Abstract
The DNA structure in phage capsids is determined by DNA–DNA interactions and bending energy. The effects of repulsive interactions on DNA interaxial distance were previously investigated, but not the effect of DNA bending on its structure in viral capsids. By varying packaged DNA length and through addition of spermine ions, we transform the interaction energy from net repulsive to net attractive. This allowed us to isolate the effect of bending on the resulting DNA structure. We used single particle cryo-electron microscopy reconstruction analysis to determine the interstrand spacing of double-stranded DNA encapsidated in phage λ capsids. The data reveal that stress and packing defects, both resulting from DNA bending in the capsid, are able to induce a long-range phase transition in the encapsidated DNA genome from a hexagonal to a cholesteric packing structure. This structural observation suggests significant changes in genome fluidity as a result of a phase transition affecting the rates of viral DNA ejection and packaging.
INTRODUCTION
Double-stranded DNA (dsDNA) viruses package their genome, a charged molecule that is microns in length, into an enclosed rigid capsid that is only a few tens of nanometers in diameter. Facilitated by a powerful motor, this packaging occurs in the majority of bacterial viruses (bacteriophages), as well as some eukaryotic viruses (e.g. human Herpes simplex virus) (1). At high packaging densities, DNA–DNA interactions are mediated mainly by electrostatic and hydration forces (2–5); the latter arising from the structuring of water molecules on the macromolecular surface. These interactions are observed to result in a spacing that ranges ∼15–20 Å in distance (6), corresponding to 3–4 water layers at each DNA surface. The interaction can be either repulsive or attractive depending on whether the hydrogen bonding of the water molecules between surfaces is disrupted or reinforced, as the molecular surfaces are brought into proximity. Within many viruses, dsDNA is packaged to a density of 50% by volume, resulting in a DNA–DNA interaxial separation of ∼27 Å (7,8) with the remaining volume filled by water. This corresponds to DNA surface separations of merely 7 Å, a distance wherein the hydration interaction is a dominant force (2).
Hydration and electrostatic forces have been measured as a function of DNA–DNA separation for free DNA condensed in a bulk solution (9). In these measurements, dsDNA arrays are pushed together by an osmotic stress polymer, e.g. polyethylene glycol (PEG), and the DNA–DNA spacing determined by X-ray scattering as a function of external osmotic pressure. This pressure can be converted to the energy of DNA–DNA interactions (10). Previous experiments, performed in the presence of mono- and polyvalent ions, nicely demonstrate how a delicate interplay between the attractive and repulsive DNA hydration interactions determines the DNA structure (6,11–15). It was shown that DNA confined in a phage capsid behaves similarly to multiple DNA arrays confined by a polymer in a bulk solution, in terms of interactions and structural ordering (8,16). However, the encapsidated DNA, unlike the condensed DNA arrays in bulk, is strongly bent by the confining capsid walls. This is due to the fact that the dsDNA persistence length (ξ ∼ 50 nm) (17,18) is of the same order of magnitude as the diameter of many viral capsids, leading to an extra bending energy term affecting the structure of the DNA in the capsid.
In this work, we used single particle cryo-electron microscopy (cryo-EM) reconstruction to analyse the encapsidated dsDNA spacing in phage λ capsids. The effects of bending and interaction energies on DNA ordering were separated by eliminating the net repulsion inside the capsid through the addition of spermine (4+) ions. This allows a switching of the DNA–DNA interaction energy from net repulsive to net attractive, with the remaining bending term determining the DNA structure in the capsid.
MATERIALS AND METHODS
Phage λ and LamB production and purification
Wild-type (wt) bacteriophage λ cI857 with genome length 48.5 kb and its shorter genome mutants with 45.7 kb (corresponding to 94% of the DNA) and with 37.7 kb DNA (corresponding to 78% of the DNA, denoted λb221) were produced by thermal induction of lysogenic Escherichia coli strains AE1, AE2 and AE3. AE strains are lysogenic E. coli strains modified to grow without LamB protein expressed on its surface to increase the yield of phage induced in the cell. The culture was then lysed by temperature induction. The phage samples were purified by CsCl equilibrium centrifugation. The samples were dialysed from CsCl against Tris-magnesium (TM) buffer [10 mM MgSO4 and 50 mM Tris–HCl (pH 7.4)]. The final titer was 1012 virions/ml, as determined by plaque assay (19).
