism known as planar cell polarity (PCP) within the imaginal disc tissue. The spindle must be controlled in both an A-B orientation, through the action of the Pins complex, as well as an A-P orientation, which is controlled by a non-canonical Frizzled/Dishevelled (Fz/Dsh) signaling pathway (David et al., 2005; Gho and Schweisguth, 1998; Roegiers et al., 2001) (Fig. 1C). Proper asymmetric division in SOPs further relies on the activity of core PCP components [a discussion of which is beyond the scope of this Review, but see Wallingford (Wallingford, 2012) for an excellent recent review]. The disruption of spindle orientation during asymmetric SOP divisions results in mis-specification of cell lineages and defects in bristle development (Bellaïche et al., 2001; Lu et al., 1999). Thus, regulated spindle orientation plays a central role in the development of multicellular sensory structures.
Basal epidermal cells contribute to the architecture of the epidermis, a stratified squamous epithelium of skin that regulates fluid and electrolyte exchange and guards against harmful or infectious substances, such as microbes. Early in development, these progenitor basal cells preferentially divide within the epithelial plane, producing two symmetric daughter basal cells to expand the undifferentiated basal layer (Fig. 1D). However, at a later developmental stage, basal cell divisions switch to an asymmetric mode, which allows the self-renewal of a proliferative basal cell while also producing a suprabasal cell that is committed to differentiating into the deep tissue layers forming the skin barrier (Lechler and Fuchs, 2005). Strikingly, this asymmetric division switch accompanies a 90° reorientation of the mitotic spindle, from parallel to perpendicular to the epithelial plane, which is dependent on the Par-aPKC and Pins-Insc-Mud polarity complexes (Fig. 1D) (Lechler and Fuchs, 2005; Poulson and Lechler, 2010). Spindle reorientation ensures asymmetric inheritance of Notch, which specifies barrier cell differentiation. Spindle misorientation mutants thus elicit impaired stratification and defects in barrier function, leading to dehydration and death (Williams et al., 2011). These results demonstrate the evolutionary conservation of coupling spindle orientation to decisions of cell fate and tissue structure, and underscore the importance of proper cell division orientation during development and in disease (see Box 1).
Linking the spindle to the cell cortex: the role of microtubule plus-end binding proteins
As highlighted above, correct orientation of the spindle with respect to cell polarity is crucial for tissue development and homeostasis. The ability to position statically the mitotic spindle, a large, otherwise dynamic cellular structure, necessitates physical connections between the cell cortex and the plus-ends of astral microtubules emanating from the spindle poles (Fig. 2). In this manner, the polarization of cortical cue(s) affords the cell the ability to position the spindle in a biased orientation via interactions between these polarized proteins and microtubule plus-end binding proteins (+TIPs). In this model, +TIPs would serve as the prey in a ‘microtubule capture’ mechanism by interacting with cortical bait.
Fig. 2.
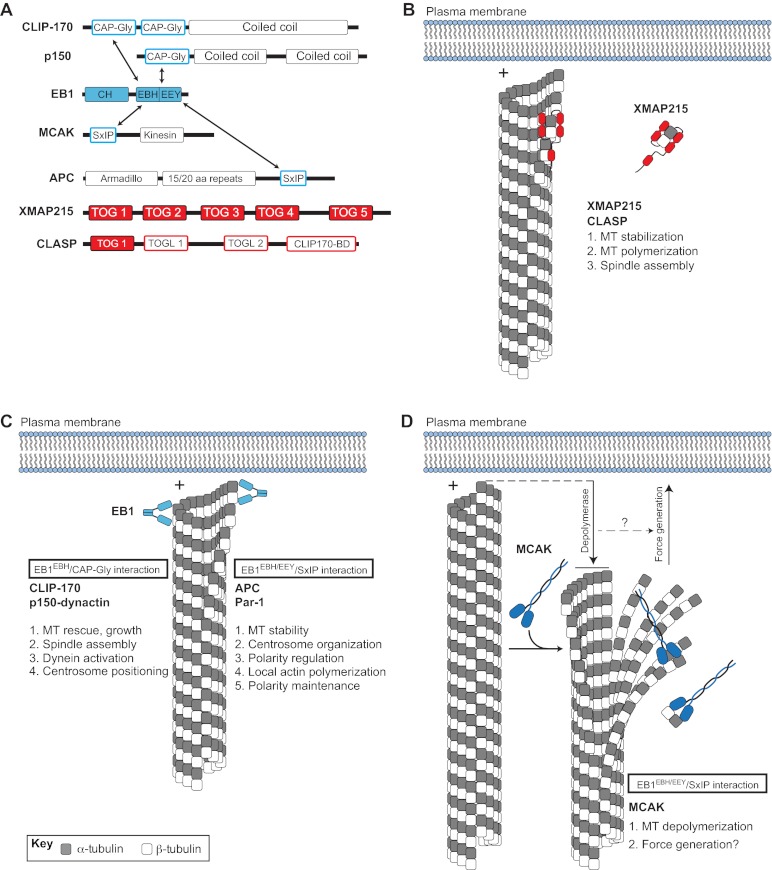
Plus-end binding proteins (+TIPs) contribute to microtubule dynamics and spindle orientation. (A) Domain architectures of various +TIPs. Arrows indicate direct interactions between +TIPs. (B) The XMAP215 and CLASP family of proteins (shown in red) are autonomously tracking +TIPs that regulate MT dynamics and spindle assembly. Interaction with microtubule (MT) plus-ends (+) occurs through direct binding of the XMAP215-CLASP TOG domain. (C) EB1 (shown in light blue) binds MT plus-ends through an N-terminal calponin homology (CH) domain and an acidic C-terminal EEY tripeptide motif. The EB1 homology domain (EBH) of EB1 also interacts with CAP-Gly domains found in CLIP-170 and p150-dynactin, thereby inducing their plus-end localization. CLIP-170 and p150-dynactin regulate MT dynamics, spindle assembly, centrosome positioning, and dynein activity. EB1 also binds a growing number of proteins containing the SxIP polypeptide motif, including APC and Par-1. The EBH/EEY domains directly bind the SxIP sequence and mediate plus-end tracking. Several members of the SxIP-containing family have been implicated in MT stability, centrosome/spindle orientation, actin polymerization, and polarity regulation/maintenance. (D) The MT depolymerase MCAK (shown in dark blue) also tracks to plus-ends through an EB1/SxIP interaction. MCAK-mediated depolymerization results in MT shortening and alteration in cortex-MT architecture (downward arrow), which may be coupled to cortical force generation, which is necessary for spindle orientation (upward arrow).
Metazoans have evolved a diverse set of +TIPs (Fig. 2A) with varied domain architectures, protein structures and biological functions (Slep, 2010). For example, members of the XMAP215/CLASP family of +TIPs contain multiple domains known as TOG or TOG-like domains, which demonstrate direct plus-end tracking activity in reconstituted systems and bind α/β-tubulin dimers directly (Fig. 2B) (Al-Bassam et al., 2007). XMAP215 members play an essential role in interphase microtubule polymerization and stabilization, which is necessary for proper formation of the mitotic spindle (Slep, 2009; Tournebize et al., 2000). ZYG-9, a C. elegans XMAP215 member, regulates spindle positioning in one-cell-stage embryos (Bellanger et al., 2007). A second autonomously tracking +TIP, end-binding protein 1 (EB1), serves as the cellular workhorse for plus-end protein localization. EB1 itself binds microtubules through an N-terminal calponin homology (CH) domain, an interaction that is enhanced by an acidic, EEY tripeptide motif at the extreme C-terminus (Fig. 2A,C). EB1 binding induces tubulin polymerization in vitro (Slep and Vale, 2007) and is necessary for proper assembly of the mitotic spindle (Rogers et al., 2002). Depletion of EB1 in Drosophila S2 cells causes a significant reduction in microtubule dynamics by inducing extended phases of no growth or of shrinkage, although the overall morphology of interphase microtubules is not disrupted. By contrast, EB1 depletion in mitotic cells induces shortened astral microtubules and fragmented microtubules at prophase along with compacted spindles, detached spindle poles and unfocused nonastral spindles at metaphase (Rogers et al., 2002). Subsequent studies indicated that EB1 stabilizes microtubules through direct interaction with Adenomatous polyposis coli (APC) and the actin polymerizing formin protein Diaphanous (Dia) (Wen et al., 2004). EB1 regulates planar spindle orientation in nonpolarized, cultured epithelial cells through a microtubule stabilization mechanism downstream of β1-integrin adhesion. This effect also requires myosin X-dependent remodeling of the actin cytoskeleton (Toyoshima and Nishida, 2007). It is also worth noting that the budding yeast homolog of EB1, Bim1, functions in spindle orientation through a complex with Kar9 and the myosin Myo2 in a process that involves the guidance of microtubules along actin filaments (Hwang et al., 2003; Korinek et al., 2000). Thus, XMAP215 (also known as Msps in Drosophila) and EB1 appear to regulate spindle orientation through the stabilization of spindle structure (Fig. 2).
