Abstract
Nucleostemin 3 (NS3) is an evolutionarily conserved protein with profound roles in cell growth and viability. Here we analyze cell-autonomous and non-cell-autonomous growth control roles of NS3 in Drosophila and demonstrate its GTPase activity using genetic and biochemical assays. Two null alleles of ns3, and RNAi, demonstrate the necessity of NS3 for cell autonomous growth. A hypomorphic allele highlights the hypersensitivity of neurons to lowered NS3 function. We propose that NS3 is the functional ortholog of yeast and human Lsg1, which promotes release of the nuclear export adapter from the large ribosomal subunit. Release of the adapter and its recycling to the nucleus are essential for sustained production of ribosomes. The ribosome biogenesis role of NS3 is essential for proper rates of translation in all tissues and is necessary for functions of growth-promoting neurons.
Keywords: Nucleostemin, ribosome biogenesis, GTPase, dopamine, serotonin
THE rate and fidelity of protein synthesis in a cell depend upon the proper assembly of the ribosome. Ribosome biogenesis begins in the nucleolus with the synthesis of ribosomal RNA (rRNA) and culminates in the creation of a fully functional ribosome competent to initiate protein synthesis in the cytoplasm. This process is impressively intricate, requiring ∼200 factors acting at various levels. Major ribosome biogenesis steps include directing rRNA post-transcriptional modification, 90S cleavage, folding/assembly of the small (40S) and large (60S) ribosomal subunits, and proper transport from the nucleus to cytoplasm where final maturation occurs (Zemp and Kutay 2007; Henras et al. 2008; Staley and Woolford 2009; Strunk and Karbstein 2009; Panse and Johnson 2010).
Export of pre-ribosomal subunits from the nucleus to the cytoplasm requires movement through the nuclear pore complex (Hurt et al. 1999; Moy and Silver 1999; Stage-Zimmermann et al. 2000; Seiser et al. 2006). The pre-60S subunit achieves nuclear export by recruiting nuclear export factors, including the nuclear export signal (NES)-bearing protein Nmd3 (Ho et al. 2000; Gadal et al. 2001). Equally essential is recycling of Nmd3 from the cytoplasm back to the nucleus for subsequent rounds of pre-60S export. The release of Nmd3 appears to be the last step of a highly ordered pathway of ribosome maturation in the cytoplasm (Lo et al. 2010). Studies in Saccharomyces cerevisiae implicated RpL10p and Lsg1p as factors critical for Nmd3 recycling. Loss of either RpL10p or Lsg1p caused accumulation of Nmd3 in the cytoplasm, which precluded its ability to return to the nucleus for additional rounds of pre-60S export (Hedges et al. 2005). Thus, the loss of Nmd3p, RpL10p, or Lsg1p through mutation causes 60S subunit deficiency in the cytoplasm.
In Drosophila, a well-known class of mutations, called Minutes affects ribosome assembly. Minutes have delayed development and small or “minute” bristles; mutations affecting 66 of the 88 predicted ribosomal proteins have this phenotype (Marygold et al. 2007). The first Minute gene was discovered by Calvin Bridges and T. H. Morgan in 1919 (Bridges and Morgan 1923). This allele of RpS8 was the founding member of what came to be a large class of genes that are often haplo-insufficient. Later, with the advent of molecular biology, it became clear that most Minute genes code for ribosomal proteins. The conclusion was that the normal rate of progression through larval development and normal bristle synthesis are highly sensitive to protein synthesis rates (Lambertsson 1998).
Loss-of-function alleles for different ribosomal protein genes can have distinctive phenotypes in addition to the shared classical Minute phenotypes. For instance, loss of RpS6, a small subunit protein, causes a counterintuitive phenotype: extensive larval overgrowth (Lin et al. 2011). Studies to determine which tissues normally require RpS6 to restrict growth showed that prothoracic glands of RpS6 mutants do not function properly. The consequent lower level of the hormone ecdysone, which promotes larval developmental progression and molts, extends the period of larval development and feeding. RpS6 mutant larvae feed longer and grow abnormally large (Lin et al. 2011). It is unclear whether the prothoracic gland is particularly sensitive to the skewed stoichiometry of ribosomal proteins in RpS6 mutants or whether RpS6 has a more specific function in this tissue, such as regulation of ecdysone synthesis directly. A specific role would be consistent with the recently proposed view that the ribosome is not a uniform constitutively active complex. Instead ribosomes are composed of variable subunits that facilitate unique translation regulation of mRNA subsets (Xue and Barna 2012). In support of this concept, mice mutant for a large subunit protein, RpL38, have specific homeotic transformations due to failed translation of, and 80S association with, a certain subset of Hox gene mRNAs (Kondrashov et al. 2011).
