Abstract
Release factor eRF1 plays a key role in the termination of protein synthesis in eukaryotes. The eRF1 consists of three domains (N, M and C) that perform unique roles in termination. Previous studies of eRF1 point mutants and standard/variant code eRF1 chimeras unequivocally demonstrated a direct involvement of the highly conserved N-domain motifs (NIKS, YxCxxxF and GTx) in stop codon recognition. In the current study, we extend this work by investigating the role of the 41 invariant and conserved N-domain residues in stop codon decoding by human eRF1. Using a combination of the conservative and non-conservative amino acid substitutions, we measured the functional activity of >80 mutant eRF1s in an in vitro reconstituted eukaryotic translation system and selected 15 amino acid residues essential for recognition of different stop codon nucleotides. Furthermore, toe-print analyses provide evidence of a conformational rearrangement of ribosomal complexes that occurs during binding of eRF1 to messenger RNA and reflects stop codon decoding activity of eRF1. Based on our experimental data and molecular modelling of the N-domain at the ribosomal A site, we propose a two-step model of stop codon decoding in the eukaryotic ribosome.
INTRODUCTION
Translation termination is mediated by the cooperative action of class 1 and class 2 polypeptide release factors (RFs) in the ribosome [reviewed in (1–5)]. When a stop codon occupies the ribosomal A site, class 1 RFs decode the stop codon and induce peptide release from the peptidyl transferase centre. In prokaryotes, two class 1 RFs, RF1 and RF2, decode UAA/UAG and UAA/UGA stop codons, respectively. In contrast, a single RF, eRF1, recognizes all three stop codons in eukaryotes. The eRF1 consists of three well-defined domains (6,7): the N-terminal domain is responsible for stop codon decoding (8–18), the M-(middle) domain is essential for peptidyl-tRNA hydrolysis at the peptidyl transferase centre (6,19,20), and the C-domain recruits the class 2 RF eRF3 (21–25). Class 2 RFs (RF3 in prokaryotes and eRF3 in eukaryotes) are translational GTPases. Although RF3 mediates recycling of class 1 RFs from post-termination complexes (26–28), eRF3 stimulates the activity of eRF1 in the presence of GTP (29–31).
Crystal structures of prokaryotic translation termination complexes have elucidated the mechanism by which RF1 (32–34) and RF2 (34,35) mediates stop codon recognition. Although crystal structures of ribosomes from lower eukaryotes have been solved (36–38), X-ray structures of eukaryotic translation termination complexes have not yet been reported. Rather, stop codon decoding in eukaryotes has been studied by a combination of kinetic, biochemical and genetic approaches. These have provided several hypotheses for the mechanism of stop codon recognition and proposed specific amino acid residues within the N-domain of eRF1 responsible for stop codon decoding (6,8–12,14–18,25,39–49).
Cross-linking experiments (39,40) and biochemical data (12,41,42) have implicated the highly conserved TASNIKS motif (positions 58–64 of human eRF1, Figure 1) in stop codon recognition. The invariant YxCxxxF motif (positions 125–131, Figure 1) has been implicated in discriminating the second and third purines of the stop codon (12,15,18,25,43). Residues of the strictly conserved GTS loop (positions 31–33 of human eRF1, Figure 1) are either in direct contact with the stop codon or implicated in its decoding (25,40,44–46). Mutagenesis based on the crystal structure of yeast eRF1•eRF3 complex with cocrystallized adenosine triphosphate (ATP) (which the authors suggest mimics a stop codon nucleotide) indicates a role for residue T32 in stop codon recognition (25). Cross-linking experiments also show that the GTx motif is proximal to the second and third guanine but more distal to the second and third adenine of stop codons, which may be compatible with alternate N-domain eRF1 conformations towards different stop codon bases within the ribosome (45).
Figure 1.
Alignment of amino acid sequences of the N-terminal domains of human, yeast and Euplotes eRF1s. Residue numbering is that of the human eRF1. Identical, conserved and semiconserved amino acid residues are black, dark grey and light grey, respectively.
The nuclear magnetic resonance solution structures of the N-domain of both the human wild-type (wt) eRF1 and UGA-only unipotent QFM_F mutant (T122Q/S123F/L124M/L126F) give evidence that these point mutations alter conformation of the strictly conserved GTS loop implicated in UGA decoding (46).
An important approach for dissecting the role of the N-domain determinants in stop codon recognition has involved the use of chimeric eRF1s, which contain the regions of the eRF1 N-domains from organisms with standard (human/yeast) and variant (ciliate) genetic codes. Functional analysis of these eRF1 chimeras has identified residues in the N-domain of ciliate eRF1s that prevent the recognition of specific stop codons. For example, an eRF1 chimera consisting of the Tetrahymena thermophila N-domain and the MC- domains of Schizosaccharomyces pombe exhibited the UGA-only specificity of T. thermophila eRF1, confirming the importance of the KATNIKD motif (18,42). In contrast, an eRF1 hybrid consisting of the T. thermophila N-domain fused to the MC- domains of Saccharomyces cerevisiae efficiently terminated at all three stop codons, when expressed in yeast cells, indicating that the eRF1 N-domain is not sufficient to determine UGA-only specificity in this species (47). The motifs sufficient for the UGA-only specificity of Paramecium tetraurelia and Stylonychia mytilus eRF1s were identified (SIKD/DPQ and QFM, respectively) by creating chimeric constructs (ciliate/human) in combination with site-directed mutagenesis (17). A similar approach identified a single A70 amino acid residue that determined the UAR-only specificity of Euplotes eRF1 (48).
To compare the relative importance of the TASNIKS and YxCxxxF motifs in stop codon decoding, these motifs from six variant code organisms—Euplotes and Blepharisma (UAR-only specificity) and Paramecium, Tetrahymena, Oxytrichia and Loxodes (UGA-only specificity)—were introduced into eRF1 of S. cerevisiae either separately or together (18). It was shown that differences between the YxCxxxF motifs of investigated species correlate with stop codon specificity typical for variant code organism.
As summarized earlier in the text, a variety of different approaches have together implicated a several eRF1 N-domain motifs in stop codon recognition. Although there is often coherence between independent studies, there is also disagreement and contradiction. This could be due to the use of diverse systems for assaying of eRF1 RF activity, including the simplified in vitro fMet release assay (49,50), a fully reconstituted in vitro eukaryotic translation system (31) and two in vivo assays—the dual gene reporter system in HeLa cells (17) and yeast cells with homologous yeast eRF1 (30,47).
In the current work, we have attempted to identify the critical stop codon recognition residues of eRF1 by comprehensively screening mutants of all N-domain invariant and conserved positions using a single physiological assay, the fully reconstituted in vitro eukaryotic translation system (31). Using this approach, we have selected 15 amino acid residues within the N-domain of human eRF1 important for stop codon decoding. Furthermore, we have demonstrated the ability of eRF3•GTP to improve the efficiency of mutant eRF1s in stop codon recognition. Based on our observations and molecular modelling of eRF1 at the ribosomal A site, we propose a two-step model for stop codon decoding by eRF1 in the eukaryotic ribosome.
MATERIALS AND METHODS
Cloning and mutagenesis of human eRF1
Plasmids with mutant eRF1 genes were obtained by site-directed mutagenesis using the polymerase chain reaction (PCR)-based ‘megaprimer’ method as described previously (15). The resulting PCR products were inserted into the NdeI–Bst98I sites of the pERF4b plasmid. The sequences of the PCR primers used for the generation of the eRF1 mutants are available in Supplementary Data (Supplementary Table S1).