In addition to its role in the formation and stabilization of spindle microtubules through direct plus-end binding, EB1 recruits an expanding list of indirect +TIPs. Several of these have been shown to regulate spindle orientation, and they fall into two groups that act through distinct EB1-interacting motifs (Kumar and Wittmann, 2012; Slep, 2010). The first group, which consists of the proteins CLIP-170 (also known as CLIP1; CLIP-190 in Drosophila) and p150-dynactin (also known as DCTN1; Glued in Drosophila), bind homodimeric EB1 C-termini through a conserved N-terminal CAP-Gly domain (Fig. 2A,C). CLIP-170 promotes microtubule growth, which has been shown to occur selectively towards the cell cortex and may contribute to directional migration of motile cells (Akhmanova et al., 2001; Komarova et al., 2002). The budding yeast homolog of CLIP-170, Bik1p, regulates spindle orientation, possibly through Num1p-mediated dynein activation and/or asymmetric polarization of Kar9 (Miller et al., 2006; Sheeman et al., 2003). As clear orthologs of Num1p and Kar9 are nonexistent or as yet undiscovered, the role of CLIP-170 in spindle orientation in higher eukaryotes remains unclear. A role for p150-dynactin in spindle orientation, however, has been demonstrated. In C. elegans zygotes, p150 (DNC-1) is necessary for proper centrosome rotation along the longitudinal axis and thus spindle orientation (Skop and White, 1998). A subsequent study demonstrating a more robust reduction of p150 expression revealed additional roles in pronuclear migration and centrosome separation (Gönczy et al., 1999). The Drosophila ortholog, Glued, has been shown to regulate both spindle assembly (Siller et al., 2005) and orientation (Siller and Doe, 2008) in larval neuroblasts. These functions of Glued occur via enhanced processivity of dynein, an essential spindle orientation component (discussed in detail below) (Culver-Hanlon et al., 2006). Notably, alterations in Huntingtin (Htt), the protein mutated in the neurodegenerative Huntington’s disease, result in mislocalization of p150 and subsequent spindle misorientation. Htt associates with p150 at spindle poles during mitosis (Gauthier et al., 2004); knockdown of Htt results in a partial loss of spindle-associated p150 along with shortened spindles and unfocused spindle poles (Godin et al., 2010). This loss of Htt-mediated spindle orientation in vivo causes mis-specification of neural progenitor cells in mice, providing evidence for a potential link between spindle orientation, cell fate and disease (see Box 1) (Godin et al., 2010).
Members of the second family of EB1-interacting +TIPs contain the short SxIP polypeptide motif (Fig. 2A,C). The SxIP motif binds in a slightly bent conformation to a hydrophobic cavity and adjacent ‘polar rim’ formed at the EB1 C-terminus (Honnappa et al., 2009). This mode of EB1 interaction, via a short polypeptide motif, allows for a vast diversity of otherwise unrelated proteins to localize to microtubule plus-ends (Kumar and Wittmann, 2012). Recent evidence suggests this interaction might be regulated by phosphorylation of the SxIP motif or flanking residues, pointing to a potential for cell cycle-dependent regulation (Buey et al., 2012; Honnappa et al., 2009). As noted above, EB1 binds the tumor suppressor protein APC, which contains a clearly defined SxIP motif, and this interaction is required for plus-end APC localization (Fig. 2A,C) (Honnappa et al., 2009). APC has a defined spindle orientation role in several model systems. Firstly, in distal tip cells of the C. elegans gonad, APC functions upstream of Rac-mediated spindle orientation, which is required for proper gonadal development (Cabello et al., 2010). APC may also control the timing of spindle rotation in the ABar, a cell present at the eight-cell stage, during C. elegans blastomere development (Walston et al., 2004). Secondly, in Drosophila, male germline stem cells (GSCs) adhere to a hub cell within the testis niche and undergo asymmetric divisions to produce a proximal self-renewed stem cell and a differentiated spermatagonial cell. Local signals from the niche induce self-renewal only in the proximal cell, thus necessitating precise orientation of division perpendicular to the niche cell. Mutations in APC result in mispositioning of the mother centrosome and subsequent defects in spindle orientation (Yamashita et al., 2003). Neuroepithelial cells in the developing Drosophila brain also rely on APC for proper spindle orientation; APC RNA interference (RNAi) results in altered planar spindle orientation and defects in proper symmetric divisions of these cells (Lu et al., 2001). Finally, several recent studies have demonstrated a role for APC-mediated spindle orientation in mammalian small intestinal crypt stem cells. For example, an APC truncation mutant causes spindle misorientation that manifests as a switch from A-B to planar orientation (Quyn et al., 2010). Moreover, analysis of intestinal tumors in APCmin (multiple in neoplasia; containing a nonsense truncation mutation at codon 850 in the APC N-terminus) mice demonstrate more severe spindle orientation phenotypes in intestinal crypt cells than in ‘normal-appearing’ heterozygous cells (although these cells show misorientation relative to wild type) (Fleming et al., 2009), suggesting the potential role of proper spindle orientation as a tumor suppressor mechanism (see Box 1). These studies highlight a key role for the tumor suppressor protein APC in spindle positioning and suggest that its EB-1 plus-end association is instrumental for promoting interactions with cortical orientation cues (Fig. 2; Table 1).
A recent proteomic study identified microtubule affinity-regulating kinase 1 (MARK1; also known as PAR-1), a serine/threonine kinase, as another SxIP-containing partner of EB1 (Fig. 2A,C) (Jiang et al., 2012). In C. elegans, PAR-1 regulates the cortical polarization of factors involved in positioning of the mitotic spindle. Loss of PAR-1 results in embryos that fail to undergo posterior displacement of the mitotic spindle and do not maintain proper A-P spindle orientation (Wu and Rose, 2007). It should be noted, however, that whether the role of C. elegans PAR-1 in these processes occurs through regulation of microtubule dynamics per se has not been firmly established (Pellettieri and Seydoux, 2002). Studies in the Xenopus embryo found that PAR-1 induces ‘vertical’ spindle orientation in the neuroepithelium, which promotes the generation of deep, differentiated neuronal progeny through asymmetric division patterns (Tabler et al., 2010). Interestingly, PAR-1 has also been shown to function in centrosome orientation checkpoint, a crucial process that ensures the proper oriented division of Drosophila male GSCs (Yuan et al., 2012). Although the precise mechanism through which PAR-1 acts in these systems remains unclear, these studies suggest that it may achieve a similar spindle orientation outcome through diverse molecular mechanisms. Moreover, whether EB1 binding and plus-end localization is required for PAR-1 function in spindle orientation has not been examined.
Finally, the kinesin-13 MCAK (also known as KIF2C), belonging to the kinesin family of microtubule motor proteins that typically move toward plus-ends (Hirokawa et al., 2009), also localizes to MT plus-ends via an EB1-SxIP interaction (Fig. 2A,D) (Honnappa et al., 2009; Tanenbaum et al., 2011). MCAK functions as a potent microtubule depolymerase, promoting catastrophe events and thus regulating microtubule length and morphology. Interestingly, a recent report using an elegant reconstitution system demonstrated that the plus-end depolymerase activity of MCAK was coupled to local force production (Oguchi et al., 2011), raising the intriguing possibility that depolymerases could participate in spindle force generation involved in spindle orientation (Fig. 2D) (Grill et al., 2001) (discussed below). This EB1-dependent localization and depolymerase activity is conserved in invertebrates, such as with the Drosophila ortholog, Klp10A (Mennella et al., 2005). Loss of Klp10A exacerbates microtubule polymerization, which results in increased density, abundance and length of spindle astral microtubules (Morales-Mulia and Scholey, 2005). Notably, proper regulation of astral microtubules is essential for mitotic spindle orientation in Drosophila neuroblasts (Siller and Doe, 2008). Although no evidence yet exists for a direct role of Kinesin-13 depolymerases in spindle orientation, the results discussed above suggest these microtubule enzymes, along with other related proteins (Loughlin et al., 2011), might serve an essential regulatory function at the interface between plus-ends and cortical spindle orientation complexes. Future studies using high resolution, real-time imaging to examine astral MT dynamics directly at the cortex during oriented cell division will be extremely valuable in understanding this process. One potential model would be that shortening of astral MT ends by depolymerase enzymes is necessary to generate gaps that cortical force generators then close through spindle movements coupled to reorientation (Fig. 2D).