The receptor was the LamB protein purified from pop 154, a strain of E. coli K12 in which the lamB gene has been transduced from Shigella sonnei 3070 (20,21). Purified LamB was solubilized from the outer membrane of E. coli cells with 1% by volume of the non-ionic surfactant n-octylpolyoxyethylene (n = 2–9).
Cryo-EM
All lambda phage samples were preserved in vitreous ice on 400-mesh C-flat grids (Protochips Inc.) containing 2 µm holes at a spacing of 2 µm. Before the application of sample, the grids were cleaned in an atmosphere of 75% argon and 25% oxygen for 25 s with a plasma cleaner (Fischione Instruments, Inc). A 3 µl aliquot of sample was applied, and subsequent blotting and rapid-freeze plunging into liquid ethane was performed with a Vitrobot (FEI co). The Vitrobot environment chamber was set to maintain a temperature of 4°C and 100% humidity, and grids were double blotted for 7 s using an offset of −2. Grids were stored in liquid nitrogen until being loaded into a Gatan CT3500 single tilt cryotransfer holder for data collection.
Frozen grids were inserted into a Tecnai F20 Twin transmission electron microscope operating at 200 keV, and data were collected on a Gatan 4kx4k CCD using the Leginon automated electron microscopy package (22). For each sample, ∼1000 images were acquired at 50 000× magnification (a pixel size of 2.26 Å) with a dose of 20e−/Å2 and using a randomized defocus ranging from −1.0 to −2.5 µm. Preliminary image analysis was performed within the Appion processing environment (23). Concurrent with data collection, particles were automatically selected using a template-based particle picker (24), and the contrast transfer function (CTF) of each image was automatically estimated (25). Only the images yielding highly accurate CTF estimations were considered for further processing. The phase information of the images was corrected based on the estimated CTF, and the particles were extracted using a box size of 300 pixels. The particles were then centered using the ‘cenalignint’ function from the EMAN software package (26). From each data set, 5000 particles were randomly selected for 3D reconstruction to maintain a consistent level of signal between each sample. The reconstructions were performed using a standard projection matching routine in EMAN, iterating through 20 rounds of refinement starting at an angular increment of 5° and decreasing to 1° at four-iteration intervals. For each refinement, a previously determined icosahedral reconstruction of phage lambda (23) was low-pass filtered to 40 Å resolution and used as a starting model. All reconstructions all resolved to ∼10 Å resolution, according to the 0.5 Fourier shell correlation. The central slice along the 5-fold axis of each reconstruction was extracted and radially averaged using EMAN (26).
RESULTS AND DISCUSSION
Cryo-EM reconstructions were performed to determine the average interaxial distance between the hexagonally ordered DNA strands. Icosahedral symmetry was imposed during the reconstruction procedure to improve the signal-to-noise ratio to more accurately determine the average interstrand spacings. As previous osmotic stress bulk DNA measurements provide an accurate description of the DNA–DNA interactions as a function of interstrand separations in a phage capsid (8,16), these interaxial separations can be directly compared with our data for the variably encapsidated DNA. Although DNA strands are pushed together by an inert osmotic stress polymer in the bulk measurements, the DNA in phage is compressed by the capsid walls. Bulk measurements revealed that by combining the variation in the external osmotic pressure (directly varying the interstrand separation) and affecting the type of intermolecular interaction using the multivalent ions, long-range phase transitions are observed in the DNA arrangement (13).
The interstrand separation can also be varied in encapsidated DNA by packaging different lengths of λ-DNA. As all viral capsids are permeable to water and ions, the DNA–DNA interactions are affected by the presence of multivalent ions. We use spermine (4+) ions to introduce an attractive component to the otherwise repulsive interactions between the DNA helices inside the capsid. DNA interstrand distances are varied by packaging 78, 94 and 100% of the wt λ-DNA length of 48 500 bp. For these three lengths of packaged λ-DNA, the genome remains stressed in the capsid with and without spermine, and exerts pressure on the capsid walls (8). However, the internal capsid pressure can be reduced to zero by opening the capsid with λ receptor LamB and allowing the DNA to freely eject until it reaches equilibrium. Without spermine ions, the ejection is complete leaving the capsid empty. However, in the presence of spermine the ejection is incomplete with a fraction of λ-DNA remaining in the capsid at equilibrium. We analyse interaction and bending effects on the resulting DNA structures for all these cases.