Here we report detailed genetic and biochemical studies of another protein that regulates ribosome assembly and has a distinctive phenotype beyond reduced growth at the cell-autonomous level. This study follows from our previous characterization of flies carrying an allele of Drosophila Nucleostemin 3 (ns3G0431) (Kaplan et al. 2008). These flies are developmentally delayed during larval stages, have partial recessive lethality, eclose late, and are small. Nucleostemin 3 (NS3) is closest in sequence to yeast Lsg1p, the protein necessary for recycling the 60S export protein Nmd3 to the nucleus (Kallstrom et al. 2003; Hedges et al. 2005). It therefore seemed reasonable that the ns3 loss-of-function phenotypes are due to an overall reduction in protein synthesis like a typical Minute. However, we found that homozygous mutant clones of ns3 were not small, indicating non-cell autonomous rescue of mutant cells by cells elsewhere that have functional NS3. Remarkably, expression of ns3-YFP in neurons using a elav-gal4 driver was capable of nearly completely rescuing the phenotype (Kaplan et al. 2008). Here we show that two newly analyzed larval lethal alleles of l(1)2Ad are strong loss-of-function alleles of ns3. The lethality of these alleles, and ns3-RNAi studies, indicates additional cell-autonomous growth roles of NS3. The hypomorphic allele ns3G0431 reveals critical growth-promoting roles of neurons. We conclude that ns3 has both cell-autonomous and non-cell-autonomous growth-promoting roles.
Materials and Methods
Percentage protein similarity and phylogenetic analyses
Percentage protein similarity over the entire length of individual fly NS members vs. S. cerevisiae Lsg1p was determined using EMBOSS Stretcher. A phylogenetic tree of fly NS members and yeast and human Lsg1p proteins was generated by ClustalW2 (Larkin et al. 2007). The distance value of each branch shows the number of substitutions as a proportion of the length of the alignment (gaps excluded).
Yeast rescue experiments
NS3 DNA was amplified by PCR from UAST–NS3–YFP using oligos: 5-GCCACTAGTACCATGGGCAAAAAGAACAAGGG and 5′-TTTTAAGCTTCTAGTTAATTAAGTGCTCGTCCAGGTGCGAGA for cloning into pRS415–GPD or 5′-GGCACTAGTAAAATGGGCAAAAAGAACAAGGGC and 5′- GCCAAGCTTCAGTGCTCGTCCAGGTGCG for cloning into pRS425–GPD. NS1 DNA was PCR amplified from complete cDNA AT23067 using primers 5′-GGCACTAGTAAAATGGCTTTAAAAAGGTTGAAGACC and 5′-GCCAAGCTTATTCAATCACATAGTCC. NS2 DNA was PCR amplified from complete cDNA SD10213 with primers 5′-GGCACTAGTAAAATGCCAAAGGTACGCAGCACC and 5′-GCCAAGCTTACGCGTCCTTCTTTTTGTTTCTATTGC. NS4 DNA was PCR amplified from complete cDNA LD10773 with primers 5′-GGCACTAGTAAAATGCCACAGCAACGGCGC and 5′-GCCAAGCTTAATCGTCCTCCAGCAGAGC. The products were digested with SpeI and HindIII and cloned into pRS415–GPD or pRS425–GPD (Mumberg et al. 1995). The various NS clones, wild-type yeast LSG1, or empty vector was transformed into AJY1171 [MATalpha lsg1Δ::KanMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 containing plasmid pAJ626 (LSG1 URA3 CEN)]. Transformants were selected on Leu− medium and restreaked onto Leu− medium containing 5-FOA and incubated for 5 days at 30°.
Drosophila culture and strains utilized
Drosophila were cultured on standard molasses media at 25° except where indicated. The following fly lines were acquired from the Bloomington Drosophila Stock Center: ddc-gal4 (Li et al. 2000), ple/th-gal4 (Friggi-Grelin et al. 2003), l(1)2Ad1, l(1)2Ad3, Df(1)BSC719, ns3G0431, P{GAL4-Act5C.FRT.P}, P{UAS-mCD8::GFP.L}LL4, P{hsFLP}22; Pin[1]/CyO, c061-gal4, and uas-lamin-gfp. The dtrh-gal4 was a gift from Dr. Ed Kravitz (Alekseyenko et al. 2010). The isoD1 line was a gift from Drs. Daryl Gohl and Tom Clandinin (Gohl et al. 2011).
Generation of expression vectors and transgenic flies
For the K350N antimorphic version of ns3, a Gateway entry clone pDONR207 (Invitrogen) containing ns3 was mutated with point mutations (K to N) using a QuikChange XL site-directed mutagenesis kit (Stratagene). LSG1 cDNA was amplified from vector pAJ2229 (Arlen Johnson) and cloned into entry clone pDONR207. Ns3(K350N) and LSG1 were then cloned into pTVW and pTW, respectively, using site-specific recombination according to the manufacturer’s Gateway cloning protocol. The resulting expression vectors were injected into yw embryos to generate transgenic fly lines.
