Abstract
The node and the notochord are important embryonic signaling centers that control embryonic pattern formation. Notochord progenitor cells present in the node and later in the posterior end of the notochord move anteriorly to the generate notochord. To understand the dynamics of cell movement during notochord development and the molecular mechanisms controlling this event, analyses of cell movements using time-lapse imaging and conditional manipulation of gene activities are required. To achieve this goal, we generated two knock-in mouse lines that simultaneously express nuclear enhanced green fluorescent protein (EGFP) and tamoxifen-inducible Cre, CreERT2, from two notochord gene loci, Foxa2 and T (Brachury). In Foxa2nEGFP-CreERT2/+ and TnEGFP-CreERT2/+ embryos, nuclei of the Foxa2 or T-expressing cells, which include the node, notochord, and endoderm (Foxa2) or wide range of posterior mesoderm (T), were labeled with EGFP at intensities that can be used for live imaging. Cre activity was also induced in cells expressing Foxa2 and T one day after tamoxifen administration. These mice are expected to be useful tools for analyzing the mechanisms of notochord development.
Keywords: node, notochord, Foxa2, T (Brachyury), dual labeling, nuclear EGFP, inducible Cre
During mouse embryogenesis, the organizer and its descendant signaling centers play central roles in the establishment of the correct body plan (reviewed in (Davidson and Tam, 2000; Niehrs, 2004; Tam and Behringer, 1997). The midline mesodermal tissue, the notochord, is the major signaling center that controls trunk and tail development. The notochord expresses both signaling molecules and their antagonists, thereby regulating the differentiation, growth, and patterning of the surrounding tissues (see review by (Cleaver and Krieg, 2001). Development of the notochord proceeds from an anterior to posterior direction through continuous addition of the more-posterior cells in coordination with the posterior extension of the body axis. The notochord develops from notochord progenitor cells (NPCs), which are initially specified in the ventral layer of the node at the late gastrula stage (Kinder et al., 2001; Ukita et al., 2009), and form the entire notochord. At later stages, the NPCs reside in the posterior end of the notochord in the tail bud to support posterior extension up to E13.5 (Cambray and Wilson, 2002; Tam et al., 1997; Ukita et al., 2009). Formation of the trunk notochord involves convergent extension movements, which are regulated by planar cell polarity signaling (Lu et al., 2004; Wang et al., 2006; Yamanaka et al., 2007).