Repulsive interactions and structure of encapsidated λ-DNA
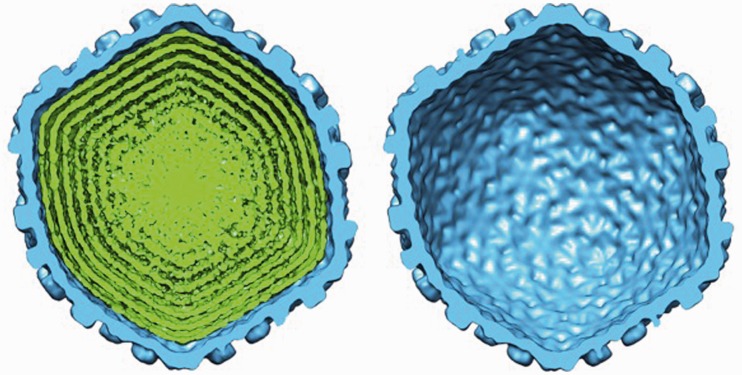
Icosahedral reconstructions were performed for λ-phages containing three different genome lengths, as well as for emptied capsid particles. No spermine was present to analyse the effect of DNA packaging density on the DNA–DNA spacing and overall structure under net repulsive conditions. Figure 1 shows a cutaway view of a wt λ-DNA length filled capsid and an empty capsid. The cryo-EM structure reveals that the entire capsid volume is filled with DNA extending all the way to the center of the capsid. Along the capsid wall, there are well ordered, multiple concentric DNA layers (∼ 4–5 layers in all cases). The layers are evenly spaced, indicating that DNA has adopted a liquid crystalline state. However, towards the centre of the capsid, this ordering disappears. Similar dsDNA distributions within the capsids were also observed for other viruses (27–31). The figure also shows a separation between the capsid inner walls and the outermost DNA layer. This suggests that there is no attractive interaction between the DNA and the inner capsid wall. Indeed, it was shown for a lambdoid phage HK97 that the inner capsid surface is negatively charged (32), leading to repulsion between the DNA and the inner capsid wall.
Figure 1.
Cutaway views of the wt lambda phage cryo-EM reconstructions on complete DNA packaging (left) and after ejection of the DNA (right). The size and structure of the capsid shell (blue) remains unchanged after DNA ejection. Spacing between the outermost layers of the DNA (green) can be observed in the fully packaged phage reconstruction, and the DNA becomes more disordered closer to the centre of the capsid. The d-spacings between the DNA layers inside the capsid were determined by computing 3D cryo-EM reconstructions of the phage particles (see ‘Materials and Methods’ section). Owing to the icosahedral symmetry imposed during the reconstruction, concentrically packed DNA within the capsid becomes shells of density. The central slice of each reconstruction was extracted along the 5-fold symmetric axis, providing a cross-section of density in which the capsid and packaged genome appear most circular.
X-ray scattering analysis of DNA filled phage λ capsids has demonstrated hexagonal short-range DNA ordering for all three packaged DNA lengths studied here (8,33). This allows us to calculate the interaxial distance between the DNA strands in the ordered peripheral phase inside the capsid. The interaxial distance is ah = (2/√3) d, where d is the distance between the centres of the concentric DNA layers derived from our cryo-EM cross-section analysis.
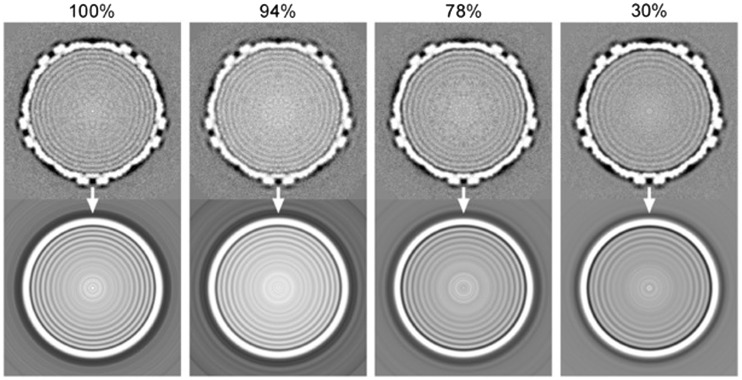
The d-spacings between the DNA layers inside the capsid were determined by computing 3D cryo-EM reconstructions of the phage particles (see ‘Materials and Methods’ section). The central slice of each reconstruction was extracted along the 5-fold symmetric axis, where the capsid and packaged genome adopt a circular organization. These slices were then radially averaged to further increase the signal-to-noise ratio of the genomic density, allowing accurate measurement of the DNA d-spacing. The variably filled capsids and their corresponding radial averages are shown in Figure 2. A 1D plot of the well-ordered DNA rings from the radially averaged central slice was plotted for each of the packaging variants (Figure 3), and the d- and interaxial spacings (ah) are summarized in Table 1.