Ribosomal subunits and recombinant proteins
The 40S and 60S ribosomal subunits, as well as eukaryotic translation factors eIF2, eIF3, eIF4F, eEF1H and eEF2, were purified from a rabbit reticulocyte lysate as described (31). The eukaryotic translation factors eIF1, eIF1A, eIF4A, eIF4B, eIF5B, eIF5, wt eRF1, mutant eRF1s and eRF3c lacking the N-terminal 138 amino acid residues were produced as recombinant proteins in Escherichia coli strain BL21 with subsequent protein purification on Ni-NTA agarose and ion-exchange chromatography (19,31).
Messenger RNA transcripts
Messenger RNA (mRNA) was transcribed by T7 RNA polymerase from MVHL-stop plasmids, contained T7 promoter, four CAA repeats, the β-globin 5′-untranslated region (UTR) and Met, Val, His and Leu codons followed by one of the three stop codons (UAA, UAG or UGA),and a 3′-UTR comprising the rest of the natural β-globin coding sequence (51). For run-off transcription, all plasmids were linearized with XhoI.
Pretermination complex assembly and purification
Pretermination complexes (preTC) were assembled as described (31). Briefly, 37 pmol of MVHL-stop mRNAs were incubated for 30 min in buffer A (20 mM Tris acetate (pH 7.5), 100 mM KAc, 2.5 mM MgCl2, 2 mM dithiothreitol (DTT)) supplemented with 400 U RNase inhibitor (RiboLock, Fermentas), 1 mM ATP, 0.25 mM spermidine, 0.2 mM GTP, 75 μg of total tRNA (acylated with Val, His, Leu and [35S]Met), 75 pmol 40S and 60S purified ribosomal subunits, 125 pmol eIF2, eIF3, eIF4F, eIF4A, eIF4B, eIF1, eIF1A, eIF5, eIF5B each, 200 pmol eEF1H and 50 pmol eEF2 and then centrifuged in a Beckman SW55 rotor for 95 min at 4°C and 50 000 rpm in a 10–30% (w/w) linear sucrose density gradient prepared in buffer A with 5 mM MgCl2. Fractions corresponding to preTC complexes according to optical density and the presence of [35S]Met were combined, diluted 3-fold with buffer A containing 1.25 mM MgCl2 (to a final concentration of 2.5 mM Mg2+) and used in peptide release assay and termination efficiency assays or conformational rearrangement analysis.
Peptide release assay
The peptide release assay was conducted as described (31) with minor modifications, as follows. Aliquots containing 0.015 pmol of the preTC assembled in the presence of [35S]Met-tRNA were incubated at 37°C for 15 min with 10 pmol of wt eRF1 or mutant eRF1s. Ribosomes and tRNA were pelleted with ice-cold 5% trichloroacetic acid (TCA) supplemented with 0.75% (w/v) casamino acids and centrifuged at 14 000g at 4°C. The amount of released [35S]-containing peptide was determined by scintillation counting of supernatants using an Intertechnique SL-30 liquid scintillation spectrometer.
Termination efficiency assay
Termination efficiency was determined as described (48). Briefly, 50 µl of aliquots containing 0.0125 pmol of preTC assembled in the presence of [35S]Met-tRNA were incubated at 37°C with 2.5 pmol of eRF1 for 0–15 min or with 0.125 pmol eRF1 in presence of 0.125 pmol eRF3 and 0.2 mM GTP, 0.2 mM MgCl2 for 0–3 min. Ribosomes and tRNA were pelleted with ice-cold 5% TCA supplemented with 0.75% (w/v) casamino acids and centrifuged at 4°C and 14 000g. The amount of released [35S]Met-containing tetrapeptide, which indicated the efficiency of peptidyl-tRNA hydrolysis, was determined by scintillation counting of supernatants. Linear increase of kobs versus eRF1 concentration is shown in Supplementary Figure S1.
eRF3 GTPase activity assay
This method is based on the measurement of [32P]Pi release, using a modified charcoal precipitation method (52). The incubation mixture (12.5 μl) contained 20 mМ Tris–HCl (pH 7.5), 30 mМ NH4Cl, 15 mМ MgCl2, 0.16 μМ ribosomes, 0.16 μМ human eRF3c and 0.5 μМ [γ-32P]GTP (10 000 cpm/pmol); the human wt eRF1or its mutants were added to give 0.04, 0.08, 0.12 and 0.16 μМ final concentrations. The reactions were run at 30°C for 20 min and terminated by mixing with 0.5 ml of a 5% (w/v) activated charcoal suspension in 50 mМ NaH2PO4, cooled on ice. The mixture was vortexed and centrifuged at 16 000g for 10 min at 4°C. Aliquots of the supernatants (0.375 ml) were counted on a scintillation counter. Values of eRF3 GTPase activity and corresponding error limits were estimated from three experiments carried out for each eRF1 mutant.
Conformational rearrangement analysis
Aliquots containing 0.2 pmol of the preTC were incubated with 10 pmol of wt eRF1 or 80 pmol of its domains for 10 min at 37°C and analysed using a primer extension protocol, as described (53,54). Toe-printing analysis was performed with a 5′-FAM labelled primer 5′-FAM-GCATTTGCAGAGGACAGG-3′ complementary to β-globin mRNA nucleotides 197–214. Complementary DNAs were separated by electrophoresis using standard GeneScan® conditions on an ABI Prism® Genetic Analyser 3100 (Applera) with ILS 600 molecular weight marker (Promega).
RelE assay
RelE analysis was performed as described (55). Aliquots containing 0.2 pmol of the preTC were incubated with 10 pmol of wt eRF1 or with 80 pmol of eRF1 domains for 10 min at 37°C, and then RelE (to final concentration of 2 μM) was added for 10 min at 37°C. The RNA was purified by phenol/chloroform extraction and precipitated with ethanol, and then reverse transcription was carried out with the same primer as for toe printing.
Modelling
The cryo-EM structure of the yeast ribosome (56) and crystal structure of the N-domain of eRF1 (6) were used for modelling purposes. The positioning was performed manually based on the overall shape of the eRF1 N-domain and the ribosomal A site using PyMOL. The modelling of the stop codon binding to the N-domain was performed manually in PyMOL. Unnatural conformations were relieved using template-docking protocol of ICM software (57) and short rounds of molecular dynamics simulation performed in AMBER force field (58) as implemented in GROMACS (59). Models are available in Supplementary Data (Supplementary Files 1 and 2).
RESULTS
Based on the crystal structure of human eRF1 (6) and multiple alignments of eukaryotic class 1 RFs (Figure 1), 41 amino acid residues in the N-domain of human eRF1 were selected as potential sites, which might determine stop codon recognition. By making multiple conservative and non-conservative mutations at each residue, we created a library of >80 mutant eRF1s. The activity of these mutants was then screened using the fully reconstituted eukaryotic translation system (Supplementary Table S2), which consisted of purified individual eukaryotic translation factors, 40S and 60S rabbit ribosomal subunits, aminoacylated tRNAs and synthetic mRNA (31). As such, this system approximates the natural situation owing to the presence of all necessary translation components and a lengthy mRNA that contains UTRs. Using this approach, we have confirmed importance for the stop codon recognition of identified before 13 amino acid residues and defined their stop-codon-nucleotide specificity; also, we have identified two new significant for stop codon decoding positions S33 and S36.