To understand the dynamics of cell movement underlying these morphogenetic movements and the molecular mechanisms controlling notochord morphogenesis, analyses of cell movements by live, time-lapse imaging, lineage tracing, and conditional manipulation of gene activities are required. Currently, multiple mouse lines that express EGFP for live imaging, as well as mouse lines that express inducible Cre in the node for lineage tracing/conditional gene manipulation (Ukita et al., 2009; Yamanaka et al., 2007) and lineage tracing in the notochord (Frank et al., 2007; Park et al., 2008) are available. However, mouse lines that can be used for both live imaging and lineage tracing/conditional gene manipulation in the node and the notochord have not yet been reported.
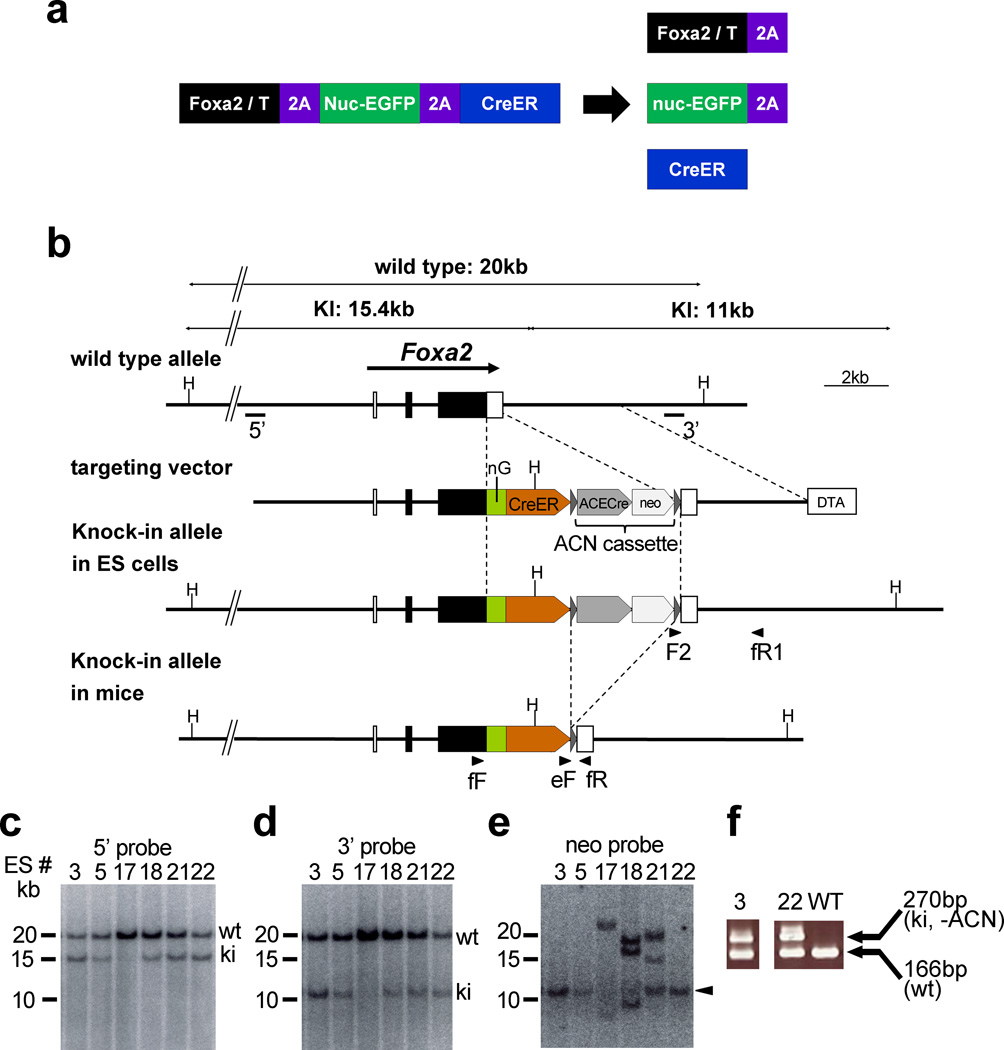
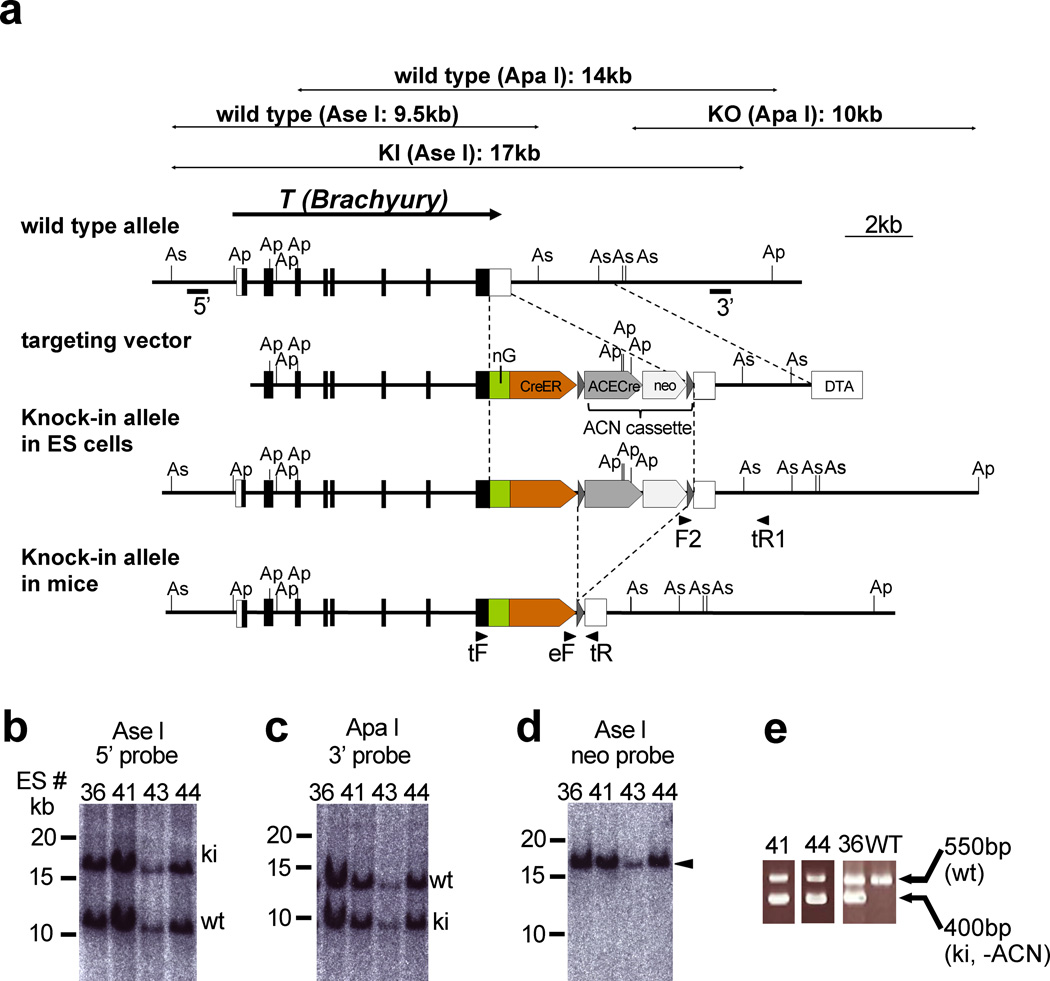
To make live imaging and conditional gene manipulation of notochord cells possible, we generated two knock-in mouse lines, Foxa2nEGFP-CreERT2 and TnEGFP-CreERT2, which express nuclear fluorescent protein (histone H2B-EGFP) (Shaner et al., 2005) and tamoxifen (Tx)-inducible Cre recombinase (CreERT2) (Feil et al., 1997) from the Foxa2 and T (encoding Brachyury) loci, by homologous recombination in embryonic stem (ES) cells. Both genes are strongly expressed in the node and notochord (Herrmann, 1991; Sasaki and Hogan, 1993). In these knock-in mouse lines, the coding sequences for nuclear fluorescent protein and CreERT2 were placed downstream of the Foxa2 or T coding sequence, with sequences for virus-derived 2A peptides (Szymczak et al., 2004) inserted between the protein coding sequences (Figs. 1a, b, 2a). Because 2A peptide-containing proteins are self-cleaved within the 2A peptide, their introduction allows the production of multiple, in this case three, proteins from a single transcript (Szymczak and Vignali, 2005; Szymczak et al., 2004) (Fig. 1a). We confirmed correct homologous recombination in the Foxa2 and T loci and deletion of the neomycin-resistance gene cassette (ACN cassette) (Bunting et al., 1999) by Southern blotting and PCR (Figs. 1c–f, and 2b–e).