Figure 2.
Central slices through the 3D cryo-EM reconstructions of the lambda phage containing different amounts of DNA (above) and the radially averaged images that were used for measuring the spacing (below). The percentage of packaged DNA relative to the wt phage is listed above each row. Only the outermost 4 or 5 rings of DNA have sufficient ordering to be used in calculating inter-DNA spacing. Note that the centre of the 30% packaged phage exhibits intensities are higher than that of the background, indicating that there is DNA present at the centre of the capsid.
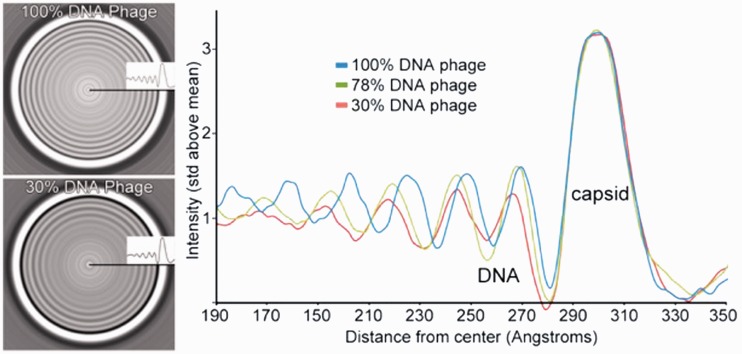
Figure 3.
Only the well-ordered peripheral rings of DNA were used to generate a 1D plot of the radially averaged central slices of the phage reconstructions. Shown are the radially averaged cross-sections for the 100% wt DNA-length and the 30% DNA-length (left). The 1D plots for the 100, 78 and 30% DNA-length phages are shown on the right.
Table 1.
Average values of DNA–DNA d-spacings between the DNA layers and the interaxial spacings for phage λ DNA length mutants with and without spermine added
| d-spacings (in Å) | Interaxial spacings (in Å) | |
|---|---|---|
| 100% WT phage λ (48.5 kb) | 22.7 | 26.2 |
| 100% WT phage λ (48.5 kb) + 1.3 mM spermine | 22.7 | 26.2 |
| 94% DNA phage λ (46.5 kb) | 24.6 | 28.4 |
| 78% DNA phage λ (37.8 kb) | 25.7 | 29.7 |
| 78% DNA phage λ (37.8 kb) + 1.3 mM spermine | 25.2 | 29.1 |
| 30% DNA phage λ in LamB + 1.3 mM spermine | 27.6 | 31.9 |
| Bulk DNA condensed by 1.3 mM spermine in TM buffer solution | 25.2 | 29.1 |
DNA d-spacings were determined from 3D reconstructions performed using 5000 particles for each phage mutant to maintain consistency between the data. Using higher numbers of particles did not change the d-spacing values. Based on the pixel size and the Gaussian spread of the radially averaged rings of DNA density, we calculate the centre of the density of each individual DNA ring to within ∼0.4 Å (standard error). We also show d-spacing and interaxial spacing values for bulk DNA condensed by 1.3 mM spermine in TM buffer solution, measured by X-ray diffraction (13).
Table 1 shows increasing interaxial spacing with decreasing packaged λ-DNA length. The values obtained from our cryo-EM analysis are essentially the same as those reported earlier for several phage λ mutants using the solution X-ray scattering (8). For wt λ-DNA length packaged in phage, ah = 26.2 Å, with a mere 6.2 Å surface separation between the DNA strands, whereas ah = 29.7 Å for the shortest packaged 78% λ-DNA length. As shown by the X-ray analysis of bulk DNA compressed by PEG under different ionic conditions, the interaxial distance value is directly related to the structure of the long-range DNA packing (6,13). Interaxial distances <∼30 Å correspond to a long-range hexagonal order, whereas ah > ∼32 Å is associated with a cholesteric packing pattern overlaid on a short-range hexagonal structure (6,13,14). Thus, the observed ordered peripheral structures in all three λ-DNA length phage mutants without spermine show long-range hexagonal packing.