Mutations in loop 59–68 reduce termination efficiency at all three stop codons
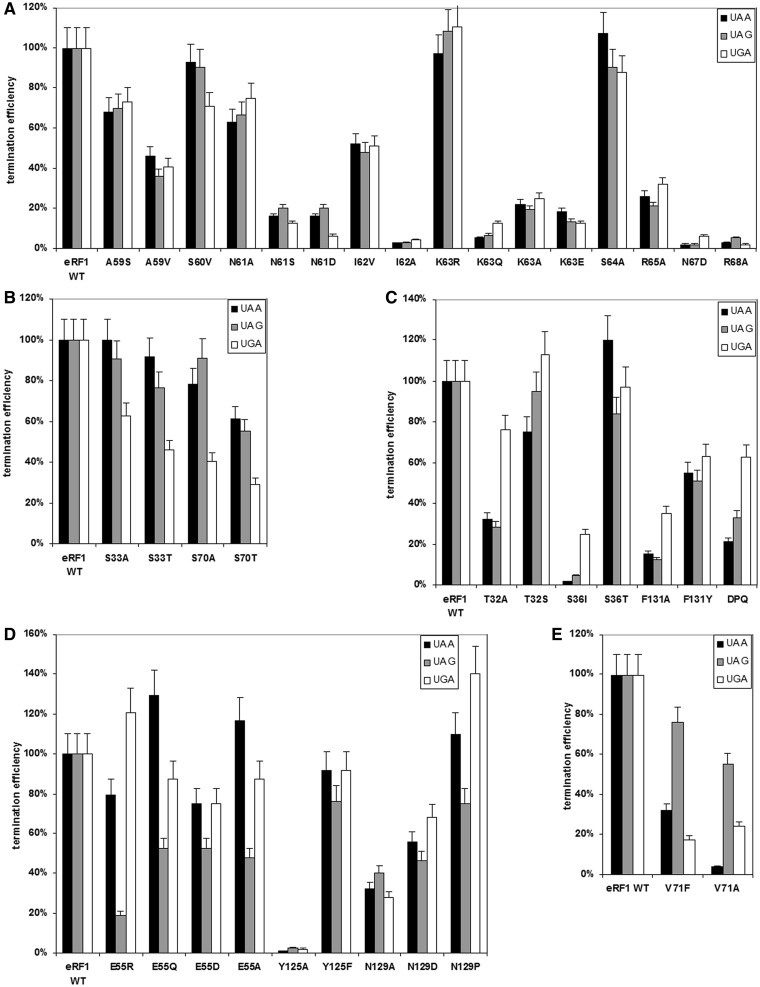
We first determined the termination efficiency of human eRF1s with mutations in the highly conserved 59–68 loop (Figure 1) that contains the NIKS motif (except one variable position at residue 66). Mutations at positions 59, 60 and 64 did not significantly affect RF activity (Figure 2A, Supplementary Table S2), implying that eRF1 function is tolerant to the nature of amino acids at these positions. In contrast, mutations at positions 61, 62, 63, 65, 67 and 68 led to a considerable reduction in termination efficiency at all three stop codons (Figure 2A, Supplementary Table S2). As expected, substitutions by amino acid residues with similar physical–chemical properties (e.g. I62V, K63R) are better tolerated than those with different size or charge (e.g. N61S, N61D, I62A, K63Q, K63A, K63E, R65A, N67D, R68A). The most significant effects on peptide release were observed for I62A, K63Q, N67D and R68A mutants, which showed almost complete loss of termination efficiency at all three stop codons.
Figure 2.
Termination efficiencies (kcat/KM (M−1 s−1)) of the wt eRF1 and mutant eRF1s at different stop codons. Mutations (A) in the loop 59–68, (B) at positions 33 and 70, (C) at positions 32, 36 and 131, (D) at positions 55, 125 and 129 and (E) at position 71. The termination efficiency of the human wt eRF1 was considered 100%.
Several lines of evidences implicate the NIKS motif in stop codon recognition (10,39,40), specifically its close proximity to the first nucleotide (uridine, U1). Therefore, residues at positions 61, 62, 63, 65, 67 and 68 of the human eRF1 N-domain have been selected as likely candidates for decoding of stop codon (U1).
Mutations at positions 33 and 70 decrease peptide release at UGA stop codon
Previously, we have found that a S70A mutation prevents UGA recognition by human eRF1 (48). In the current work, we have also tested the ability of S70T mutant with a more conservative mutation to recognize stop codons (Figure 2B, Supplementary Table S2) and confirm reduced RF activity of this mutant at UGA stop codon relative to the other two stop codons. As threonine, like serine, contains an OH group, these data highlight the importance of both the nature and size of amino acid chain at this position in UGA recognition.
In accordance with the X-ray structure of eRF1, the highly conserved serine residue at position 33 is located close to S70 (6). Therefore, to test whether S33 is also involved in decoding the UGA stop signal, the RF activities of S33A and S33T mutants were measured. Interestingly, the results were similar to that observed for S70A and S70T mutants (Figure 2B, Supplementary Table S2). Therefore, we conclude that both S33 and S70 of the human eRF1 N-domain participate in UGA recognition. As the UGA stop codon differs from UAA and UAG codons by the presence of G at the second position, we propose that S33 and S70 recognize the second guanine (G2).
Mutations at positions 32, 36 and 131 reduce RF activity at UAA and UAG stop codons
Among all the tested eRF1 mutants, positions 32, 36 and 131 significantly influence UAA and UAG decoding (Supplementary Table S2). These positions are invariant (T32 and F131) or highly conserved (S36) among eukaryotic eRF1s (Figure 1). Although an F131A mutation leads to a significant decrease of peptide release at all three stop codons, this mutant still exhibits moderate RF activity towards UGA (Figure 2C). In contrast, the F131Y mutant (which retains the aromatic ring) exhibits similar and substantial RF activity at all three stop codons (Figure 2C). This may be consistent with a requirement for an aromatic ring at the position 131 to maintain the optimal conformation of eRF1 for UAA and UAG decoding.
Residue S36 is located close to F131 in the crystal structure of eRF1 (6). Replacing serine with the bulkier isoleucine at this position leads to a complete loss of RF activity at UAA and UAG stop codons, whereas RF activity is partially retained in the presence of UGA (Figure 2C). As anticipated, a conservative S36T mutation does not affect UAG and UGA recognition, and RF activity increases in the presence of UAA stop codon (∼20%). It is possible therefore that an interaction between S36 and F131 preserves the proper structure of the N-domain required for UAA and UAG recognition.
Finally, the termination efficiency of T32A mutant is preferentially reduced at UAA and UAG and is not changed at UGA (Figure 2C). As anticipated, the RF activity of a T32S mutant with conservative amino acid substitution is largely unaffected at all three stop codons (Figure 2C). Thus, the presence of the hydroxyl group at position 32 is crucial for UAA and UAG stop codon decoding by human eRF1. As UAA and UAG stop codons share adenine at the second position (A2), we propose that T32, S36 and F131 are important for decoding of this nucleotide.
Mutations at positions 55, 125 and 129 reduce peptide release at UAG stop codon
Perhaps consistent with E55 being the highly conserved amino acid residue among eRF1s (Figure 1), both conservative and non-conservative E55 mutations exhibited a preferential decrease in UAG recognition (Figure 2D). Therefore, the glutamate residue at this position is important for UAG decoding.
Interestingly, removal of the aromatic ring at position 125 (Y125A) results in complete loss of termination activity at all three stop codons, whereas a conservative Y125F substitution only causes a modest reduction in termination efficiency at the UAG stop codon (Figure 2D). These results are in agreement with the data published earlier (15) and indicate the importance of the aromatic ring at position 125 for termination activity of eRF1.
Among the N129 mutants, the most significant effect was observed for N129P (Figure 2D), which resulted in a 10 and 40% termination efficiency increase at UAA and UGA, respectively. In contrast, the termination efficiency of this mutant at UAG was decreased by up to 20%. Two other mutants, N129A and N129D, exhibit moderately reduced RF activity that was similar at all three stop codons (Figure 2D). Y125 and N129 are part of the highly conserved YxCxxxF motif of human eRF1 (Figure 1).