Figure 1. Production of the Foxa2nEGFP-CreERT2/+ mouse.
(a) Strategy for production of three proteins from a single transcript. 2A-peptide-containing proteins are self-cleaved within of the 2A peptide. Therefore, the modified Foxa2 (or T)-locus, which has a single coding sequence for a FOXA2 (or T)-2A-nuclear EGFP-2A-CreERT2 protein, will produce three proteins: FOXA2 (or T)-2A, nuclear EGFP-2A, and CreERT2.
(b) Targeting strategy. The Foxa2nEGFP-CreERT2 allele was generated by homologous recombination in ES cells. Exons are indicated by filled and open boxes, and coding regions by filled boxes. Triangles represent loxP sites. 5’ and 3’ indicate the approximate positions of 5’ and 3’ probes, respectively, for Southern hybridization. Abbreviations: H, Hind III; nEGFP, ERAV 2A peptide followed by nuclear (histone H2B) EGFP; CreER, TaV 2A peptide followed by CreERT2; ACE-Cre, a cassette carrying the testis-specific ACE promoter-Cre-poly A signal; neo, an expression cassette for the neomycin-resistance gene; DT-A, MC1-DT-A-poly A signal cassette. The ACN cassette consists of tACE-Cre and neo cassettes flanked by loxP sites, and this cassette is removed in the mouse testis during germline transmission. Arrowheads indicate the approximate positions of PCR primers. F2, SVloxP-3’-F2; fR1, Foxa2-PC-3’-R1; fF, Foxa2 genotype F; eF, ERT2 genotype F; fR, Foxa2 genotype R.
(c–e) Southern blot analysis of homologous recombination in ES cells. Hind III-digested ES cell genomes were separated on a 0.3% agarose gel and blotted to nylon membranes. Hybridization with the 5’ probe (c) and 3’ probe (d) produced two bands that corresponded to the predicted lengths of the wild-type (wt) and knock-in (ki) alleles. Hybridization with the internal (neo) probe (e) gave a single band (arrowhead) corresponding to the knock-in allele.
(f) Genotype determination of Foxa2nEGFP-CreERT2/+ mice by PCR. Note that the ACN cassette was removed in knock-in mice #3 and #22
Figure 2. Production of the TnEGFP-CreERT2/+ mouse.
(a) Targeting strategy. The TnEGFP-CreERT2 allele was generated by homologous recombination in ES cells. Exons are indicated by filled and open boxes, and coding regions by filled boxes. Triangles represent loxP sites. 5’ and 3’ indicate the approximate positions of the 5’ and 3’ probes, respectively, for Southern hybridization. Abbreviations: As, Ase I; Ap, Apa I; nEGFP, ERAV 2A peptide followed by nuclear (histone H2B) EGFP; CreER, TaV 2A peptide followed by CreERT2; ACE-Cre, a cassette carrying the testis-specific ACE promoter-Cre-poly A signal; neo, an expression cassette for the neomycin-resistance gene; DT-A, MC1-DT-A-poly A signal cassette. The ACN cassette consists of the tACE-Cre and neo cassettes flanked by loxP sites. Arrowheads indicate the approximate positions of the PCR primers. F2, SVloxP-3’-F2; tR1, T-PC-3’-R1; tF, T genotype F; eF, ERT2 genotype F; tR, T genotype R.
(b–d) Southern blot analysis of homologous recombination in ES cells. Ase I-digested (b, d) or Apa I-digested (c) ES cell genomes were separated on a 0.3% agarose gel and blotted onto nylon membranes. Hybridization with the 5’ probe (b) and the 3’ probe (c) yielded two bands that corresponded to the predicted lengths of the wild-type (wt) and knock-in (ki) alleles. Hybridization with the internal (neo) probe (d) gave a single band (arrowhead) corresponding to the knock-in allele. (e) Genotype determination of TnEGFP-CreERT2 mice by PCR. Note that the ACN cassette was removed in knock-in mice #36, 41 and 44.
The Foxa2nEGFP-CreERT2 mouse lines were produced from two independent ES cell lines (#3, #22). Because both knock-in lines exhibited the same phenotype, we used the line derived from ES cell line #22 for most of the analyses. Similarly, TnEGFP-CreERT2 mouse lines were produced from three independent ES cell lines (#36, #41, #44), and the line derived from ES cell line #41 was used for most of the analyses. Foxa2nEGFP-CreERT2/nEGFP-CreERT2 mice could be maintained as homozygotes, suggesting that introduction of the transgene did not significantly affect Foxa2 function. In contrast, although we designed the allele to express T, some of the TnEGFP-CreERT2/+ mice showed various degrees of tail defects (curled, short, or no tail, data not shown) similar to heterozygous T mutants (Chesley, 1935), and TnEGFP-CreERT2 homozygotes were not obtained (data not shown). Therefore, loss of or significant reduction in T gene activity should be considered in order to use the TnEGFP-CreERT2 mouse. Perhaps the addition of the 2A peptide to the C-terminus interfered with the activity of the T protein or the cleavage of the 2A sequence between T and the nuclear EGFP was incomplete. It is also possible that insertion of the additional sequences altered T RNA stability. Knock-in mice that were similarly designed for a homeobox gene locus, Noto, (NotonmCherry-CreERT2/nmCherry-CreERT2) also showed a mutant phenotype (Ukita et al., 2009).