In summary, a vast number of diverse protein families localize to plus-ends through distinct mechanisms. This network of protein complexes appears to regulate spindle orientation by different mechanisms, most of which are unknown and will require further studies to resolve completely. More importantly, understanding how these varied mechanisms function together at the cortex/+TIP interface will be an important future research focus. For example, do components of the cortical spindle orientation complex affect the activity of MT stabilizing agents at the +TIP? Conversely, do stable MTs provide regulatory feedback for activity of cortical protein complexes? Does the cortical actin cytoskeleton play an intervening role between the cortex and MTs, perhaps by both maintaining cortical polarity and interacting with spindle MTs? The actin cytoskeleton has been shown to regulate leading edge MT turnover in migratory cells (Gupton et al., 2002). The mechanism(s) by which the actin cytoskeleton contributes to spindle orientation in animal cells remains an area of emerging investigation. Local changes in cortical actin polymerization, induced by the activity of +TIPs, such as APC and Dia, could provide a site for MT capture and spindle stabilization. In turn, stable MTs might provide a site for actin-MT crosslinking protein interaction (Rodriguez et al., 2003). Alternatively, +TIP-induced actin regulation could play an important role in stabilizing the cortical localization and polarity of other spindle orientation regulators (Li and Gundersen, 2008). Regulation of the cortical actin network through external forces (Fink et al., 2011) or integrin-mediated adhesion (Toyoshima and Nishida, 2007) can bias spindle orientation in cultured cells. Also, the Spire and Formin families of actin nucleators have been shown to cooperate in the asymmetric positioning of the meiotic spindle during oocyte division by establishing a cortical actin network necessary for spindle movement (Pfender et al., 2011). Finally, how do MT enzymes that regulate growth and catastrophe dynamics contribute to +TIP architecture, and how do MT dynamics regulate coupling to cortical complexes? It seems plausible that the geometry of the MT plus-ends relative to the cell cortex is vital for establishing productive interactions that promote spindle movements (Su et al., 2012). We suggest that a significant role of +TIPs is to shape MT architecture dynamically to provide effective cortical coupling necessary for spindle orientation, possibly through enhancement of cortical force generators (Fig. 2D) (Kozlowski et al., 2007). Although a significant effort has been made to understand the cortical cues involved in spindle orientation, further studies that address the role of the microtubule plus-end will help to elucidate the molecular interplay involved in cortical coupling.
Bridging the gap between polarity and spindle positioning: Pins-Dlg-Khc-73, an MT-capturing complex
One fundamental aspect of spindle orientation is the ability of cortical cues to capture the spindle physically, probably as an initial step in dictating its ultimate position. Our discussion of +TIPs above demonstrates the variety of potential microtubule prey in this capture event. What then is the cortical bait that secures the spindle connection? How are these cortex-microtubule interactions established, maintained and regulated? What are the functional consequences of spindle capture on other aspects of the spindle orientation process? Static spindle-cortex connections could allow for proper localization and/or activation of pathways that act in subsequent steps; for example, the microtubule-mediated offloading of force-generating molecules to the cortex (Markus and Lee, 2011; Pecreaux et al., 2006). Below, we discuss the regulation of a recently characterized cortical spindle-capturing complex and present a model for how it cooperates with an evolutionarily conserved force-generating complex.
As discussed earlier, the evolutionarily conserved polarity protein Pins regulates spindle orientation in a variety of cell types within diverse organisms to achieve distinct biological outcomes (David et al., 2005; Du et al., 2001; Schaefer et al., 2001). Recently, we identified a ‘microtubule capture’ pathway in Drosophila that involves a Pins-Dlg-Khc-73 complex (Fig. 3A). Discs large (Dlg) binds directly to a short, linear motif in the ‘Linker’ domain of Pins and is required for proper spindle orientation in both an ‘induced cell polarity’ S2 cell system and neuroblasts (Johnston et al., 2009; Siegrist and Doe, 2005). Interestingly, this interaction is dependent upon phosphorylation of Pins by the mitotic, centrosomal kinase Aurora-A (Aurka in mammals; Aur in Drosophila), as is Pins-mediated spindle orientation (Johnston et al., 2012; Johnston et al., 2009; Lee et al., 2006a). In this model, Dlg associates with the cell cortex and localizes with polarized Pins (Siegrist and Doe, 2005). Dlg then interacts with Khc-73, a kinesin motor protein that moves towards and localizes at microtubule plus-ends (Hanada et al., 2000; Huckaba et al., 2011; Siegrist and Doe, 2005); Dlg binding appears to enhance Khc-73 activity in vitro (Yamada et al., 2007). Dlg binding occurs through a unique domain of Khc-73, termed the maguk binding stalk (MBS). Thus, Dlg serves as an adaptor protein that links polarized Pins at the cell cortex to the microtubule plus-end motor, Khc-73, and loss of any of these three components results in spindle orientation defects (Fig. 3A). Interestingly, both the Pins Linker and Khc-73 MBS domains bind to the guanylate kinase (GUK) domain of Dlg (Johnston et al., 2009; Siegrist and Doe, 2005), suggesting that they bind at distinct sites within the GUK domain. The structural basis for phosphorylation-dependent Pins-Dlg complex formation has recently been detailed (Johnston et al., 2012; Zhu et al., 2011a) but a structural analysis of the MBS domain alone or in complex with Dlg is lacking. It remains to be determined whether Khc-73 motor activity per se is required for spindle orientation, although misexpression of the MBS domain alone functions as a dominant negative, suggesting that other domains are indeed required (Johnston et al., 2009; Siegrist and Doe, 2005).
Fig. 3.