As the UAG codon is unique with respect to the third guanine (G3), our data suggest that positions 55, 125 and 129 of the human eRF1 N-domain are important for decoding of this nucleotide.
Mutations at position 71 decrease peptide release at UAA and UGA stop codons
Mutations of the invariant valine at position 71 in human eRF1 (Figure 1) inhibit UAA and UGA stop codon recognition whilst retaining activity towards UAG (Figure 2E). Both V71A and V71F mutants, which contain substitutions for the small and bulk residues, respectively, exhibit the reduced termination efficiency at UAA and UGA. Thus, an appropriately sized residue at position 71 is likely required for UAA and UGA decoding, specifically for recognition of the third adenine (A3).
Binding of eRF1 mutants with the ribosome
As binding of eukaryotic RFs (eRF1 and eRF3) to the ribosome results in activation of the eRF3 GTPase (52), GTP hydrolysis in eRF1•eRF3•GTP•ribosome complex is a measure of efficiency of the eRF1-ribosome interaction. Therefore, to determine whether the mutational effects detailed earlier in the text could be caused by changes in the efficiency of eRF1 binding to the ribosome, eRF3 GTPase assays were performed in the presence of several mutant eRF1s (Table 1).
Table 1.
Stimulation of eRF3 GTPase activity by mutant eRF1s
| Position | Mutation | GTPase, % |
|---|---|---|
| eRF1 WT | 100 | |
| T32 | S | 133 ± 2 |
| A | 128 ± 12 | |
| S33 | A | 124 ± 11 |
| T | 121 ± 7 | |
| S36 | I | 52 ± 8 |
| T | 125 ± 10 | |
| E55 | R | 163 ± 15 |
| Q | 166 ± 6 | |
| D | 157 ± 13 | |
| A | 106 ± 1 | |
| A59 | S | 99 ± 2 |
| V | 101 ± 3 | |
| S60 | V | 122 ± 20 |
| N61 | A | 106 ± 9 |
| S | 102 ± 2 | |
| D | 89 ± 4 | |
| I62 | V | 104 ± 4 |
| A | 93 ± 13 | |
| K63 | R | 180 ± 24 |
| Q | 104 ± 8 | |
| A | 104 ± 14 | |
| E | 104 ± 14 | |
| S64 | A | 141 ± 11 |
| R65 | A | 48 ± 2 |
| N67 | D | 81 ± 6 |
| R68 | A | 35 ± 5 |
| S70 | A | 93 ± 7 |
| T | 83 ± 5 | |
| V71 | F | 78 ± 9 |
| A | 106 ± 6 | |
| Y125 | A | 106 ± 6 |
| F | 150 ± 12 | |
| C127 | A | 161 ± 12 |
| S | 132 ± 9 | |
| D128 | G | 159 ± 13 |
| R | 149 ± 6 | |
| N129 | A | 89 ± 3 |
| D | 120 ± 12 | |
| P | 134 ± 12 | |
| F131 | A | 96 ± 7 |
| Y | 105 ± 11 | |
| N129, K130 | PQ | 89 ± 2 |
Mutations within the N-domain leading to a decrease in eRF1 stimulating activity towards eRF3 GTPase are marked in bold.
Interestingly, most of the examined eRF1 mutants stimulated eRF3 GTPase activity to a similar extent as wt eRF1 or even better (Table 1). Consequently, the overall structure of these eRF1 mutants and their ability to bind the ribosome are preserved. For S36I, R65A and R68A mutants, the ability to stimulate eRF3 GTPase activity was decreased however. The very low RF activities of mutant eRF1s at these positions and weak stimulation of eRF3 GTPase activity could be due to the disrupted rRNA binding following substitution of the positive charged arginine by uncharged alanine (R65A and R68A) or due to the partial damage of the overall eRF1 structure (S36I).
The effect of eRF3 on stop codon recognition
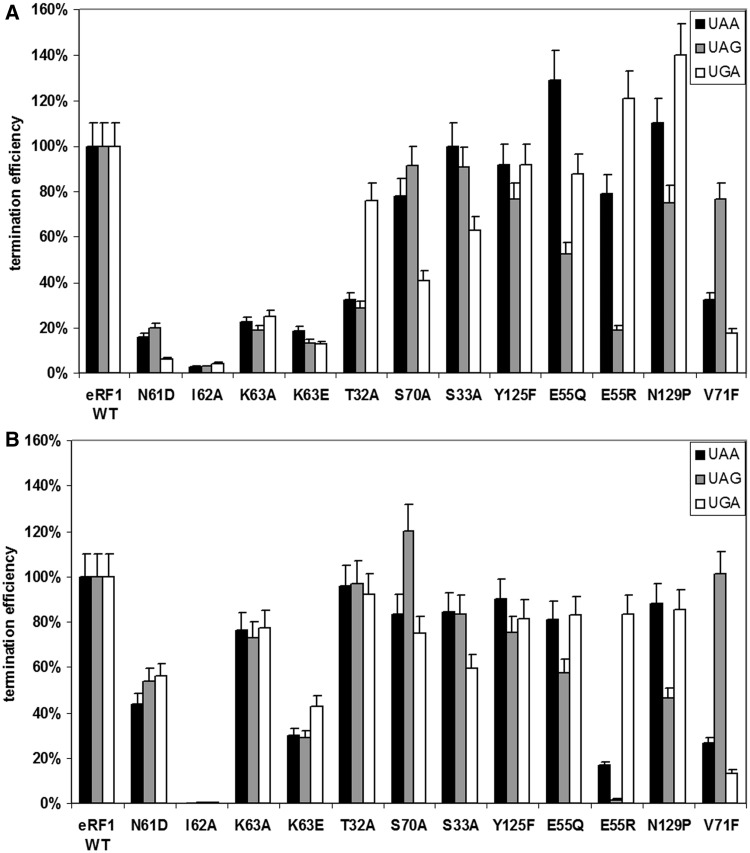
Next, we assessed the influence of eRF3 on the most meaningful eRF1 mutants by repeating the RF assays in the absence or presence of eRF3-GTP. As shown in Figure 3, termination activity is almost completely restored for T32A, K63A, S70A eRF1 mutants and partially restored for N61D and K63E mutants in the presence of eRF3-GTP. In contrast, the remaining eRF1 mutants exhibited similar patterns of stop codon recognition in the absence or presence of eRF3-GTP. Together, these data suggest that in some cases, eRF3 can improve the recognition function of eRF1.
Figure 3.
Termination efficiencies (kcat/KM (M−1 s−1)) of the wt eRF1 and mutant eRF1s at different stop codons (A) in the absence and (B) in the presence of eRF3-GTP. The termination efficiency of the human wt eRF1 was considered as 100%.