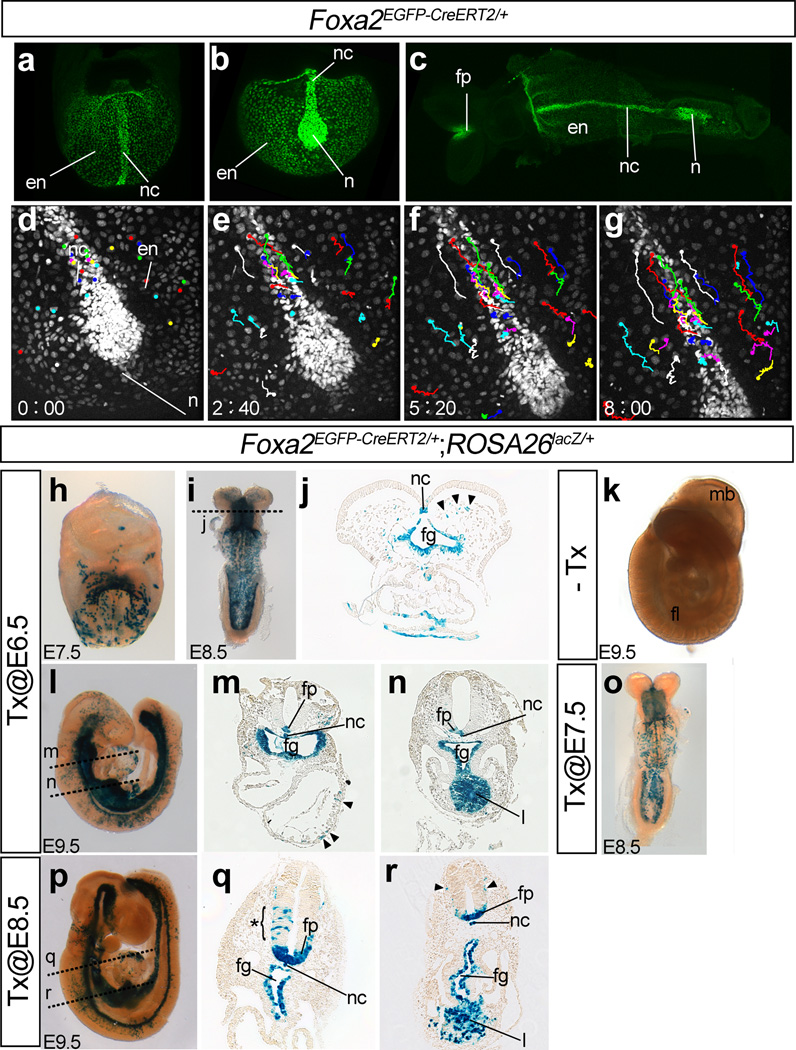
In Foxa2nEGFP-CreERT2/+ embryos, consistent with expression of Foxa2 in the node, the notochord, the floor plate of the neural tube, and the definitive endoderm (Monaghan et al., 1993; Sasaki and Hogan, 1993), GFP signals were observed in the nuclei of these tissues between E7.5 and E8.5 (Fig.3a–c). Reflecting weaker expression of Foxa2 in the endoderm, GFP signals in the endoderm were weaker than those of the notochord. Both higher and lower GFP signals were strong enough to perform time-lapse imaging. Time-lapse imaging of Foxa2nEGFP-CreERT2/+ embryos with a confocal microscope between E7.5 and E8.25 recorded dynamic movements of the cells in the notochord and the endoderm (Fig. 3d–g). Detailed analysis of cell behaviors in the notochord and the endoderm will be described elsewhere.
Figure 3. Characterization of Foxa2nEGFP-CreERT2/+ embryos.
(a–c) Expression of nuclear EGFP in Foxa2nEGFP-CreERT2/+ embryos. Frontal (a) and ventral (b) views of an E7,75 embryo, and ventral view of an E8.5 embryo (c). The EGFP signal was observed in the nuclei of the cells in the node, the notochord, the floor plate of the neural tube, and the definitive endoderm.
(d–g) Snapshot images of a time-lapse movie of EGFP expression of a Foxa2nEGFP-CreERT2/+ embryo between E7.75 and E8.25. The number in each panel indicates the elapsed time (hour: minutes). Color lines indicate traces of cellular movements.
(h–r) Induction of Cre recombinase activity in Foxa2nEGFP-CreERT2/+;ROSA26lacZ/+ embryos by Tx administration. Tx administration at E6.5 induced Cre-mediated recombination in the node, the notochord, floor plate of the neural tube and the definitive endoderm. β-galactosidase activity at E7.5 (h, frontal view), E8.5 (i, ventral view; j, a cross section through foregut) and E9.5 (l, right side view; m, n, cross sections through the heart [m] or the liver primordium [n]). β-galactosidase activity was also induced in similar patterns at one day after Tx administration at E7.5 (o, ventral view) or E8.5 (p, right side view; q, r, cross sections through the heart [q] or the liver primordium [r]). Arrowheads in panels j, m, and r and the asterisk in panel q indicate examples of β-galactosidase–positive cells that do not express endogenous Foxa2.
No β-galactosidase activity was detected without Tx (k). Approximate positions of the sections are shown in panels i, l and p with broken lines.