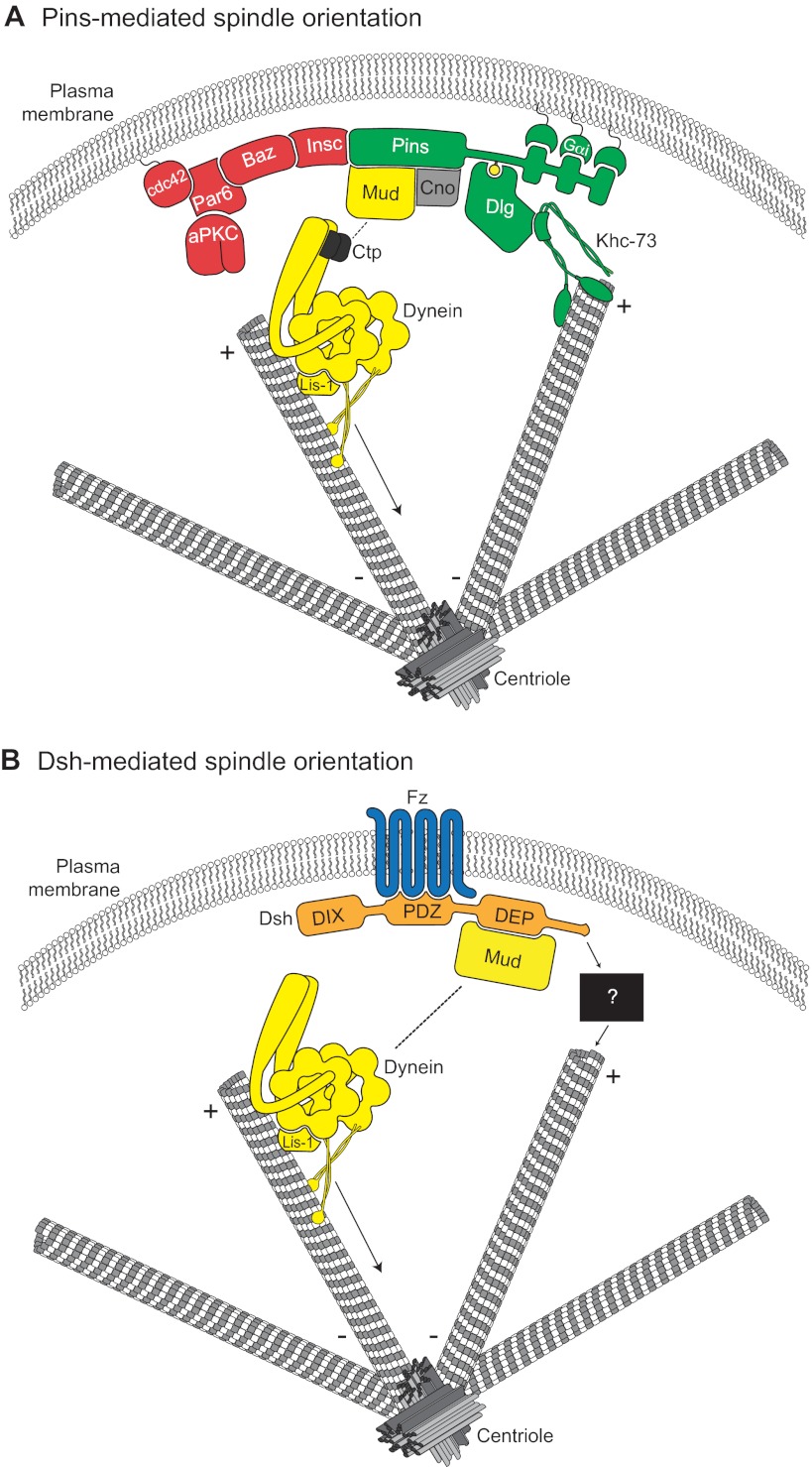
Molecular models for Pins- and Dsh-mediated spindle orientation. (A) The Cdc42-Par6-aPKC-Baz polarity complex associates with Pins through the adapter protein Insc. Pins cortical association is mediated by direct binding between three consecutive GoLoco domains and the heterotrimeric G-protein Gαi. Pins serves as the ‘hub’ for two distinct spindle orientation pathways in Drosophila neuroblasts. First, Pins (via its TPR domain, not shown) binds directly to Mud, which associates with cytoplasmic dynein through Ctp. Lis-1 functions as an activator of dynein microtubule motor activity. The scaffold protein Cno is required for maintenance of the apical Pins-Mud complex. The dynein-dynactin complex moves processively towards MT minus-ends (arrow) and generates pulling forces on the mitotic spindle. Second, the Pins linker domain is phosphorylated (yellow circle) by Aurora-A kinase. This phosphorylation allows direct binding with Dlg, which then associates with the kinesin motor protein Khc-73; the plus-end binding capacity of Khc-73 is thought to provide an MT capture or attachment mechanism. Together, the MT-capture and force-generation pathways elicit robust spindle alignment. (B) The Frizzled receptor (Fz, blue) is planar polarized in many cell types. Following Wnt activation, Fz recruits the cytoplasmic scaffold protein Dishevelled (Dsh, orange) to the cortex via its PDZ domain. The Fz-Dsh complex regulates spindle orientation in Drosophila sensory organ precursor cells through association (via the Dsh DEP domain) with Mud. Similar to Pins, Mud associates with the dynein motor complex. Whether the Dsh-Mud pathway is sufficient for spindle orientation or if additional pathways (black box) are required remains to be investigated.
Using the induced polarity assay in Drosophila S2 cells, we found that although full-length Pins could robustly orient the mitotic spindle, the Linker domain alone was sufficient for a ‘partial orientation’ phenotype. We found that this intermediate activity of the isolated Linker domain typically oriented spindles precisely at the distal edge of the cortical Pins crescent (as opposed to the center of the crescent with full-length Pins). Live-cell imaging experiments revealed that mitotic spindles with poles contacting non-Pins-expressing regions of the cell cortex rotated slowly before becoming secured at the edge of the Linker crescent (Johnston et al., 2009). These results suggest that the Pins-Dlg-Khc-73 complex functions to capture the mitotic spindle statically by establishing a physical connection between cortical polarity and microtubule plus-ends (Fig. 3A, Fig. 4).
Fig. 4.
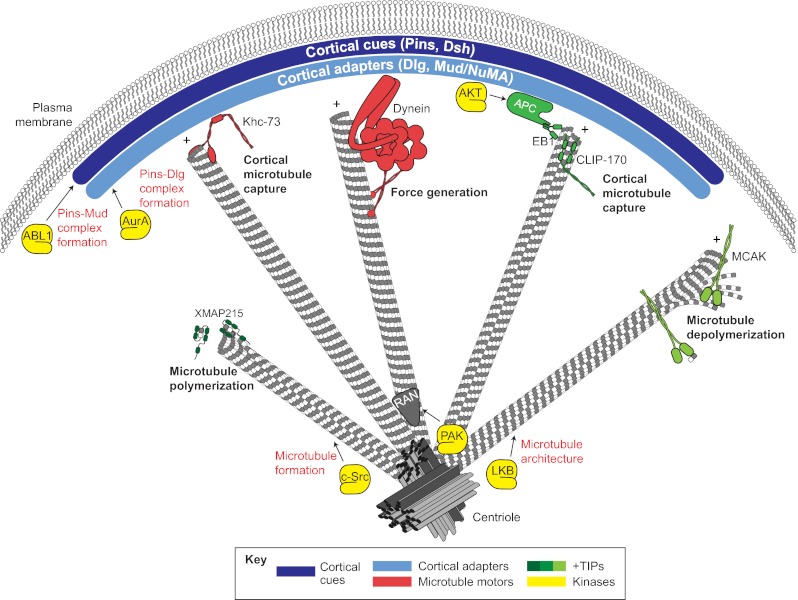
A summary of the mechanisms that contribute to spindle orientation. Cortical spindle orientation cues, such as Pins and Dsh (dark blue), polarize to specific regions of the cell cortex. Cortical adapters, such as Dlg and Mud/NuMA (light blue), are then recruited by direct interactions with cortical cues. Polarized cortical cues and adapters interface with spindle microtubules through interactions with various plus-end localized microtubule-binding proteins, including motor proteins (e.g. Khc-73 and Dynein; red) and additional regulatory complexes (e.g. EB1-APC; green). These elements provide microtubule capture and forge generation effects that coordinate spindle positioning. Additional plus-end binding proteins aid in microtubule dynamics, such as polymerases (e.g. XMAP215, dark green) and depolymerases (e.g. MCAK, light green), which are necessary for proper spindle assembly and function. An assortment of protein kinases (yellow) have been shown to regulate spindle orientation through various mechanisms, including regulation of microtubule architecture (e.g. LKB and c-Src), regulation of cortical cue-adapter complexes (e.g. ABL1 and AurA) and localization of microtubule regulators such as Ran (gray) (e.g. PAK).
Whether such a capture mechanism occurs with other polarity proteins remains to be determined; the plethora of microtubule plus-end binding proteins discussed above suggests this may indeed be a widespread component of spindle orientation. Also unclear is the precise function of this spindle-capturing activity. Does static spindle capture allow for activation of additional pathways at specific sites? Are other regulators localized or offloaded to the cortex more efficiently from the astral microtubules of a stationary spindle? Do cortical force generators act with higher fidelity on a ‘captured’ spindle? Alternatively, the capture of microtubules at the edge of a cortical polarity complex may be sufficient for spindle orientation in certain cellular contexts.