The toe-print shift occurs at all three stop codons during stop codon recognition by eRF1
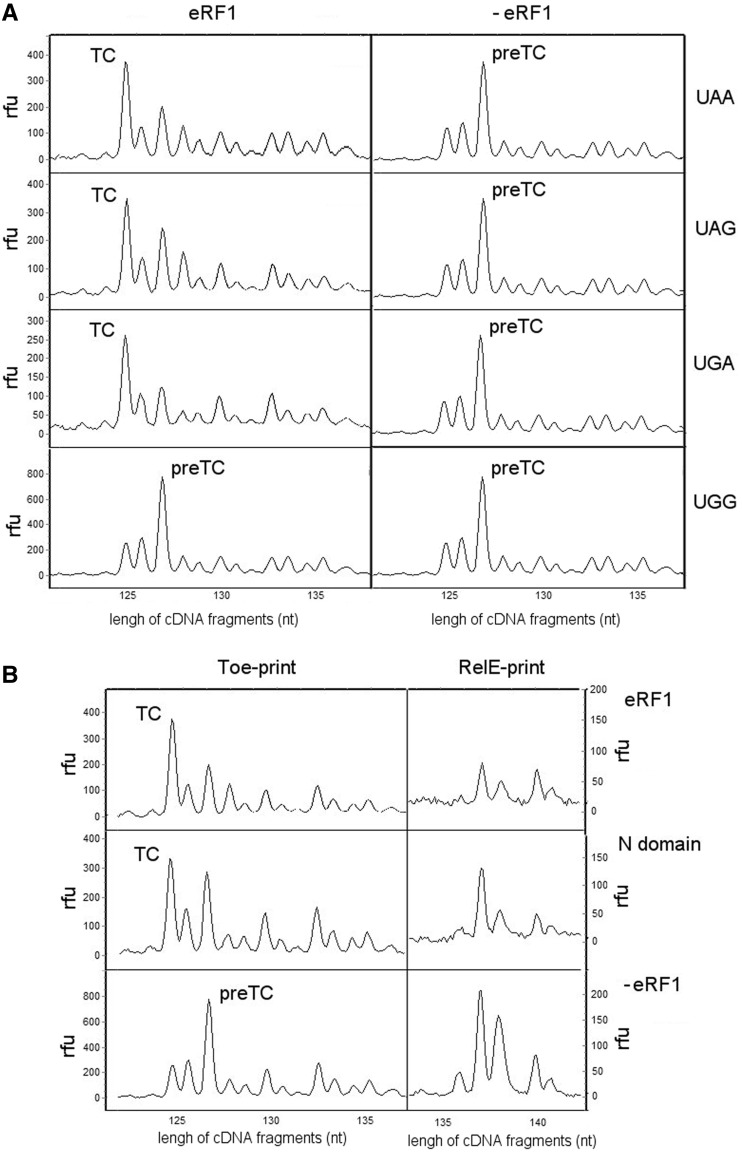
We have previously observed that binding of eRF1 alone to the preTC induces a major conformational change characterized by a 2 nt forward shift of the toe-print peak attributed to the preTC to a position +15 nt downstream of the first nucleotide of the UAA stop codon, and that addition of eRF3-GTP increases the yield of shifted complex (54). To extend this finding, we assessed the ability of eRF1 to induce the toe-print shift in the presence of the remaining stop codons, UGA and UAG. Interestingly, and consistent with UAA, eRF1 also induces the toe-print shift in preTC at UAG and UGA, but does not change the position of the ribosomal complex at a UGG sense codon (Figure 4A). This suggests that conformational rearrangement of the ribosomal complex is a distinctive feature of stop codon recognition by eRF1.
Figure 4.
Conformational rearrangement of ribosomal complexes during termination of translation. (A) Toe-prints with the human wt eRF1 at different stop codons and UGG codon, (B) Toe-prints and corresponding RelE-prints with the human wt eRF1 or its N-domain at UAA stop codon. The cDNAs corresponding to preTC, termination complex and after RelE hydrolysis of mRNA at the ribosomal A site have the 127, 125 and 138 nt length, respectively. RFU, relative fluorescence unit.
It has been shown that RelE toxin hydrolyses mRNA in the 80S ribosome when the A site is vacant (55). Therefore, we used this strategy to examine the occupancy of the ribosomal A site in the shifted complexes. Following eRF1 binding and ribosomal rearrangement, the RelE peak practically disappeared (Figure 4B), indicating that eRF1 occupancy is maintained in the ribosomal A site after toe-print shift. Unfortunately, limitations of the assay prevent us from determining whether the stop codon fully remains in the A site of the ribosome after conformational rearrangement of the preTC or partially translocates to the P site, as in both cases, eRF1 would occupy the ribosomal A site.
To determine which part of eRF1 is involved in induction of the conformational change within the ribosome, the individual domains of human eRF1 were tested in toe-print assay at the UAA stop codon. Figure 4B indicates that the N-terminal 145 amino acids (N-domain) are sufficient to induce the toe-print shift, but the yield of shifted complex is lower than that with the full-length eRF1. Addition of the M (146-276 aa), C (276-437 aa) or MC domains to the preTC does not induce the toe-print shift (Supplementary Figure S2A), whereas addition of the N-domain in combination with either the M- or MC-domain causes the same effect as the N-domain alone (Supplementary Figure S2B). Next, we measured the ability of the N-domain to induce the toe-print shift at UAG and UGA stop codons and a UGG sense codon. As expected, the same strong stop codon selectivity of the toe-print shift was observed for the N-domain, as for the full-length eRF1 (Supplementary Figure S2C). Furthermore, RelE poorly hydrolysed mRNA following the toe-print shift induced by the N-domain, thereby confirming its occupancy in the ribosomal A site (Figure 4B). Taken together with data on a crucial role of the N-domain in stop codon decoding, these results indicate that the observed conformational rearrangement is an integral part of the stop codon decoding function of eRF1.
Modelling of the N-domain positioning at the A site of the ribosome
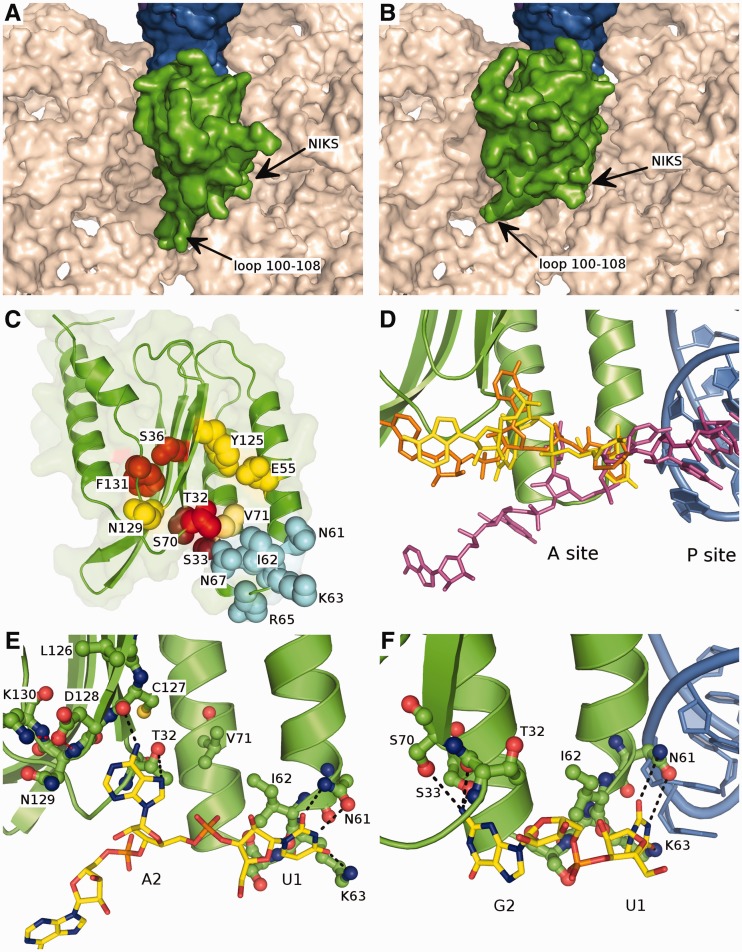
To attempt to interpret the mutagenesis results in the 3D context, we have modelled the orientation of the N-domain of eRF1 at the A site of the ribosome. The eRF1 NIKS motif should be located close to the A site stop codon, as it is believed that these residues are directly involved in stop codon recognition (39,40). This is consistent with the cryo-EM structure of the complex, which consists of Dom34-Hbs1 no-go mediated decay factors bound to the ribosome, and is structurally similar to eRF1•eRF3 (60). Two options for the N-domain position were selected that are rotated relative to each other by ≈30° (Figure 5A and B, Supplementary Files 1 and 2). In the first case, loop 100–108 of eRF1 is placed between hairpin 560–580 and helix 1265–1280/1432–1442 of the 18S rRNA with α-helix 2 of the N-domain contacting the P site tRNA (Figure 5A). In the second case, loop 100–108 of eRF1 lies between helix 1265–1280/1432–1442 and helix 1202–1212/1448–1455 of the 18S rRNA, whereas the P site tRNA is facing α-helices 2 and 3 of eRF1 (Figure 5B). The second option for orientation of the N-domain makes loop 100–108 partially clash with the extended C-terminal arm of ribosomal protein S30e. However, the suggested models are only approximate, and the possible backbone flexibility of the eRF1 N-domain has not been taken into account. The precise contacts of the eRF1 N-domain with the ribosome can only be deduced when the crystal structure of a eukaryotic translation termination complex has been solved.