Abbreviations: en, endoderm; fg, foregut; fp, floor plate of the neural tube; l, liver primordium; n, node; nc, notochord.
When Cre activity was induced by oral administration of Tx to pregnant female mice at E6.5, the cells in the node, notochord, floor plate, and endoderm of Foxa2nEGFP-CreERT2/+;ROSA26RLacZ/+ embryos expressed β-galactosidase at E7.5, E8.5 and E9.5 (Fig. 3h–j, l-n). At E9.5, the liver primordium, which expresses Foxa2, is also labeled. The frequency of β-galactosidase-positive cells at E8.5 was higher than that at E7.5, perhaps because Cre activity is induced more than one day after Tx administration. No labeled cells were observed without administration of Tx (Fig. 3k). Similar Foxa2-like patterns of β-galactosidase-positive cells were also obtained by Tx administration at E7.5 and E8.5 (Fig. 3o–r). Scattered distribution of β-galactosidase–positive cells was also observed in the cells that were expected to be Foxa2-negative (Sasaki and Hogan, 1993) (examples for the same are indicated using arrowheads and an asterisk in Fig. 3j, m, q, r), suggesting that Foxa2 was expressed in the progenitors of these cells and/or that the CreERT2 protein was more stable than the FOXA2 protein.
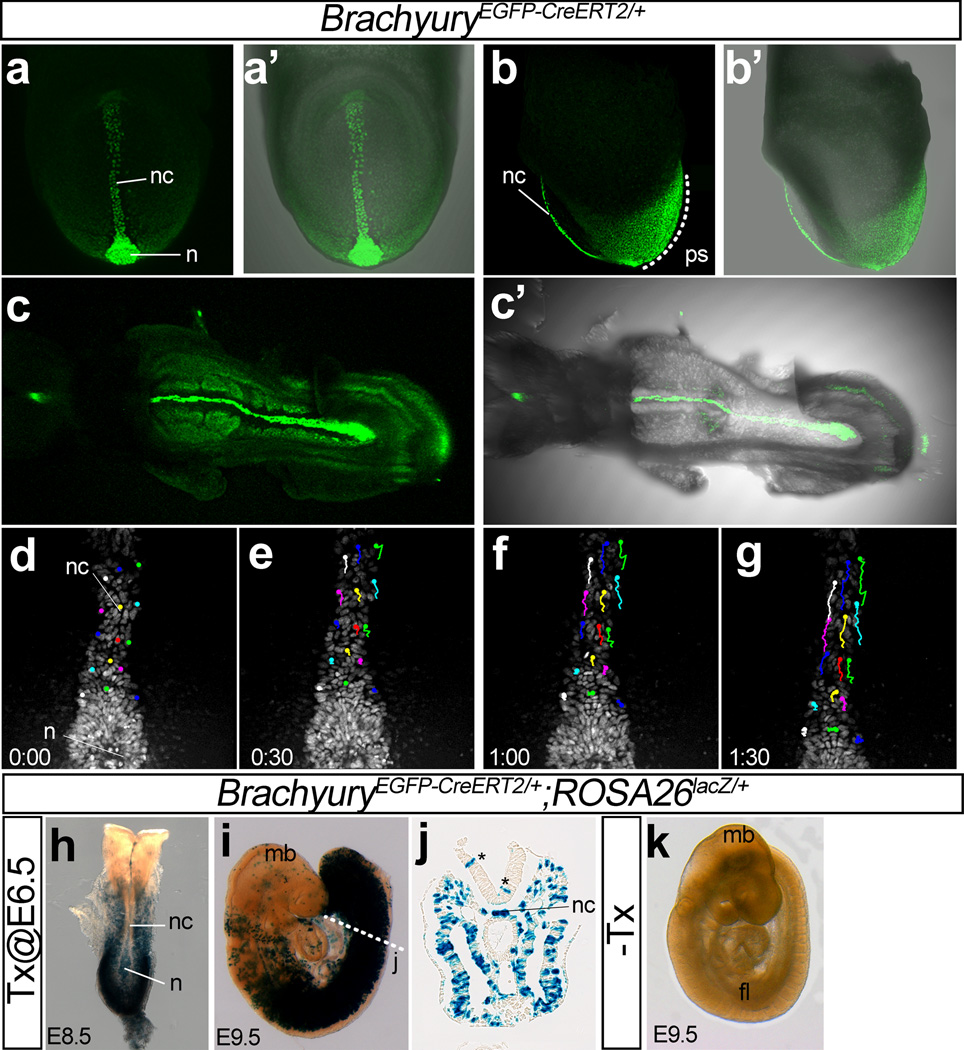
In TnEGFP-CreERT2/+ embryos, consistent with the findings that T is expressed in the node, notochord, and the nascent mesoderm around the primitive streak (Kispert and Herrmann, 1994; Wilkinson et al., 1990), EGFP signals were observed in the nuclei of these tissues between E7.5 and E8.5 (Fig. 4a–c). GFP expression in the nascent mesoderm around the primitive streak was much wider and expanded more anteriorly than the area of T expression that may be caused by a longer half-life of H2B-EGFP than T protein. Although the notochordal GFP fluorescence was slightly weaker than that of Foxa2nEGFP-CreERT2/+ mice, this mouse can also be used for time-imaging. Time-lapse imaging of TnEGFP-CreERT2/+ embryos with a confocal microscope starting from E7.75 recorded dynamic movements of notochord cells (Fig. 4d–g).