Linking polarity to spindle force generation: the role of the Mud-dynein complex
Perhaps one of the most well characterized effectors in spindle orientation pathways is the Mud-dynein complex (Figs 2, 4) (Gönczy, 2008; Siller and Doe, 2009). Drosophila Mud is a large (>2000 amino acid) coiled-coil protein originally identified as a regulator of neuroblast proliferation (Guan et al., 2000). Mud shares overall weak sequence homology with the C. elegans and mammalian functional orthologs LIN-5 and NuMA, respectively (Siller et al., 2006). However, these proteins share a relatively higher conservation in a small C-terminal domain termed the Pins-binding domain (PBD). The PBD directly interacts with the Pins tetratricopeptide repeat (TPR) domain, and this interaction requires prior Gα protein-mediated relief of an autoinhibitory conformation between the TPR and GoLoco domains of Pins (Du and Macara, 2004; Nipper et al., 2007). This interaction recruits Mud to the Pins cortical crescent, thereby polarizing Mud localization (Fig. 3A). Notably, Mud also localizes to spindle poles and microtubules, independent of Pins, where it functions in the establishment and proper maintenance of focused microtubules (Bowman et al., 2006; Izumi et al., 2006; Silk et al., 2009; Siller et al., 2006). Additional studies have shown that the scaffold protein, Canoe (Cno), promotes formation of the cortical Pins-Mud complex through several small GTPases (e.g. Ran) and is important for proper spindle orientation (Fig. 3A) (Speicher et al., 2008; Wee et al., 2011).
The function of Mud in Pins-mediated spindle orientation appears to be that of a cortex-to-microtubule adaptor protein, analogous to the role played by Dlg (Fig. 4). Mud associates with cytoplasmic dynein, which serves as the sole minus-end-directed microtubule motor protein during mitosis. Dynein is a large protein complex consisting of core light, intermediate and heavy chains, the latter of which possesses the ATPase activity necessary for movement along microtubules. The non-catalytic subunits serve as binding sites for additional regulatory proteins, some of which are essential for dynein activity and, thus, spindle orientation (e.g. p150-dynactin and LIS-1) (Kardon and Vale, 2009; Siller and Doe, 2008). Precisely how Mud interacts with dynein remains unclear, although recent evidence suggests that Mud associates with the dynein light chain subunit, Cut up (Ctp), which is required for centrosomal localization of Mud and spindle orientation in neuroblasts (Fig. 3A) (Wang et al., 2011). Recent evidence also suggests that a cortically localized component of dynein, via specific interaction with Mud, is required for proper spindle orientation (Kotak et al., 2012). Thus, Mud facilitates a link between the dynein motor complex and cortically polarized Pins.
In contrast to the ‘spindle capture’ mechanism proposed for Khc-73, dynein is thought to provide the ‘force generation’ required for rapid spindle repositioning (Gönczy, 2008). The minus-end directional movement of dynein along astral microtubules (towards the spindle pole) results in net pulling forces on the spindle towards cortical Pins tethered by Mud (Gusnowski and Srayko, 2011) (Fig. 3A). Spindle-severing experiments in C. elegans embryos have established a role for the Pins-Mud-dynein complex in generating the force necessary for spindle positioning (Colombo et al., 2003; Nguyen-Ngoc et al., 2007). Additionally, the polarized localization of the Pins-Mud complex results in unequal pulling forces at one spindle pole, which probably contributes to the asymmetry in spindle morphology and subsequent size of daughter cells (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006).
The requirement of the Mud-dynein complex for proper spindle positioning has been firmly established in several systems. However, with our ‘induced polarity’ assay, we found that this complex was not sufficient to regulate spindle orientation in isolation (Johnston et al., 2009). Instead, Mud-dynein functioned synergistically with Dlg-Khc-73 downstream of Pins. Whether direct crosstalk exists between these two complexes, or if they perform otherwise independent functions that diverge below Pins, remains to be determined. Kinesin family members play an important role in plus-end delivery of various cargo proteins (Hirokawa et al., 2009), suggesting a model in which Khc-73 aids in localization of Pins-Mud pathway components. Live-cell analysis revealed that the PinsTPR-Mud-dynein component was required the for rapid, dynamic spindle movement necessary for robust orientation, further strengthening the model of Mud-dynein complex-mediated force generation. An important question for future investigation will be how the minus-end Mud-dynein pathway synergizes with that of the plus-end Dlg/Khc-73. Interestingly, cooperative involvement between opposite end-directed motor protein pathways has also been seen in meiotic spindle positioning (Ellefson and McNally, 2009).
Recently, the Mud-dynein complex was shown to regulate spindle orientation mediated through the Wnt-Frizzled (Fz) effector Dishevelled (Dsh) in both Drosophila and zebrafish (Fig. 3B) (Ségalen et al., 2010). The Fz-Dsh complex had long been known to influence planar spindle orientation in the pI division of Drosophila SOPs; however, a bona fide downstream pathway was unknown (Bellaïche et al., 2001; Gho and Schweisguth, 1998). Both genetic and biochemical evidence demonstrated that Mud acts directly downstream of the Dishevelled-Egl10-Pleckstrin (DEP) domain of Dsh (Fig. 3B). Although this model is analogous to that of Mud acting downstream of the Pins TPR domain, the DEP and TPR domains share no similarities in primary sequence or tertiary structure. These results, therefore, highlight a striking example of convergent evolution of a shared signaling pathway downstream of divergent upstream activators. Whereas the minimal TPR-binding peptide has been identified in Mud and its structure determined (Smith and Prehoda, 2011; Zhu et al., 2011b), the precise molecular basis for the Dsh-Mud complex remains to be determined. Whether additional pathways act downstream of Dsh is unclear, and a complete molecular model for Dsh-mediated spindle orientation remains to be described (Fig. 3B).
Despite ample research establishing an important role for the Mud-dynein complex in spindle orientation, many questions remain regarding its molecular function. How does Mud binding affect dynein activity? Structural and biophysical data are beginning to illuminate the conformational changes dynein might undergo to achieve its movement along microtubules (Carter et al., 2011; Redwine et al., 2012). Does Mud actively alter these structural transitions to enhance dynein function? How is synergy achieved between Mud-dynein and Dlg-Khc-73 complexes downstream of Pins? Despite diverse sequence determinants for binding, do Pins and Dsh regulate Mud-dynein function in similar ways? Does the Mud-dynein pathway function together with a secondary pathway in Dsh-mediated spindle orientation?
Phosphoregulation of spindle orientation: an assortment of kinases regulates spindle orientation
Intense efforts have recently been focused on identifying additional regulators of spindle orientation. Notably, a network of protein kinases has now been shown to influence spindle positioning in various systems. Although an in depth discussion of each kinase is beyond the scope of this Review, a cursory examination is warranted and highlights the level of complexity to which this biological process has evolved.
As discussed above, APC localizes at plus-ends and regulates MT stability and spindle orientation (Fleming et al., 2009; Wen et al., 2004; Yamashita et al., 2003). In early Drosophila embryos, the kinase Akt localizes to the cell cortex and is necessary for cortical localization of APC, and reduction of Akt results in improper centrosome positioning, bent mitotic spindles, and spindle misorientation in epithelial cells (Buttrick et al., 2008). The p21-activated kinase (PAK) regulates astral microtubule formation and controls mitotic spindle orientation and cortical anchoring, possibly by regulating localization of the dynein-dynactin complex (Bompard et al., 2013). PAK-mediated phosphorylation of the Ran GTPase promotes active Ran at the centrosome (Bompard et al., 2010). Interestingly, Ran activation has been shown to promote cortical Mud localization in Drosophila (Speicher et al., 2008; Wee et al., 2011). Thus, PAK phosphorylation may act to initiate Ran-mediated formation of the Pins-Mud spindle orientation complex.
The energy-regulated AMP-activated kinase (AMPK; SNF1A in Drosophila; PRKAA1 in mammals) localizes to spindle poles during mitosis, and a recent study of human cell culture systems demonstrated that loss of AMPK results in astral microtubule abnormalities and spindle misorientation (Thaiparambil et al., 2012). This study identified the myosin regulatory light chain (MRLC) as an essential AMPK target, although how phosphorylated MRLC regulates spindle orientation remains unknown. Additionally, the spindle orientation activity of AMPK is dependent upon activation by the upstream kinase LKB1 (Thaiparambil et al., 2012; Wei et al., 2012). Cell culture studies have also demonstrated a role for the tyrosine kinase c-Src (also known as Csk) in spindle orientation. Spindle misorientation is seen in early prometaphase upon loss of c-Src, which localizes to spindle poles; however, no apparent c-Src target has been identified (Nakayama et al., 2012). Cells deficient in c-Src demonstrate severely reduced astral microtubules, suggesting the role of c-Src is to promote spindle pole development. Notably, an earlier study in C. elegans showed that c-Src (CSK-1) is necessary for proper spindle rotation in the EMS cell division and proposed that the kinase functions downstream of Wnt signaling, which is necessary for proper endoderm specification (Bei et al., 2002).