Figure 5.
Modelled orientation of eRF1 at the A site of the ribosome. (A and B) The first and second options of eRF1 N-domain (green) orientation at the ribosomal A site. The surface of 18S rRNA is shown in beige, tRNA in the P site is in blue. (C) The overall structure of the eRF1 N-domain. The amino acid residues important for the stop codon decoding are shown in spheres; the residues involved in the recognition of U1 are coloured in light blue, those important for the decoding of the second nucleotide are coloured in red, and those important for the third nucleotide recognition are coloured in yellow. (D) Comparison of the position of mRNA in the ribosome A site from the cryo-EM structure (56) (purple) and the modelled conformations of the mRNA (UGA and UAA codons are in yellow and orange, respectively) bound to eRF1: the bound mRNA is pulled onto the eRF1 and thus into the ribosome. (E) The proposed mode of binding of the UAR first two nucleotides. Stop codon is shown in yellow, eRF1 N-domain is green, black dotted-lines depict the possible contacts between eRF1 and the stop codon, red dotted lines correspond to the inter-eRF1 contacts. (F) 90° rotation of (E) with the first two nucleotides of UGA codon bound to eRF1, tRNA in the P site is shown in blue.
DISCUSSION
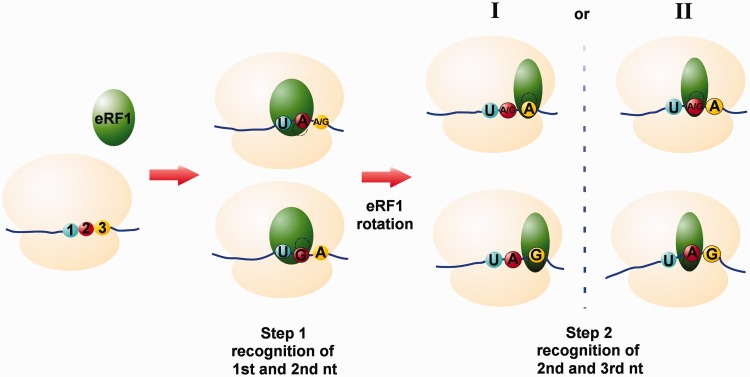
In this study, we have shown importance for stop codon decoding of the 15 amino acid residues within the N-domain of human eRF1 (T32, S33, S36, E55, N61, I62, K63, R65, N67, R68, S70, V71, Y125, N129 and F131). Amino acid residues that appear to be important for the recognition of the second and third nucleotides of the stop codon are situated close to each other in the structure of the eRF1 N-domain (Figure 5C). However, the approximate size of purine is ≈7–8 Å, making it difficult to imagine the binding of all three nucleotides simultaneously. For example, residue T32, which is important for decoding of A2 in UAA/UAG codons, is located at a distance of ≈5 Å from V71, which specifically influences the recognition of the third nucleotide of UAA/UGA codons (6). These observations could be reconciled by sequential, as opposed to simultaneous, recognition of the three stop codon nucleotides (Figure 6). In this model, the first and second nucleotides of the stop codon would be recognized initially, followed by a conformational rearrangement of eRF1 in the ribosome that permits decoding of the third or second and third nucleotides. If this model is correct, the two proposed options for the orientation of the N-domain in the ribosomal A site could correspond to these two steps of stop codon recognition. In fact, a two-step model of stop codon decoding was first proposed by Chavatte et al. (13) based on experimental data. In agreement with our two-step model, human eRF1 cross-links with all three stop codons and the UGG sense codon, whereas Euplotes eRF1, which does not recognize the UGA stop codon, does not cross-link with the UGA/UGG codons (61). Cross-linking of wt eRF1 with the UGG sense codon could be explained if we propose that only two nucleotides of the stop codon (UG or UA) are recognized at the first step. When decoding of G2 is impaired, as in the case of Euplotes eRF1, UGG does not cross-link with factor.
Figure 6.
Models for stop codon recognition by eRF1. (Step 1) Binding of eRF1 to the A site of the ribosome. Recognition of the first and second nucleotides of stop codon (UA or UG). Second A and G are decoded at different sites of eRF1. (Step 2) Rotation of eRF1 within the ribosome and decoding of the second and third nucleotides of stop codon (AG, AA or GA). Two possible modes of stop codon interactions. (I) The different amino acid residues of eRF1 directly decode the third A or G, and the ribosome recognizes the second A/G or A according to the third nucleotide. (II) eRF1 decodes the second nucleotide (A/G or A), and the ribosome interacts with the third A or G, respectively. The ribosome is shown in beige, eRF1 is in green, the first, second and third nucleotides of stop codon are in blue, red and yellow, respectively.
Recognition of the first stop codon nucleotide (U1)
The data in Figure 5 suggest that recognition of the first nucleotide of a stop codon is mainly mediated by an interaction with the side chain of K63, which is a part of the universally conserved NIKS motif (Figure 1). This is further supported by finding that K63 cross-links with the S4 atom of a modified stop codon U1 nucleotide (39). The O4 atom of U1 could interact with the amino group of K63, whereas the O2 and N3 atoms could form hydrogen bond with the carboxamide group of N61. The importance of a positive charge at position 63 for U1 recognition is supported by the observation that replacing K63 for the negatively charged glutamate or uncharged glutamine or alanine leads to a significant loss of RF activity (Figure 2A), whereas K63R mutation does not. The ability of eRF1 to discriminate against cytosine may be due to inability of the N4 atom of cytosine to form a hydrogen bond with the K63 amino group. Discrimination against purines may be explained by their larger size and inability of even the long side chain of lysine to reach the hydrogen acceptor atoms of purines.
Substitution of N61 by aspartate or serine results in almost complete loss of RF activity (Figure 2A), likely owing to inability of these residues to form the same interactions with U1 as asparagine. Other residues in the NIKS motifs also likely play a role in stop codon recognition. I62 is most important for maintaining the proper conformation of the NIKS loop, as its side chain is stacked against V71, A59 and R68 (Figure 5E). Consistent with an important structural requirement, substituting isoleucine at this position with a smaller residue (alanine) causes complete elimination of RF activity (Figure 2A). Other conserved amino acid residues R65, N67 and R68 from the NIKS loop are likely necessary for the proper positioning of eRF1 at the ribosomal A site, as eRF3 GTPase stimulating activity of these mutants are reduced (Table 1).