Figure 4. Characterization of TnEGFP-CreERT2/+ embryos.
(a–c) Expression of nuclear EGFP in TnEGFP-CreERT2/+ embryos. Frontal (a, a’) and left side (b, b’) views of an E7,75 embryo, and ventral views of an E8.5 embryo (c, c’). The EGFP signal was observed in the nuclei of the cells in the node, the notochord, and wide range of the posterior mesoderm.
(d–g) Snapshot images of a time-lapse movie of EGFP expression of a TnEGFP-CreERT2/+ embryo starting from E7.75. The number in each panel indicates the elapsed time (hour: minutes). Color lines indicate traces of cellular movements.
(h–k) Induction of Cre recombinase activity in TnEGFP-CreERT2/+;ROSA26lacZ/+ embryos by administration of Tx. Tx administration at E6.5 induced Cre-mediated recombination in the node and wide range of posterior mesoderm. β-galactosidase activity at E8.5 (h, dorsal view) and E9.5 (i, left side view; j, a cross section close to the posterior end of the notochord). Asterisks in panel j indicate examples of β-galactosidase–positive cells that do not express endogenous T. No β-galactosidase activity was observed without Tx (k).
Abbreviations: n, node; nc, notochord; ps, primitive streak.
When Cre activity was induced by oral administration of Tx to pregnant female mice at E6.5, the cells in the node, notochord, and a wide area of the posterior mesoderm of TnEGFP-CreERT2/+;ROSA26RLacZ/+ embryos expressed β-galactosidase at E8.5 and E9.5 (Fig. 4f–j). No labeled cells were observed without induction by Tx (Fig. 4k). The wide distribution of β-galactosidase-positive cells in the posterior mesoderm suggests that this mouse can also be used to label and/or genetically manipulate a wide range of mesodermal cells.
In conclusion, we established two knock-in mouse lines that can be used for live and time-lapse imaging, lineage tracing and conditional gene manipulation in the notochord. It should be noted that expression of Foxa2 and T are not restricted to the node and the notochord. For example, Foxa2 is expressed widely in tissues derived from endoderm (liver, lung, pancreas, stomach, intestine, prostate and bladder), ectoderm (several brain structures and olfactory epithelium) and mesoderm (kidney, vagina, uterus, and seminal and coagulating glands) during development (Besnard et al., 2004). T is transiently expressed in developing heart, yolk sac, and limb (Inman and Downs, 2006; Liu et al., 2003a). T/Brachyury expression in cancer promotes epithelial-mesenchymal transition and confers cancer stem cell characteristics (Fernando et al., 2010; Palena et al., 2007; Sarkar et al., 2012). Therefore, the mice described here should be also useful for studies of other tissues and/or cancers that express these genes.
METHODS
Mouse lines
The Foxa2nEGFP-CreERT2/+ (Acc. No. CDB0601K: http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) mouse was generated at the RIKEN CDB by homologous recombination in ES cells using the standard protocol described in http://www.cdb.riken.jp/arg/Methods.html. Briefly, the C57BL/6J mouse BAC genomic DNA clone RP24-386G1, which contains the Foxa2 gene, was obtained from BACPAC Resources, Children’s Hospital Oakland Research Institute (CHORI), and the gene with its surrounding genomic material was subcloned into MC1-DTA-pA/pMW118 (Nishioka et al., 2008; Sawada et al., 2008) using homologous recombination in E. coli, a recombineering method (Liu et al., 2003b).
The Foxa2 targeting vector was designed to remove the termination codon of the FOXA2 protein, and introduced upstream of the following sequences: equine rhinitis A virus (ERAV) 2A peptide (Szymczak and Vignali, 2005), a fusion protein of human histone H2B and EGFP (Shaner et al., 2005), Thosea asigna virus (TaV) 2A peptide (Szymczak and Vignali, 2005), and CreERT2 (Feil et al., 1997). We used 2A peptide sequences of ERAV and TaV because both sequences have previously been used successfully (Szymczak and Vignali, 2005). The neomycin-resistance gene was supplied as the ACN cassette, which is self-excised in the germ line (Bunting et al., 1999). The targeting vector was generated using a recombineering method (Liu et al., 2003b).
TT2 ES cells (Yagi et al., 1993) were transformed by electroporation with linearized targeting vector, followed by positive and negative selection with G418 and DT-A, respectively. The ES cell clones were screened for homologous recombination by long polymerase chain reaction (PCR) using LA-Taq (TaKaRa, Japan). To screen for the Foxa2 locus, the following primer pair was used: SVloxP-3’-F2 (F2, 5’-AGGCCCAGGGCTCGCAGCCAACGTCG-3’) and Foxa2-PC-3’-R1 (fR1, 5’-CCTGAGCTGGGTGCACCTCCCAGTCC-3’). The positions of these primers are indicated in Fig. 1b. Correct homologous recombination of the Foxa2 locus and the absence of randomly inserted targeting vectors were confirmed in the PCR-positive clones by Southern hybridization (Fig. 1c–e). Among the 96 clones examined using PCR, 54 clones showed positive results. Among the PCR-positive clones, 20 clones were analyzed further by using Southern hybridization, and among these, 17 clones were confirmed as correct homologous recombinants. The confirmed ES clones were injected into 8-cell embryos of ICR to produce chimeric mice. Chimeric founders from two independent ES cell lines (#3, #22) were crossed with C57BL/6J mice, and the resulting knock-in mouse lines were maintained with the ICR background. Confirmation that the ACN cassette had been removed and genotype determination were performed by PCR with primers that produced both knock-in and wild-type bands: Foxa2 genotype F (fF, 5’-CCAGGCCTATTATGAACTCATCC-3’), Foxa2 genotype R (fR, 5’-ACACAGACAGGTGAGACTGCTC-3’), ERT2 genotype F (eF, 5’-GGGCTCTACTTCATCGCATTCC-3’). The positions of these primers are indicated in Fig. 1b. A 166-bp product is generated from the wild-type allele with fF and fR, and a 270-bp product is generated from the knock-in allele with eF and fR. The PCR conditions were 95 °C for 1 min, 30 cycles of 95 °C for 30 seconds, 58 °C for 30 seconds, 72 °C for 1 min, followed by 72 °C for 5 min.