Using a HeLa cell model, and translating findings in vivo to mouse epidermis, Matsumura and colleagues recently performed a kinase-targeted RNAi screen for regulators of spindle orientation and identified the Abelson kinase (ABL1) (Matsumura et al., 2012). Loss of spindle orientation relative to the cell-substrate adhesion plane resulted from delocalization of the human Pins homolog LGN (also known as GPSM2) from polarized to uniform cortical. Moreover, ABL1 phosphorylation of the Mud homolog NuMA was necessary for maintenance of the LGN-NuMA cortical complex. It remains to be determined precisely how ABL1 phosphorylation regulates localization of the LGN-NuMA complex. A similar delocalization of LGN was recently shown in an epithelial cell system upon loss of aPKC (Hao et al., 2010). aPKC phosphorylates LGN at the apical surface, which promotes 14-3-3 binding and removal of LGN from this region of the cell cortex. In the absence of aPKC-mediated phosphorylation, LGN accumulates at the apical cortex and promotes the loss of planar spindle orientation. Interestingly, this phosphorylation occurs at the conserved Aurora-A phosphorylation site previously shown to promote the Pins-Dlg pathway (Johnston et al., 2012; Johnston et al., 2009). This suggests that diverse cell types might use different kinase-dependent pathways phosphorylating the same position to achieve unique spindle orientation outcomes.
Collectively, these studies have revealed an important role for phosphoregulation of spindle orientation pathways (Fig. 4). A common theme for kinase function appears to be regulating subcellular localization of specific spindle orientation components. In some cases, phosphorylation promotes cortical localization, whereas it results in cortical removal in other cases. Many unanswered questions await further research in this area. What are the molecular mechanisms involved in phosphorylation-dependent localization patterns? How does phosphorylation affect direct protein-protein interactions underlying the macromolecular complexes involved in spindle orientation? What specific residues are phosphorylated in each of these components? What are the spatiotemporal constraints of phosphoregulation? Finally, what are the phosphatases that direct dephosphorylation and how are they regulated throughout the cell cycle?
Conclusions
The ability to orient cell division allows for the generation of cellular diversity and for regulated spatial placement of daughter cells within tissues. Nature has evolved sophisticated signaling pathways that allow cortical cues to communicate with the mitotic spindle to influence its position prior to cell division (Fig. 4). The microtubule plus-end is a node for the localization of diverse +TIPs that serve as spindle-capture prey. Whether autonomously localized proteins, such as XMAP215 and CLIP-170, or cargo of the essential +TIP EB1, such as APC, these assorted members regulate microtubule organization and aid in the cortical connection of the spindle. The cortex, by contrast, contains orientation cues, such as Pins and Dsh, that organize protein complexes to function as spindle-capturing bait as well as force generators for the rapid spindle motions required for repositioning during cell division. In this Review, we have summarized many of the seminal findings describing the molecular basis for spindle orientation. Although significant progress has been made on this front, it is apparent that the surface has merely been scratched. What additional +TIPs might serve as spindle-capturing prey? How do microtubule dynamics and the regulatory proteins involved relate to spindle capture and force generation? What other cortical complexes serve as spindle positioning cues, and in what cell types do these function? Is dynein the lone force generator involved in spindle orientation, and how is the dynein complex regulated at the molecular level? How do divergent spindle capturing events cooperate with the conserved force-generating complex? Finally, does the association between spindle orientation defects and human disease play a causal role, and can spindle orientation regulators represent potential therapeutic targets? We excitedly await further analysis of this fascinating biological process.
Acknowledgments
The authors would like to thank Dr Chris Q. Doe for careful reading of the manuscript and insightful comments.
Footnotes
Funding
Much of the authors’ personal work discussed in this Review was funded by a National Institutes of Health Training grant [M.S.L.] and a Damon Runyon Cancer Research Foundation fellowship award [C.A.J.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Akhmanova A., Hoogenraad C. C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B. M., De Zeeuw C. I., Grosveld F., et al. (2001). Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923–935 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., Larsen N. A., Hyman A. A., Harrison S. C. (2007). Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure 15, 355–362 [DOI] [PubMed] [Google Scholar]
- Arata Y., Lee J. Y., Goldstein B., Sawa H. (2010). Extracellular control of PAR protein localization during asymmetric cell division in the C. elegans embryo. Development 137, 3337–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei Y., Hogan J., Berkowitz L. A., Soto M., Rocheleau C. E., Pang K. M., Collins J., Mello C. C. (2002). SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev. Cell 3, 113–125 [DOI] [PubMed] [Google Scholar]
- Bellaïche Y., Gho M., Kaltschmidt J. A., Brand A. H., Schweisguth F. (2001). Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat. Cell Biol. 3, 50–57 [DOI] [PubMed] [Google Scholar]
- Bellanger J. M., Carter J. C., Phillips J. B., Canard C., Bowerman B., Gönczy P. (2007). ZYG-9, TAC-1 and ZYG-8 together ensure correct microtubule function throughout the cell cycle of C. elegans embryos. J. Cell Sci. 120, 2963–2973 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 [DOI] [PubMed] [Google Scholar]
- Bompard G., Rabeharivelo G., Frank M., Cau J., Delsert C., Morin N. (2010). Subgroup II PAK-mediated phosphorylation regulates Ran activity during mitosis. J. Cell Biol. 190, 807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G., Rabeharivelo G., Cau J., Abrieu A., Delsert C., Morin N. (2013). P21-activated kinase 4 (PAK4) is required for metaphase spindle positioning and anchoring. Oncogene 32, 910–919 [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Neumüller R. A., Novatchkova M., Du Q., Knoblich J. A. (2006). The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742 [DOI] [PubMed] [Google Scholar]
- Buey R. M., Sen I., Kortt O., Mohan R., Gfeller D., Veprintsev D., Kretzschmar I., Scheuermann J., Neri D., Zoete V., et al. (2012). Sequence determinants of a microtubule tip localization signal (MtLS). J. Biol. Chem. 287, 28227–28242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttrick G. J., Beaumont L. M., Leitch J., Yau C., Hughes J. R., Wakefield J. G. (2008). Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J. Cell Biol. 180, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello J., Neukomm L. J., Günesdogan U., Burkart K., Charette S. J., Lochnit G., Hengartner M. O., Schnabel R. (2010). The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 8, e1000297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C., Doe C. Q. (2009). Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17, 134–141 [DOI] [PubMed] [Google Scholar]
- Carter A. P., Cho C., Jin L., Vale R. D. (2011). Crystal structure of the dynein motor domain. Science 331, 1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N. N., Kirby C. M., Kemphues K. J. (1995). Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par-2 and par-3. Genetics 139, 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo K., Grill S. W., Kimple R. J., Willard F. S., Siderovski D. P., Gönczy P. (2003). Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science 300, 1957–1961 [DOI] [PubMed] [Google Scholar]
- Conklin E. G. (1905). The organization and cell-lineage of the ascidian embryo. J. Acad. Nat. Sci. Phila. 13, 1–119 [Google Scholar]
- Culver-Hanlon T. L., Lex S. A., Stephens A. D., Quintyne N. J., King S. J. (2006). A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat. Cell Biol. 8, 264–270 [DOI] [PubMed] [Google Scholar]
- David N. B., Martin C. A., Segalen M., Rosenfeld F., Schweisguth F., Bellaïche Y. (2005). Drosophila Ric-8 regulates Galphai cortical localization to promote Galphai-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat. Cell Biol. 7, 1083–1090 [DOI] [PubMed] [Google Scholar]
- Delaval B., Bright A., Lawson N. D., Doxsey S. (2011). The cilia protein IFT88 is required for spindle orientation in mitosis. Nat. Cell Biol. 13, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. Q. (2008). Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575–1587 [DOI] [PubMed] [Google Scholar]
- Du Q., Macara I. G. (2004). Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516 [DOI] [PubMed] [Google Scholar]
- Du Q., Stukenberg P. T., Macara I. G. (2001). A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 3, 1069–1075 [DOI] [PubMed] [Google Scholar]
- Ellefson M. L., McNally F. J. (2009). Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol. Biol. Cell 20, 2722–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S., Kemphues K. J. (1995). Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83, 743–752 [DOI] [PubMed] [Google Scholar]