Recognition of the second nucleotide of UGA (G2)
The recognition of G2 in the UGA stop codon is mostly influenced by S33 and S70 mutations (Figures 1B), which may be consistent with their close proximity in a groove on the surface of eRF1. Presumably, −OH groups of these residues bind the N2 amino group of G2. This would also allow the nearby backbone oxygens of T32 and G31 to act as hydrogen acceptors and contribute to G2 binding (Figure 5F). This could explain the partial retention of termination activity for S33 and S70 mutants at UGA stop codon (42 and 63% activity for S70A and S33A, respectively). The substitution of S33 and S70 by threonine most likely disturbs the local conformation of eRF1, thus decreasing the termination efficiency.
The discrimination against adenine at this position might occur due to the absence of the hydrogen donor group in adenine in position where N2 group of guanine is situated, whereas pyrimidines are discriminated due to their smaller size.
These results are consistent with our earlier report on the role of S70 in UGA recognition (48). Based on nuclear magnetic resonance studies, it has been suggested that formation of a hydrogen bond between the carbonyl oxygen of S33 and the hydroxyl group of S70 in the UGA-only QFM_F mutant of human eRF1 stabilizes the GTS loop in conformation necessary for UGA recognition (46). These data are in agreement with cross-linking studies, indicating that although the GTS loop neighbours guanines of stop signals, it is positioned further from adenines (45).
Recognition of the second nucleotide of UAA/UAG codons (A2)
Recognition of the second A of UAA/UAG codons is specifically affected by mutations of residues T32, S36 and F131 (Figure 2C). Although S36I and F131A mutants displayed substantial decreases in UAA and UAG recognition, moderate levels of UGA recognition were maintained. As residues 36 and 131 are buried within the protein core, they likely play a structural role that maintains an appropriate eRF1 conformation (Figure 5C). Therefore, we propose that among those residues affecting recognition of the second nucleotide of UAA and UAG, only T32 directly interacts with A2.
QFM_F and DPQ human eRF1 mutants (Figure 2C), eRF1s from variant-code organisms (17) and S. cerevisiae eRF1 after introduction of the YxCxxxF motifs from variant-code organisms (18) all show reduced termination efficiency at UAR stop codons without affecting activity at UGA. Interestingly, both these groups of aforementioned amino acid residues are either a part of the YxCxxxF motif (positions 125–131 of human eRF1, Figure 1) or flank it. This would suggest that the precise protein backbone conformation in this region is important for A2 decoding. Residue D128 may be a key residue in this respect, as its side chain is fixed in a specific conformation by hydrogen bonds with the backbone nitrogen of residue 129 and the side chain of K130 (Figure 5E). Proline at position 129 prevents interaction of the backbone nitrogen with other amino acid residues, whereas the K130Q mutation likely causes the reduced binding efficiency towards the carboxylic group of D128. All the mutations observed are likely to change the local orientation of the D128 side chain and the YxCxxxF loop conformation. On the other hand, residues 122, 123, 124 and 126 form a cluster of interacting and interdependent residues with the side chain of L126 in close proximity to that of D128. Substitution of L126 for the more bulky phenylalanine may change both the conformation of D128 and the local backbone conformation.
A2 could bind to the hydroxyl group of T32 and/or the backbone oxygen of C127, thus requiring the precise conformation of the backbone in this region (Figure 5C). In this case, A2 and G2 discrimination is probably due to the inability of the backbone oxygen of C127 to form a contact with the O4 atom of the guanine. Alternatively, A2 might be bound and stabilized in the region of T32, by stacking with A2256 of 28S rRNA in such a way that its phosphate group comes close to the negatively charged D128 side chain, for example. Thus, disturbances in the precise conformation of D128 would confer increased repulsive electrostatic interactions and prevent the binding to A2. It is unclear, however, whether a mere change of side chain conformation could cause an electrostatic repulsion strong enough to prevent A2 binding. Consistent with our data and the hypotheses others have reported (8,25,40,45), T32 and D128 play important role in decoding of the second nucleotide of UAA and UAG stop codons.
Recognition of the third stop codon nucleotide
Recognition of the third nucleotide of the UAG stop codon is influenced by the mutations at residues 55 and 125, and partially at residue 129 (Figure 2D). In agreement with these results, human and yeast eRF1 E55A mutants (numbering according to human eRF1) also displayed preferential loss of UAG recognition tested in an independent assay system (15,25). However, in contrast, an E55A mutant of yeast eRF1 only exhibited efficient termination at the UAA stop codon (18). Moreover, in our experiments, an E55R mutant showed a bipotent phenotype, whereas no effect on RF activity was detected in an in vitro fMet release assay (15). The reasons for these discrepancies are not known and might be partially explained by using diverse assay systems.
Residue 125 may play a structural role, as Y125A substitution prevented termination at all three stop codons, whereas Y125F mutation influenced recognition of the UAG codon to a greater extent than the other two stop codons. Among the N129 mutations, only N129P decreased termination efficiency at UAG greater than at the other stop codons, likely owing to conformation changes of the protein. Recognition of the third A in UGA/UAA codons is most influenced by mutations of residue V71 (Figure 2E). The importance of residues E55, Y125 and V71 for recognition of the third nucleotide of stop codons has been previously demonstrated (15,18). It was also proposed that residues E55 and Y125 are critical for recognition of the third adenine, whereas V71 is crucial for recognition of the second base of stop codons (25). Our two step model of stop codon recognition reconciles these apparently contradictory views.
It is well known that eRF1 is able to terminate efficiently at UAA, UAG and UGA stop codons while avoiding termination at the UGG sense codon. Taking into account the results of our mutagenesis experiments, it is difficult to propose a model where this discrimination is solely due to stop codon-binding mediated by eRF1; it is likely, therefore, that the ribosome itself may be involved in stop codon decoding. Two modes of interaction of the second and third nucleotides of stop codons can be discussed in this regard (Figure 6). According to the first mode, G3 decoding occurs through direct interaction with the side chain of E55, whereas A3 is bound in the region of V71 (Figure 6, step 2, I). At the same time, the second nucleotide is recognized by the ribosome with the recognition site depending on the binding mode of the third stop codon nucleotide with eRF1: the A2 is allowed to bind in case of the G3, whereas both G2 and A2 (or actually any nucleotide, if pyrimidines are discriminated at the first decoding step) are allowed to bind when the A is located in the third position of stop codon. Alternatively, the second mode of interaction assumes that the recognition of the third nucleotide occurs through binding to the ribosomal RNA (Figure 6, step 2, II). In this case, the second stop codon nucleotide is still bound to eRF1 but in a different manner. A2 of the UAG stop codon contacts E55 with its amino group, whereas the G3 is recognized by the ribosome. The second nucleotide of UAA and UGA binds at the region of V71 in a promiscuous way, allowing for both A and G binding with the third nucleotide bound by the ribosome in a way that only allows A binding. The putative regions of the ribosome for binding of the third nucleotide are located at the vicinity of nucleotides 577, 1274 or 1634 of 18S rRNA. Consistent with the second model, the X-ray structure of eRF1•eRF3•ATP complex shows ATP bound to the N-domain of eRF1 via hydrogen bonding to E55 (25). At the same time, this mode of binding can be reasonably simulated by the second option of eRF1 positioning at the ribosomal A site (Figure 5B).
On comparison of the modelled conformation of mRNA bound to eRF1 in the first orientation (Figure 5A), and the intact mRNA taken from the cryo-EM structure of the ribosome (56), a difference of ≈8–12 Å in the positions of the second stop codon nucleotides was revealed (Figure 5C). This difference could explain toe-printing results showing that eRF1 binding to stop codons in the preTC shifts the ribosome forward two nucleotides towards the 3′-end of the mRNA (Figure 4). Interestingly, a similar ribosome shift was detected during translation termination by ribosome profiling (62). Thus, the displacement of the ribosomal complex observed in our experiments likely reflects stop codon recognition activity of eRF1 (Figure 4).