DNA–DNA interaction and DNA bending are two main energy terms determining the structure of the encapsidated DNA (8,16,17). To reduce the bending energy, DNA will be pushed towards the capsid walls (providing lowest curvature), whereas repulsive DNA–DNA interactions will push DNA strands as far from each other as possible, filling the entire capsid space and maximizing the interstrand surface separation. Although the bending energy term cannot be neglected (8,16), the interaction energy term is dominant under the net DNA–DNA repulsion in the absence of multivalent ions. It maximizes the separation between the DNA strands so that DNA occupies the entire capsid volume, as seen in Figures 1 and 2. Therefore, the interaxial spacing is increasing with the decreasing packaged DNA length.
Repulsive and attractive interactions—bending induced DNA phase transition
It was shown that at interaxial DNA–DNA separations of ah≲ 26 Å, there is a dominant short-range repulsion interaction (11,13,34). In TM buffer, interhelical forces are repulsive at all separations (8). Adding 1 mM spermine to TM buffer causes DNA in solution to precipitate to an equilibrium interaxial spacing of 29.1 Å [determined using X-ray diffraction, as described in (13)]. However, at the 26.2 Å spacing characteristic of wt λ-DNA phage, even in spermine, DNA–DNA forces are highly repulsive (12) and should still dominate bending. We tested this assumption by adding 1 mM spermine to the wt λ-DNA phage solution. Data shown in Table 1 confirm that the interaxial spacing is not affected by the addition of spermine ions. Thus, although 1 mM spermine concentration is sufficient to induce a net attractive interaction between the DNA strands, confinement within capsid causes the interhelical interactions to remain net repulsive. This suggests that the length of packaged wt λ-DNA in a phage capsid has been evolutionarily optimized to result in DNA interstrand separations and interactions that are not strongly affected by the ionic conditions in the cellular and extracellular environments. This increases the robustness of both DNA packaging and ejection, minimizing the influence of external ionic fluctuations. This hypothesis has been recently verified by our measurements of the efficiency of genome packaging in phage λ as a function of salt and packaged DNA length (35).
Next, we investigated the interplay between the attractive and repulsive interstrand interactions in phage λ capsids and how the type of interactions affects the long-range DNA structure. The shorter 78% wt λ-DNA length λ mutant provides a good experimental system for such analysis, as the interaxial spacing is ah = 29.7 Å, which is larger than the spacing in the dominantly repulsive regime where the interactions are independent of the ionic conditions (at ah≲ 26 Å). Moreover, 78% λ-DNA length phage also has larger interstrand spacing than the DNA condensed by 1 mM spermine in the bulk TM buffer solution, where ah = 29.1 Å. However, with addition of 1 mM spermine to the 78% λ-DNA length packaged capsids, we found the interaxial spacing to decrease from 29.7 Å to 29.1 Å. Thus, the interstrand DNA interactions in 78% λ-DNA length packaged phage are changing from net repulsive to net attractive when spermine is added. The interaxial spacing within the capsid is now the same as found for free DNA condensed by 1 mM spermine in the solution without an external osmotic pressure. Interestingly, although DNA is bent inside the capsid, while it is not in the bulk, bending does not seem to influence the DNA spacing inside the capsid. This could be due to cancelling effects; the energy owing to DNA bending decreases with smaller spacings, whereas the energy owing to DNA packing defects or deviations from local hexagonal order in toroidal or spooled condensates increases with smaller spacings (36). However, even with net DNA–DNA attractive interaction in the 78% λ-DNA length phage in 1 mM spermine, the encapsidated DNA is still under stress.
We demonstrate this by opening the 78% λ-DNA length phage capsid with LamB receptor and allowing the encapsidated DNA to freely eject into the 1 mM spermine solution. DNA ejection will proceed until full equilibrium is established, and DNA in the capsid is no longer under stress (37). To ensure that the ejected fraction of the DNA is not influenced by the condensation of DNA by spermine after ejection, we added the endonuclease DNase I before LamB addition, which immediately digests the DNA on ejection. In the presence of DNase I and 1 mM spermine, 30% of wt λ-DNA length remains in the opened capsid at equilibrium, as determined by ultraviolet absorbance measurements (38,39). As there are no net DNA repulsive interactions present, the driving force for this DNA ejection is the bending stress caused by the capsid confinement. The bending stress is relieved, as DNA is ejected from the capsid. The ejection, however, will not continue to completion, as the remaining portion of the bending energy that would drive the DNA from the capsid is counter balanced by the attractive interaction between the DNA strands induced by spermine ions.