- Fink J., Carpi N., Betz T., Bétard A., Chebah M., Azioune A., Bornens M., Sykes C., Fetler L., Cuvelier D., et al. (2011). External forces control mitotic spindle positioning. Nat. Cell Biol. 13, 771–778 [DOI] [PubMed] [Google Scholar]
- Fischer E., Legue E., Doyen A., Nato F., Nicolas J. F., Torres V., Yaniv M., Pontoglio M. (2006). Defective planar cell polarity in polycystic kidney disease. Nat. Genet. 38, 21–23 [DOI] [PubMed] [Google Scholar]
- Fleming E. S., Temchin M., Wu Q., Maggio-Price L., Tirnauer J. S. (2009). Spindle misorientation in tumors from APC(min/+) mice. Mol. Carcinog. 48, 592–598 [DOI] [PubMed] [Google Scholar]
- Gauthier L. R., Charrin B. C., Borrell-Pagès M., Dompierre J. P., Rangone H., Cordelières F. P., De Mey J., MacDonald M. E., Lessmann V., Humbert S., et al. (2004). Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118, 127–138 [DOI] [PubMed] [Google Scholar]
- Gho M., Schweisguth F. (1998). Frizzled signalling controls orientation of asymmetric sense organ precursor cell divisions in Drosophila. Nature 393, 178–181 [DOI] [PubMed] [Google Scholar]
- Godin J. D., Colombo K., Molina-Calavita M., Keryer G., Zala D., Charrin B. C., Dietrich P., Volvert M. L., Guillemot F., Dragatsis I., et al. (2010). Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron 67, 392–406 [DOI] [PubMed] [Google Scholar]
- Goldstein B. (1995). Cell contacts orient some cell division axes in the Caenorhabditis elegans embryo. J. Cell Biol. 129, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2008). Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355–366 [DOI] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., Hyman A. A. (1999). Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147, 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. (2007). Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8, 462–472 [DOI] [PubMed] [Google Scholar]
- Grill S. W., Gönczy P., Stelzer E. H., Hyman A. A. (2001). Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409, 630–633 [DOI] [PubMed] [Google Scholar]
- Guan Z., Prado A., Melzig J., Heisenberg M., Nash H. A., Raabe T. (2000). Mushroom body defect, a gene involved in the control of neuroblast proliferation in Drosophila, encodes a coiled-coil protein. Proc. Natl. Acad. Sci. USA 97, 8122–8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S. L., Salmon W. C., Waterman-Storer C. M. (2002). Converging populations of f-actin promote breakage of associated microtubules to spatially regulate microtubule turnover in migrating cells. Curr. Biol. 12, 1891–1899 [DOI] [PubMed] [Google Scholar]
- Gusnowski E. M., Srayko M. (2011). Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J. Cell Biol. 194, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T., Lin L., Tibaldi E. V., Reinherz E. L., Chishti A. H. (2000). GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. J. Biol. Chem. 275, 28774–28784 [DOI] [PubMed] [Google Scholar]
- Hao Y., Du Q., Chen X., Zheng Z., Balsbaugh J. L., Maitra S., Shabanowitz J., Hunt D. F., Macara I. G. (2010). Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 20, 1809–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig O. (1884). Das problem der befruchtung und der isotropie des eies. eine theorie der vererbung. Jena. Z Med. Naturwiss. 18, 276–318 [Google Scholar]
- Hildebrandt F., Otto E. (2005). Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 6, 928–940 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y., Niwa S. (2009). Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J., Buey R. M., Lawera A., et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366–376 [DOI] [PubMed] [Google Scholar]
- Huckaba T. M., Gennerich A., Wilhelm J. E., Chishti A. H., Vale R. D. (2011). Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J. Biol. Chem. 286, 7457–7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. (2003). Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161, 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y., Ohta N., Hisata K., Raabe T., Matsuzaki F. (2006). Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8, 586–593 [DOI] [PubMed] [Google Scholar]
- Jiang K., Toedt G., Montenegro Gouveia S., Davey N. E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L. B., Bezstarosti K., et al. (2012). A proteome-wide screen for mammalian sxip motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800–1807 [DOI] [PubMed] [Google Scholar]
- Johnston C. A., Hirono K., Prehoda K. E., Doe C. Q. (2009). Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 138, 1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C. A., Doe C. Q., Prehoda K. E. (2012). Structure of an enzyme-derived phosphoprotein recognition domain. PLoS ONE 7, e36014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen J. A., SanAgustin J., Baker S. P., Pazour G. J. (2012). Disruption of IFT complex A causes cystic kidneys without mitotic spindle misorientation. J. Am. Soc. Nephrol. 23, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon J. R., Vale R. D. (2009). Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10, 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311–320 [DOI] [PubMed] [Google Scholar]
- Komarova Y. A., Akhmanova A. S., Kojima S., Galjart N., Borisy G. G. (2002). Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159, 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T., Matsuzaki F. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93–101 [DOI] [PubMed] [Google Scholar]
- Korinek W. S., Copeland M. J., Chaudhuri A., Chant J. (2000). Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science 287, 2257–2259 [DOI] [PubMed] [Google Scholar]
- Kotak S., Busso C., Gönczy P. (2012). Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski C., Srayko M., Nedelec F. (2007). Cortical microtubule contacts position the spindle in C. elegans embryos. Cell 129, 499–510 [DOI] [PubMed] [Google Scholar]
- Kraut R., Chia W., Jan L. Y., Jan Y. N., Knoblich J. A. (1996). Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, 50–55 [DOI] [PubMed] [Google Scholar]
- Kumar P., Wittmann T. (2012). +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22, 418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A., Knoblich J. A. (2012). Spindle orientation in mammalian cerebral cortical development. Curr. Opin. Neurobiol. 22, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Andersen R. O., Cabernard C., Manning L., Tran K. D., Lanskey M. J., Bashirullah A., Doe C. Q. (2006a). Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20, 3464–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Robinson K. J., Doe C. Q. (2006b). Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594–598 [DOI] [PubMed] [Google Scholar]
- Li R., Gundersen G. G. (2008). Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9, 860–873 [DOI] [PubMed] [Google Scholar]
- Loughlin R., Wilbur J. D., McNally F. J., Nédélec F. J., Heald R. (2011). Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Usui T., Uemura T., Jan L., Jan Y. N. (1999). Flamingo controls the planar polarity of sensory bristles and asymmetric division of sensory organ precursors in Drosophila. Curr. Biol. 9, 1247–1250 [DOI] [PubMed] [Google Scholar]
- Lu B., Roegiers F., Jan L. Y., Jan Y. N. (2001). Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature 409, 522–525 [DOI] [PubMed] [Google Scholar]
- Markowitz S. D., Bertagnolli M. M. (2009). Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 361, 2449–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S. M., Lee W. L. (2011). Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell 20, 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S., Hamasaki M., Yamamoto T., Ebisuya M., Sato M., Nishida E., Toyoshima F. (2012). ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat. Commun. 3, 626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. (2005). Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 7, 235–245 [DOI] [PubMed] [Google Scholar]
- Miller R. K., D’Silva S., Moore J. K., Goodson H. V. (2006). The CLIP-170 orthologue Bik1p and positioning the mitotic spindle in yeast. Curr. Top. Dev. Biol. 76, 49–87 [DOI] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J. M. (2005). Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell 16, 3176–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y., Matsui Y., Takeda Y., Okamoto M., Abe K., Fukumoto Y., Yamaguchi N. (2012). c-Src but not Fyn promotes proper spindle orientation in early prometaphase. J. Biol. Chem. 287, 24905–24915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T., Afshar K., Gönczy P. (2007). Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9, 1294–1302 [DOI] [PubMed] [Google Scholar]
- Nipper R. W., Siller K. H., Smith N. R., Doe C. Q., Prehoda K. E. (2007). Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc. Natl. Acad. Sci. USA 104, 14306–14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi Y., Uchimura S., Ohki T., Mikhailenko S. V., Ishiwata S. (2011). The bidirectional depolymerizer MCAK generates force by disassembling both microtubule ends. Nat. Cell Biol. 13, 846–852 [DOI] [PubMed] [Google Scholar]
- Pease J. C., Tirnauer J. S. (2011). Mitotic spindle misorientation in cancer - out of alignment and into the fire. J. Cell Sci. 124, 1007–1016 [DOI] [PubMed] [Google Scholar]
- Pecreaux J., Roper J. C., Kruse K., Julicher F., Hyman A. A., Grill S. W., Howard J. (2006). Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 16, 2111–2122 [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Seydoux G. (2002). Anterior-posterior polarity in C. elegans and Drosophila - PARallels and differences. Science 298, 1946–1950 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J. A. (2001). DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3, 43–49 [DOI] [PubMed] [Google Scholar]