Here, we have also demonstrated that class 2 RF eRF3 can improve the RF activity some of eRF1 mutants (Figure 3). A similar effect was shown for some chimeric eRF1s (48). We propose that eRF3 can ‘rescue’ those mutations, which do not induce a conformational rearrangement in eRF1 and only influence the ability of eRF1 to interact with the stop codon. Obviously, eRF1 uses a network of amino acid residue for recognition of each stop codon nucleotide; therefore, disturbances in a single residue could theoretically be corrected by increasing termination efficiency of eRF1 by eRF3. Indeed, all the eRF1 mutants whose termination efficiency was improved by eRF3 exhibited damaged hydrogen bonding with the stop codon nucleotides (T32 with А2, К63 with U1 and S70 with G2).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1 and 2, Supplementary Files 1 and 2.
FUNDING
Russian Foundation for Basic Research [12-04-00147, 12-04-33085 to E.A., 11-04-00840-a to L.F. and 11-04-91340 to A.G.]; Program on Molecular and Cellular Biology of the Russian Academy of Sciences (to L.F. and to E.A.). Funding for open access charge: Personal funds of the authors.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to P. Kolosov and A. Seit-Nebi for participation in some experiments of eRF1 mutagenesis, to V. Hauryliuk for kindly providing RelE toxin and to T. Pestova and C. Hellen who provided with recombinant plasmids encoding initiation factors. They gratefully thank M. Coleman for critical reading of this manuscript. Sequencing of mutant eRF1s and cDNA fragment analyses were performed by the Center of the collective use ‘Genome’ of EIMB RAS.
REFERENCES
- 1.Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr Opin. Struct. Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 4.Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Ann. Rev. Biophys. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 5.Klaholz BP. Molecular recognition and catalysis in translation termination complexes. Trends Biochem. Sci. 2011;36:282–292. doi: 10.1016/j.tibs.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 7.Frolova LY, Merkulova TI, Kisselev LL. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muramatsu T, Heckmann K, Kitanaka C, Kuchino Y. Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett. 2001;488:105–109. doi: 10.1016/s0014-5793(00)02391-7. [DOI] [PubMed] [Google Scholar]
- 10.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki Y, Blouin C, Doolittle WF, Roger AJ. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res. 2002;30:532–544. doi: 10.1093/nar/30.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavatte L, Frolova L, Laugâa P, Kisselev L, Favre A. Stop codons and UGG promote efficient binding of the polypeptide release factor eRF1 to the ribosomal A site. J. Mol. Biol. 2003;331:745–758. doi: 10.1016/s0022-2836(03)00813-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim OT, Yura K, Go N, Harumoto T. Newly sequenced eRF1s from ciliates: the diversity of stop codon usage and the molecular surfaces that are important for stop codon interactions. Gene. 2005;346:277–286. doi: 10.1016/j.gene.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 15.Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang H, Wong JY, Bao Q, Cavalcanti AR, Landweber LF. Decoding the decoding region: analysis of eukaryotic release factor (eRF1) stop codon-binding residues. J. Mol. Evol. 2005;60:337–344. doi: 10.1007/s00239-004-0211-8. [DOI] [PubMed] [Google Scholar]
- 17.Lekomtsev S, Kolosov P, Bidou L, Frolova L, Rousset JP, Kisselev L. Different modes of stop codon restriction by the Stylonychia and Paramecium eRF1 translation termination factors. Proc. Natl Acad. Sci. USA. 2007;104:10824–10829. doi: 10.1073/pnas.0703887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conard SE, Buckley J, Dang M, Bedwell GJ, Carter RL, Khass M, Bedwell DM. Identification of eRF1 residues that play critical and complementary roles in stop codon recognition. RNA. 2012;18:1210–1221. doi: 10.1261/rna.031997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito K, Ebihara K, Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eurwilaichitr L, Graves FM, Stansfield I, Tuite MF. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- 24.Merkulova TI, Frolova LY, Lazar M, Camonis J, Kisselev LL. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 27.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 28.Gao N, Zavialov AV, Ehrenberg M, Frank J. Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. J. Mol. Biol. 2007;374:1345–1358. doi: 10.1016/j.jmb.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;12:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 32.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petry S, Brodersen DE, Murphy FV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 38.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 39.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulygin KN, Khairulina YS, Kolosov PM, Ven'yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Kisselev LL, Karpova GG. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010;16:1902–1914. doi: 10.1261/rna.2066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson KS, Ito K, Noller HF, Nakamura Y. Functional sites of interaction between release factor RF1 and the ribosome. Nat. Struct. Biol. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 42.Ito K, Frolova L, Seit-Nebi A, Karamyshev A, Kisselev L, Nakamura Y. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl Acad. Sci. USA. 2002;99:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternative eRF1 conformations modulate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Chai B, Wang W, Liang A. Functional characterization of polypeptide release factor 1b in the ciliate Euplotes. Biosci. Rep. 2010;30:425–431. doi: 10.1042/BSR20090154. [DOI] [PubMed] [Google Scholar]
- 45.Bulygin KN, Khairulina YS, Kolosov PM, Ven'yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Karpova GG. Adenine and guanine recognition of stop codon is mediated by different N domain conformations of translation termination factor eRF1. Nucleic Acids Res. 2011;39:7134–7146. doi: 10.1093/nar/gkr376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong LE, Li Y, Pillay S, Frolova L, Pervushin K. Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1. Nucleic Acids Res. 2012;40:5751–5765. doi: 10.1093/nar/gks192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salas-Marco J, Fan-Monogue H, Kalmeyer AK, Klobutcher LA, Farahaugh PI, Bedwell DM. Distinct paths to stop codon assignment by the variant-code organisms Tetrahymena and Euplotes. Mol. Cell Biol. 2006;26:438–447. doi: 10.1128/MCB.26.2.438-447.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eliseev B, Kryuchkova P, Alkalaeva E, Frolova L. A single amino acid change of translation termination factor eRF1 switches between bipotent and omnipotent stop-codon specificity. Nucleic Acids Res. 2011;39:599–608. doi: 10.1093/nar/gkq759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caskey CN, Beaudet AL, Tate WP. Mammalian release factor: in vitro assay and purification. Methods Enzymol. 1974;30:293–303. doi: 10.1016/0076-6879(74)30032-8. [DOI] [PubMed] [Google Scholar]
- 50.Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 51.Alkalaeva E, Eliseev B, Ambrogelly A, Vlasov P, Kondrashov FA, Gundllapalli S, Frolova L, Söll D, Kisselev L. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009;583:3455–3460. doi: 10.1016/j.febslet.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 53.Gould PS, Bird H, Easton AJ. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques. 2005;38:397–400. doi: 10.2144/05383ST02. [DOI] [PubMed] [Google Scholar]
- 54.Shirokikh NE, Alkalaeva EZ, Vassilenko KS, Afonina ZA, Alekhina OM, Kisselev LL, Spirin AS. Quantitative analysis of ribosome-mRNA complexes at different translation stages. Nucleic Acids Res. 2010;38:e15. doi: 10.1093/nar/gkp1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreev D, Hauryliuk V, Terenin I, Dmitriev S, Ehrenberg M, Shatsky I. The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA. 2008;14:233–239. doi: 10.1261/rna.693208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5 Å resolution. Proc. Natl Acad. Sci. USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 58.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 59.Hess B, Kutzner C, van der Spoel D. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 60.Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 61.Chavatte L, Kervestin S, Favre A, Jean-Jean O. Stop codon selection in eukaryotic translation termination: comparison of the discriminating potential between human and ciliate eRF1s. EMBO J. 2003;22:1644–1653. doi: 10.1093/emboj/cdg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.