The radially averaged cryo-EM reconstruction cross-section of the 30% λ-DNA length phage shows that the interaxial spacing significantly increases from 29.1 Å to 31.9 Å with this decrease in packaged DNA length (Figure 3 and Table 1). The 31.9 Å spacing represents a free energy minimum that is more stable than the 29.1 Å minimum. The 31.9 Å minimum is not accessible before ejection owing to the capsid volume constraint with fixed DNA size. For bulk DNA condensed in spermine solution, the interaxial spacing of 29.1 Å corresponds to a long-range hexagonal order (34). The 31.9 Å interaxial distance is characteristic of a long-range less ordered cholesteric packing structure (12–14,34). A hexagonal—cholesteric phase transition was demonstrated by several independent measurements on DNA condensed by PEG and/or multivalent ions in bulk solution using X-ray diffraction to determine DNA structure (12–14,34). It is likely, in similarity to the bulk measurements, that there is a transition in the long-range DNA arrangement inside the capsid from a hexagonal to a cholesteric state when DNA is allowed to eject from phage λ in 1 mM spermine until it reaches equilibrium (40–46). The cholesteric packing arrangement has weaker interstrand attractions than hexagonally ordered DNA, resulting in larger spacings and higher levels of disorder, marked by an increase in the X-ray scattering peak width (12–14,34). The attractive interaction between the DNA strands in the capsid in 1 mM spermine is weaker than that for DNA condensed by the same amount of spermine in the bulk, where DNA is hexagonally ordered. Optimal attraction requires a phosphate–phosphate correlation between the DNA helices that minimizes DNA–DNA interaction. When DNA is bending inside the capsid, the initial correlation between two helices that have slightly different radii of curvature is lost, and the mutual orientation between helices must be re-established. This leads to the so-called ‘packing defect’ that weakens the attraction between the encapsidated DNA strands favouring cholesteric packing, compared with the unconstrained hexagonally packed DNA arrays condensed by spermine in solution.
Even if the long-range genomic structure is cholesteric, we can still use the expression ah = (2/√3) d to calculate the interaxial spacing ah from the d-spacings between the concentric DNA layers determined by cryo-EM particle reconstructions. It was previously shown for DNA condensed in bulk (34,47) that the local short-range DNA structure remains hexagonally ordered, whereas the long-range structure has a cholesteric pattern. In a cholesteric state, the DNA is organized in parallel planes with a continuous rotation of the orientation of the DNA molecules from plane to plane. However, locally, the DNA double helices will have six neighbouring strands, corresponding to a local short-range hexagonal arrangement with a long-range cholesteric texture superimposed on it. Figure 2 shows central cross-section from the reconstructions of a receptor-open λ-capsid containing 30% of wt λ-DNA in 1 mM spermine at equilibrium. The figure shows that even in the case of a 30% λ-DNA filled capsid, the entire capsid volume is occupied by the DNA, as the intensities observed at the centre of the capsid are >1 standard deviation higher than the mean background intensity. However, DNA is ordered in concentric layers only in the periphery of the capsid. Assuming the same packing density throughout the entire capsid volume, the change in interaxial distance ah from wt to 78% packaged λ-DNA length corresponds well to that expected maintaining constant volume (26.2/29.7)2 ∼ 78%. The change in ah adding 1 mM spermine to the 78% and ejecting is different from that expected for volume filling with 30% of the wt λ-DNA left, namely (26.2/31.9)2 ∼ 67%. This calculation indicates that DNA must have lower packing density in the centre of the capsid.
CONCLUSION
In conclusion, we have observed for the first time a bending-induced phase transition of encapsidated DNA. Such a phase transition leads to higher disorder and a significant increase in the genome fluidity. In a hexagonal phase, DNA motion is limited, leading to reduced interstrand fluidity. The observation of a cholesteric structure at lower DNA filling fractions suggests that a cholesteric-to-hexagonal phase transition occurs during the packaging process in vivo, as the packaged DNA fraction is increasing to interstrand separations of 30–34 Å. This should significantly affect the packaging rate by slowing it down. In parallel, during the ejection process, an increased fluidity of the genome will speed up the ejection rate, improving the efficiency of viral replication.