- Peyre E., Jaouen F., Saadaoui M., Haren L., Merdes A., Durbec P., Morin X. (2011). A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 193, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. (2011). Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21, 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson N. D., Lechler T. (2010). Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol. 191, 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quyn A. J., Appleton P. L., Carey F. A., Steele R. J., Barker N., Clevers H., Ridgway R. A., Sansom O. J., Näthke I. S. (2010). Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6, 175–181 [DOI] [PubMed] [Google Scholar]
- Redwine W. B., Hernández-López R., Zou S., Huang J., Reck-Peterson S. L., Leschziner A. E. (2012). Structural basis for microtubule binding and release by dynein. Science 337, 1532–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez O. C., Schaefer A. W., Mandato C. A., Forscher P., Bement W. M., Waterman-Storer C. M. (2003). Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5, 599–609 [DOI] [PubMed] [Google Scholar]
- Roegiers F., Younger-Shepherd S., Jan L. Y., Jan Y. N. (2001). Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat. Cell Biol. 3, 58–67 [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. (2002). Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A., Shevchenko A., Knoblich J. A. (2000). A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353–362 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Petronczki M., Dorner D., Forte M., Knoblich J. A. (2001). Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183–194 [DOI] [PubMed] [Google Scholar]
- Schepers A. G., Vries R., van den Born M., van de Wetering M., Clevers H. (2011). Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 30, 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger A., Shelton C. A., Maloof J. N., Meneghini M., Bowerman B. (1999). Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev. 13, 2028–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M., Schaefer M., Knoblich J. A. (1999). Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402, 548–551 [DOI] [PubMed] [Google Scholar]
- Ségalen M., Johnston C. A., Martin C. A., Dumortier J. G., Prehoda K. E., David N. B., Doe C. Q., Bellaïche Y. (2010). The Fz-Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish. Dev. Cell 19, 740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M. A., Pellman D. (2003). Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13, 364–372 [DOI] [PubMed] [Google Scholar]
- Siegrist S. E., Doe C. Q. (2005). Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123, 1323–1335 [DOI] [PubMed] [Google Scholar]
- Silk A. D., Holland A. J., Cleveland D. W. (2009). Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184, 677–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K. H., Doe C. Q. (2008). Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev. Biol. 319, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K. H., Doe C. Q. (2009). Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11, 365–374 [DOI] [PubMed] [Google Scholar]
- Siller K. H., Serr M., Steward R., Hays T. S., Doe C. Q. (2005). Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell 16, 5127–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K. H., Cabernard C., Doe C. Q. (2006). The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8, 594–600 [DOI] [PubMed] [Google Scholar]
- Skop A. R., White J. G. (1998). The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 8, 1110–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep K. C. (2009). The role of TOG domains in microtubule plus end dynamics. Biochem. Soc. Trans. 37, 1002–1006 [DOI] [PubMed] [Google Scholar]
- Slep K. C. (2010). Structural and mechanistic insights into microtubule end-binding proteins. Curr. Opin. Cell Biol. 22, 88–95 [DOI] [PubMed] [Google Scholar]
- Slep K. C., Vale R. D. (2007). Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell 27, 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. R., Prehoda K. E. (2011). Robust spindle alignment in Drosophila neuroblasts by ultrasensitive activation of pins. Mol. Cell 43, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher S., Fischer A., Knoblich J., Carmena A. (2008). The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr. Biol. 18, 831–837 [DOI] [PubMed] [Google Scholar]
- Su X., Ohi R., Pellman D. (2012). Move in for the kill: motile microtubule regulators. Trends Cell Biol. 22, 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler J. M., Yamanaka H., Green J. B. (2010). PAR-1 promotes primary neurogenesis and asymmetric cell divisions via control of spindle orientation. Development 137, 2501–2505 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M. E., Medema R. H., Akhmanova A. (2011). Regulation of localization and activity of the microtubule depolymerase MCAK. BioArchitecture 1, 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiparambil J. T., Eggers C. M., Marcus A. I. (2012). AMPK regulates mitotic spindle orientation through phosphorylation of myosin regulatory light chain. Mol. Cell. Biol. 32, 3203–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma C. R., Toso A., Gutbrodt K. L., Reggi S. P., Frew I. J., Schraml P., Hergovich A., Moch H., Meraldi P., Krek W. (2009). VHL loss causes spindle misorientation and chromosome instability. Nat. Cell Biol. 11, 994–1001 [DOI] [PubMed] [Google Scholar]
- Thoma C. R., Matov A., Gutbrodt K. L., Hoerner C. R., Smole Z., Krek W., Danuser G. (2010). Quantitative image analysis identifies pVHL as a key regulator of microtubule dynamic instability. J. Cell Biol. 190, 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A. J., Rybina S., Pozniakovsky A., Mayer T. U., Walczak C. E., Karsenti E., Hyman A. A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13–19 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. (2007). Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26, 1487–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle J. A., Cooper J. A., Waterston R. H. (1994). Transient localized accumulation of actin in Caenorhabditis elegans blastomeres with oriented asymmetric divisions. Development 120, 2317–2328 [DOI] [PubMed] [Google Scholar]
- Wallingford J. B. (2012). Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28, 627–653 [DOI] [PubMed] [Google Scholar]
- Walston T., Tuskey C., Edgar L., Hawkins N., Ellis G., Bowerman B., Wood W., Hardin J. (2004). Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev. Cell 7, 831–841 [DOI] [PubMed] [Google Scholar]
- Wang C., Li S., Januschke J., Rossi F., Izumi Y., Garcia-Alvarez G., Gwee S. S., Soon S. B., Sidhu H. K., Yu F., et al. (2011). An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev. Cell 21, 520–533 [DOI] [PubMed] [Google Scholar]
- Wee B., Johnston C. A., Prehoda K. E., Doe C. Q. (2011). Canoe binds RanGTP to promote Pins(TPR)/Mud-mediated spindle orientation. J. Cell Biol. 195, 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Bhattaram V. K., Igwe J. C., Fleming E., Tirnauer J. S. (2012). The LKB1 tumor suppressor controls spindle orientation and localization of activated AMPK in mitotic epithelial cells. PLoS ONE 7, e41118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004). EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- Werts A. D., Roh-Johnson M., Goldstein B. (2011). Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development 138, 4411–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. E., Beronja S., Pasolli H. A., Fuchs E. (2011). Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., Knust E. (1999). Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402, 544–547 [DOI] [PubMed] [Google Scholar]
- Wu J. C., Rose L. S. (2007). PAR-3 and PAR-1 inhibit LET-99 localization to generate a cortical band important for spindle positioning in Caenorhabditis elegans embryos. Mol. Biol. Cell 18, 4470–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. H., Hanada T., Chishti A. H. (2007). The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry 46, 10039–10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y. M., Jones D. L., Fuller M. T. (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 [DOI] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X., Chia W. (2000). Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409 [DOI] [PubMed] [Google Scholar]
- Yuan H., Chiang C. Y., Cheng J., Salzmann V., Yamashita Y. M. (2012). Regulation of cyclin A localization downstream of Par-1 function is critical for the centrosome orientation checkpoint in Drosophila male germline stem cells. Dev. Biol. 361, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Shang Y., Xia C., Wang W., Wen W., Zhang M. (2011a). Guanylate kinase domains of the MAGUK family scaffold proteins as specific phospho-protein-binding modules. EMBO J. 30, 4986–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Wen W., Zheng Z., Shang Y., Wei Z., Xiao Z., Pan Z., Du Q., Wang W., Zhang M. (2011b). LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Gαi/LGN/NuMA pathways. Mol. Cell 43, 418–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal R. R., Ahringer J., van Luenen H. G., Rushforth A., Anderson P., Plasterk R. H. (1996). G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell 86, 619–629 [DOI] [PubMed] [Google Scholar]