The TnEGFP-CreERT2/+ (Acc. No. CDB0604K: http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) mouse was generated at the RIKEN CDB by homologous recombination in ES cells using essentially the same strategy as that of Foxa2nEGFP-CreERT2/+. Only the points that differ from the Foxa2 locus are described below. The C57BL/6J mouse BAC genomic DNA clone RP23-99E8, which contains the T gene, was used to create the T targeting vector. To screen for homologous recombination in the T locus, the following primer pair was used: SVloxP-3’-F2 and T-PC-3’-R1 (tR1, 5’-TGGAGTCTCCAAGAGGACAGGATTCC-3’). The positions of these primers are indicated in Fig. 2a. Among the 96 clones examined using PCR, 77 clones showed positive results. Among the PCR-positive clones, 11 clones were analyzed further by using Southern hybridization, and among these, 10 clones were confirmed as correct homologous recombinants. Chimeric founders were generated from three independent ES cell lines (#36, #41, #44). Genotype determination and confirmation that the ACN cassette had been removed were performed by PCR with following primers: T genotype F (tF, 5’-TATCCCAGTCTCTGGTCTGTGA-3’), T genotype R (tR, 5’-TAGGACCCTACCTAGCAAAGGA-3’), ERT2 genotype F (eF). The positions of these primers are indicated in Fig. 2a. A 550-bp product is generated from the wild-type allele with tF and tR, and a 400-bp product is generated from the knock-in allele with eF and tR.
The Cre reporter mouse line, Gt(ROSA)26Sortm1Sor (described as ROSA26RLacZ in this paper) (Soriano, 1999) was obtained from the Jackson Laboratory.
Mice were housed in environmentally controlled rooms of the Laboratory Animal Housing Facility of RIKEN CDB and the Center for Animal Resources and Development (CARD) of Kumamoto University. All experiments have been carried out following the regulations for animal and recombinant DNA experiments in the RIKEN CDB and Kumamoto University and the laws and notifications of the Japanese government.
Both Foxa2nEGFP-CreERT2/+ (Acc. No. CDB0601K) and TnEGFP-CreERT2/+ (Acc. No. CDB0604K) mice will be available for non-profit scientific research from the Laboratory for Animal Resources and Genetic Engineering, RIKEN CDB http://www.cdb.riken.jp/arg/mutant%20mice%20list.html).
Induction of Cre recombinase activity
To induce Cre recombinase activity in embryos, pregnant mice were given tamoxifen (Sigma, T5648) dissolved in peanut oil (10 mg/ml) at 0.12 mg/g body weight, by oral gavage, as described previously (Park et al., 2008).
β-galactosidase staining of embryos
Whole-mount staining of embryos for β-galactosidase activity, and paraffin sectioning of the stained embryos were performed as previously described (Wurst and Gossler, 2000).
Imaging
Images of the whole-mount embryos and sections were acquired with a Leica MZ16 or an Axioplan2 (Zeiss) microscope equipped with an AxioCam HRc (Zeiss). For some images of whole-mount embryos, all-focal-levels images were generated by merging multiple images from different focal planes using Dynamic Eye REAL software (Mitani Corporation, Japan). Confocal images of whole-mount embryos were acquired with a Nikon AZ-C1 macro-confocal microscope.
Time-lapse confocal images of live embryos were acquired with an Olympus IX81N-ZDC equipped with an incubator unit CU109 (Live Cell Instrument), a 488 nm diode laser (Olympus), a spinning disk confocal unit CSU-X1 (Yokogawa) and an EMCCD camera iXon+ DU897E (Andor). The stage and the optics carrier of the microscope were also enclosed by a custom made environmental chamber. The entire imaging system was controlled using MetaMorph software (Molecular Devices). To acquire images around the node and the notochord, embryos were immobilized using modified culture dishes as described previously (Yamanaka et al., 2007). Images were acquired every 10 minutes, with 14 (for Foxa2nEGFP-CreERT2/+) or 11 (for TnEGFP-CreERT2/+) Z-positions at each time point, and the section interval was 8 µm. Drift in embryo positioning during imaging was corrected using the iSEMS program (Kato and Hayashi, 2008). Z-stack images were generated by using MetaMorph, and analysis of cell movements was performed using Image J.