The importance of DNA bending inside the phage capsid has often been overlooked owing to the fact that the interaction energy for a fully packaged phage λ is the dominant stress contribution to the overall DNA energy state (8,17). However, our present cryo-EM analysis of the encapsidated DNA structure with the net repulsive interaction ‘turned on and off’ allows us to separate the repulsive interaction from the DNA bending-induced stress and packing defects, revealing its significant role for the long-range DNA structure inside the capsid. Thus, there are important interactions in the capsid other than DNA–DNA force that manifest themselves in the ah = 31.9 Å free energy minimum that is not seen with unconfined DNA condensates. This 31.9 Å minimum is more stable than the 29.1 Å found in bulk DNA condensed in spermine. Furthermore, as exemplified by the shorter 78% wt-DNA length λ mutant, the bending stress will play a more significant role for larger viruses with typically lower packaged DNA densities, such as Human Herpes Simplex 1.
FUNDING
Swedish Research Council, VR [622-2008-726 to A.E.]; NSF [CHE-1152770 to A.E.]. G.C.L. acknowledges support from Damon Runyon Cancer Research Foundation. NIH [R01 GM 54076-16 to J.E.J.]; NIH [R01 AI 5R01AI040101 to J.E.J.]; Intramural Research Program of the National Institutes of Health (in part), NICHD (Eunice Kennedy Shriver National Institute of Child Health and Human Development) (to D.C.R.). Funding for open access charge: NSF [CHE-1152770 to A.E.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Ting Liu and Dave Bauer for help with UV measurements, and William Gelbart, Luca Tubiana, Anton Petrov and Martin Castelnovo for important feedback and discussions. Electron microscopic imaging and reconstruction were conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health (NIH) through the National Center for Research Resources’ P41 program [RR017573].
REFERENCES
- 1.Flint SJ, Enquist LW, Krug RM, Skalka AM, Racaniello VR. Principles of Virology: Molecular Biology, Pathogenesis and Control. Washington, DC: American Society for Microbiology; 2000. [Google Scholar]
- 2.Rau DC, Parsegian VA. Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys. J. 1992;61:260–271. doi: 10.1016/S0006-3495(92)81832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez M, Rau D, O'Sullivan D, Brown AH, Papa R, Attene G. Genetic structure and linkage disequilibrium in landrace populations of barley in Sardinia. Theor. Appl. Genet. 2012;125:171–184. doi: 10.1007/s00122-012-1824-8. [DOI] [PubMed] [Google Scholar]
- 4.Strey HH, Podgornik R, Rau DC, Parsegian VA. DNA–DNA interactions. Curr. Opin. Struct. Biol. 1998;8:309–313. doi: 10.1016/s0959-440x(98)80063-8. [DOI] [PubMed] [Google Scholar]
- 5.Leikin S, Parsegian VA, Rau DC, Rand RP. Hydration forces. Ann. Rev. Phys. Chem. 1993;44:369–395. doi: 10.1146/annurev.pc.44.100193.002101. [DOI] [PubMed] [Google Scholar]
- 6.Stanley C, Rau DC. Evidence for water structuring forces between surfaces. Curr. Opin. Colloid Interface Sci. 2011;16:551–556. doi: 10.1016/j.cocis.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 8.Qiu X, Rau DC, Parsegian VA, Fang LT, Knobler CM, Gelbart WM. Salt-dependent DNA-DNA spacings in intact bacteriophage lambda reflect relative importance of DNA self-repulsion and bending energies. Phys. Rev. Lett. 2011;106:028102. doi: 10.1103/PhysRevLett.106.028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsegian VA, Rand RP, Fuller NL, Rau DC. Osmotic stress for the direct measurement of intermolecular forces. Methods Enzymol. 1986;127:400–416. doi: 10.1016/0076-6879(86)27032-9. [DOI] [PubMed] [Google Scholar]
- 10.Rau DC, Lee B, Parsegian VA. Measurement of the repulsive force between polyelectrolyte molecules in ionic solution: hydration forces between parallel DNA double helices. Proc. Natl Acad. Sci. USA. 1984;81:2621–2625. doi: 10.1073/pnas.81.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd BA, Rau DC. Interplay of ion binding and attraction in DNA condensed by multivalent cations. Nucleic Acids Res. 2008;36:501–510. doi: 10.1093/nar/gkm1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd BA, Parsegian VA, Shirahata A, Thomas TJ, Rau DC. Attractive forces between cation condensed DNA double helices. Biophys. J. 2008;94:4775–4782. doi: 10.1529/biophysj.107.127332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rau DC, Parsegian VA. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys. J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raspaud E, Durand D, Livolant F. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophys. J. 2005;88:392–403. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hud NV, Downing KH. Cryoelectron microscopy of lambda phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Natl Acad. Sci. USA. 2001;98:14925–14930. doi: 10.1073/pnas.261560398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evilevitch A, Fang LT, Yoffe AM, Castelnovo M, Rau DC, Parsegian VA, Gelbart WM, Knobler CM. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophys. J. 2008;94:1110–1120. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc. Natl Acad. Sci. USA. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silhavy TJ. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 20.Graff A, Sauer M, Van Gelder P, Meier W. Virus-assisted loading of polymer nanocontainer. Proc. Natl Acad. Sci. USA. 2002;99:5064–5068. doi: 10.1073/pnas.062654499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roa M, Scandella D. Multiple steps during interaction between coliphage-lambda and its receptor protein Invitro. Virology. 1976;72:182–194. doi: 10.1016/0042-6822(76)90322-6. [DOI] [PubMed] [Google Scholar]
- 22.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roseman AM. FindEM–a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 2004;145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 27.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 30.Huet A, Conway JF, Letellier L, Boulanger P. In vitro assembly of the T=13 procapsid of bacteriophage T5 with its scaffolding domain. J. Virol. 2010;84:9350–9358. doi: 10.1128/JVI.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Rau DC. Incomplete ion dissociation underlies the weakened attraction between DNA helices at high spermidine concentrations. Biophys. J. 2005;89:1932–1940. doi: 10.1529/biophysj.105.065060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurmemmedov E, Castelnovo M, Medina E, Catalano CE, Evilevitch A. Challenging packaging limits and infectivity of phage lambda. J. Mol. Biol. 2012;415:263–273. doi: 10.1016/j.jmb.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Park SY, Harries D, Gelbart WM. Topological defects and the optimum size of DNA condensates. Biophys. J. 1998;75:714–720. doi: 10.1016/S0006-3495(98)77561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evilevitch A. Effects of condensing agent and nuclease on the extent of ejection from phage lambda. J. Phys. Chem. B. 2006;110:22261–22265. doi: 10.1021/jp060573j. [DOI] [PubMed] [Google Scholar]
- 38.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl Acad. Sci. USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evilevitch A, Gober JW, Phillips M, Knobler CK, Gelbart WM. Measurements of DNA lengths remaining in a viral capsid after osmotically suppressed partial ejection. Biophys. J. 2005;88:751–756. doi: 10.1529/biophysj.104.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornyshev AA, Leikin S, Malinin SV. Chiral electrostatic interaction and cholesteric liquid crystals of DNA. Eur. Phys. J. E. 2002;7:83–93. [Google Scholar]
- 41.Cherstvy AG, Kornyshev AA, Leikin S. Temperature-dependent DNA condensation triggered by rearrangement of adsorbed cations. J. Phys. Chem. B. 2002;106:13362–13369. [Google Scholar]
- 42.Leforestier A, Livolant F. Supramolecular ordering of DNA in the cholesteric liquid crystalline phase: an ultrastructural study. Biophys. J. 1993;65:56–72. doi: 10.1016/S0006-3495(93)81063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leforestier A, Livolant F. Cholesteric liquid crystalline DNA; a comparative analysis of cryofixation methods. Biol. Cell. 1991;71:115–122. doi: 10.1016/0248-4900(91)90058-u. [DOI] [PubMed] [Google Scholar]
- 44.Livolant F, Levelut AM, Doucet J, Benoit JP. The highly concentrated liquid-crystalline phase of DNA is columnar hexagonal. Nature. 1989;339:724–726. doi: 10.1038/339724a0. [DOI] [PubMed] [Google Scholar]
- 45.Livolant F. Cholesteric organization of DNA in the stallion sperm head. Tissue Cell. 1984;16:535–555. doi: 10.1016/0040-8166(84)90029-6. [DOI] [PubMed] [Google Scholar]
- 46.Livolant F. Cholesteric organization of DNA in vivo and in vitro. Eur. J. Cell Biol. 1984;33:300–311. [PubMed] [Google Scholar]
- 47.Lepault J, Dubochet J, Baschong W, Kellenberger E. Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J. 1987;6:1507–1512. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]