Supplementary Material
Acknowledgments
We thank Ms. H. Sato, K. Ukita, and M. Shibata for excellent technical assistances; the Laboratory for Animal Resources and Genetic Engineering of RIKEN CDB for generating the mutant mice and housing the mice; the CARD of Kumamoto Univ. for housing the mice; Drs. K. Kato and S. Hayashi for help with the iSEMS program; Dr. Y. Saijoh for advice on Cre induction by Tamoxifen administration; Dr. H. Enomoto for advice on knock-in strategy and plasmids; Dr. I. Matsuo for sharing mice; Dr. N. Copeland for the recombineering system; Dr. K.R. Thomas for the ACN cassette plasmid; and Dr. P. Chambon for the CreERT2 plasmid. These studies were supported by grants from RIKEN and the Uehara Memorial Foundation to H.S., and by Grants-in-Aid for Scientific Research (KAKENHI) from MEXT (21116003) and JSPS (23247036) to H.S., and by a National Institutes of Health grant (HD30284) to R. R. B.
References
- Besnard V, Wert SE, Hull WM, Whitsett JA. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr Patterns. 2004;5:193–208. doi: 10.1016/j.modgep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes & development. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray N, Wilson V. Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development (Cambridge, England) 2002;129:4855–4866. doi: 10.1242/dev.129.20.4855. [DOI] [PubMed] [Google Scholar]
- Chesley P. Development of the short-tailed mutant in the house mouse. J Exp Zool. 1935;70:429–459. [Google Scholar]
- Cleaver O, Krieg PA. Notochord patterning of the endoderm. Developmental biology. 2001;234:1–12. doi: 10.1006/dbio.2001.0214. [DOI] [PubMed] [Google Scholar]
- Davidson BP, Tam PP. The node of the mouse embryo. Curr Biol. 2000;10:R617–R619. doi: 10.1016/s0960-9822(00)00675-8. [DOI] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Elliott SA, Park EJ, Hammond J, Saijoh Y, Moon AM. System for inducible expression of cre-recombinase from the Foxa2 locus in endoderm, notochord, and floor plate. Dev Dyn. 2007;236:1085–1092. doi: 10.1002/dvdy.21093. [DOI] [PubMed] [Google Scholar]
- Herrmann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development (Cambridge, England) 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- Inman KE, Downs KM. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene Expr Patterns. 2006;6:783–793. doi: 10.1016/j.modgep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Kato K, Hayashi S. Practical guide of live imaging for developmental biologists. Dev Growth Differ. 2008;50:381–390. doi: 10.1111/j.1440-169X.2008.01029.x. [DOI] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Wakamiya M, Sasaki H, Behringer RR, Nagy A, Tam PP. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development (Cambridge, England) 2001;128:3623–3634. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- Kispert A, Herrmann BG. Immunohistochemical analysis of the Brachyury protein in wild-type and mutant mouse embryos. Developmental biology. 1994;161:179–193. doi: 10.1006/dbio.1994.1019. [DOI] [PubMed] [Google Scholar]
- Liu C, Nakamura E, Knezevic V, Hunter S, Thompson K, Mackem S. A role for the mesenchymal T-box gene Brachyury in AER formation during limb development. Development (Cambridge, England) 2003a;130:1327–1337. doi: 10.1242/dev.00354. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003b;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Borchers AG, Jolicoeur C, Rayburn H, Baker JC, Tessier-Lavigne M. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430:93–98. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kaestner KH, Grau E, Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development (Cambridge, England) 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Regionally specific induction by the Spemann-Mangold organizer. Nature reviews. 2004;5:425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mechanisms of development. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Palena C, Polev DE, Tsang KY, Fernando RI, Litzinger M, Krukovskaya LL, Baranova AV, Kozlov AP, Schlom J. The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell-mediated cancer immunotherapy. Clin Cancer Res. 2007;13:2471–2478. doi: 10.1158/1078-0432.CCR-06-2353. [DOI] [PubMed] [Google Scholar]
- Park EJ, Sun X, Nichol P, Saijoh Y, Martin JF, Moon AM. System for tamoxifen-inducible expression of cre-recombinase from the Foxa2 locus in mice. Dev Dyn. 2008;237:447–453. doi: 10.1002/dvdy.21415. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Shields B, Davies ML, Muller J, Wakeman JA. BRACHYURY confers cancer stem cell characteristics on colorectal cancer cells. Int J Cancer. 2012;130:328–337. doi: 10.1002/ijc.26029. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development (Cambridge, England) 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Molecular and cellular biology. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Vignali DA. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert opinion on biological therapy. 2005;5:627–638. doi: 10.1517/14712598.5.5.627. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nature biotechnology. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mechanisms of development. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Tam PP, Steiner KA, Zhou SX, Quinlan GA. Lineage and functional analyses of the mouse organizer. Cold Spring Harb Symp Quant Biol. 1997;62:135–144. [PubMed] [Google Scholar]
- Ukita K, Hirahara S, Oshima N, Imuta Y, Yoshimoto A, Jang CW, Oginuma M, Saga Y, Behringer RR, Kondoh H, et al. Wnt signaling maintains the notochord fate for progenitor cells and supports the posterior extension of the notochord. Mechanisms of development. 2009 doi: 10.1016/j.mod.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hamblet NS, Mark S, Dickinson ME, Brinkman BC, Segil N, Fraser SE, Chen P, Wallingford JB, Wynshaw-Boris A. Dishevelled genes mediate a conserved mammalian PCP pathway to regulate convergent extension during neurulation. Development (Cambridge, England) 2006;133:1767–1778. doi: 10.1242/dev.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Wurst W, Gossler A. Gene trap strategies in ES cells. In: Joyner AL, editor. Gene Targeting A Practical Approach. New York: Oxford University Press Inc; 2000. [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Analytical biochemistry. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Developmental cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.