Fly weight, developmental time to eclosion, and percentage viability studies
The Gal4 drivers (dtrh-gal4, ddc-gal4, th-gal4) were all backcrossed five times with the isogenic line isoD1 to reduce background genetic effects (Gohl et al. 2011), and a recombinant dtrh-gal4, th-gal4 line was subsequently created. Males of this strain were crossed to virgin females of the genotype ns3G0431/FM7i, Act > GFP; uas-ns3-YFP for 6–8 hr in collection bottles, where embryos were laid on standard molasses/agar plates. The embryos were aged for 24 hr at 29° until hatching, then 40 non-GFP (ns3/Y and ns3/+) larvae were transferred to vials containing standard molasses medium, in triplicate. Vials were placed at 29° and the number of flies eclosing daily was scored. Fly weight measurements were carried out as previously described (Kaplan et al. 2008). Survival to adulthood of ns3G0431/Y males with and without expression of ns3-yfp in DA, serotonergic (5-HT), and DA plus 5-HT neurons was quantified by dividing the total ns3G0431/Y male flies that eclosed over the number expected to eclose (40, the number of ns3G0431/+ females eclosed) from three replicates of vials that carried 40 ns3G0431/Y and ns3G0431/+ larvae.
Ns3 allele molecular mapping
ns3l(1)2Ad1 is an allele containing a deletion of 8 bp in the second exon (the deleted nucleotides are flanked by dashes: TACG-TAAAGGAG-GTGG). This is predicted to result in a truncation after 198 of the 606 amino acids (33%), followed by three missense amino acids and an early stop. The truncated protein would not contain the putative GTP-binding domain (as predicted by InterPro Scan), which includes amino acids 349–396. The ns3l(1)2Ad3 allele has a deletion of 46 bp in the second exon (the deleted nucleotides are flanked by dashes: GCCG-AGGAGCTAGTGCGAGCCGAGAACGAGGCATTTCTCGACTGGCGCCG-CGACCTGGCG). This may result in a truncation after 126 of the 606 amino acids (21%), followed by 12 missense amino acids and an early stop. This predicted truncated protein would not contain the putative GTP-binding domain. ns3G0431 is an insertion of P{lacW} into the 5′-UTR (insertion exists in the following sequence at the site of the slash: TGATTGTCAAAAACACGTGCAATGTTTGCCC/G).
Creation of recombinant NS3–maltose-binding protein and GTPase assay
Full-length ns3 was cloned into a pMal–p5X vector that contains a maltose-binding protein (MBP) tag. MBP-tagged NS3 protein (NS3–MBP) was expressed in BL21 codon+DE competent cells and purified under the following conditions. The culture was grown at 37° in LB amp supplemented with glucose until an OD600 nm of 0.5 was reached. The flask was then cooled to 16° and induced with 0.1 mM IPTG overnight. The cells were pelleted and resuspended in 25 ml column buffer [20 mM Hepes, 200 mM NaCl, 1 mM EDTA, protease inhibitors (Sigma P8465), and 1 mM DTT]. Cell lysis was achieved by the addition of lysozyme (0.2 mg/ml) for 30-min on ice. The lysate was then sonicated, Triton X-100 was added to a final concentration of 1%, and the lysate was centrifuged for 30 min at 48,384 × g at 4°. The supernatant was then applied to a column containing 1 ml of Amylose Resin (New England BioLabs) and gently rotated overnight at 4°. The supernatant was then allowed to flow through the column (Kontes flex column 1 × 10 cm) by gravity, the amylose NS3–MBP carrying resin bed was washed with column buffer four times, and NS3–MBP was eluted with multiple 500-μl aliquots of column buffer containing 10 mM maltose. The various elutions were tested for the presence of NS3–MBP via SDS–PAGE analysis, and the elutions containing NS3–MBP were pooled. Two major bands were seen, 112 (NS3–MBP) and 42 kDa (MBP). Two fractionation steps were then applied. First, an Amicon Ultra100 was used to separate NS3–MBP (112 kDa) from MBP (42 kDa). Second, the <100-kDa flow-through from the Amicon Ultra100 fraction was further purified with an Amicon Ultra30. The ≥100-kDa, 112-kDa NS3–MBP containing retentate, from fractionation 1 and the ≥30-kDa, 42-kDa MBP containing retentate from frationation 2 were used for the GTPase assay. NS3–MBP and MBP concentrations were measured by Bradford protein assay. The GTPase activities of NS3–MBP and MBP were determined using Innova Biosciences GTPase Kit following the manufacturer’s protocol. A standard curve was made using Pi standards and was used for calculation of amount of released Pi and GTPase activity.
NS3 antibody creation, purification, and Western analysis
Two rabbits were immunized against the C terminus of NS3 (amino acids 306–606) (Josman Labs). Raw serum from the rabbit recognizing the 70-kDa NS3 via Western analysis was then purified with a NS3–MBP bound column (AminoLink Plus Immobilization Kit, Pierce) and subsequently used for the immunolocalization (1:250) and Western analyses herein (1:250). For Western analyses, standard procedures were applied using embryonic protein extracts obtained via microcentrifuge tube–pestle homogenization in PIPES buffer [80 mM PIPES, pH 7.0, 5 mM EGTA, 5 mM MgCl2, and a tablet of protease inhibitors (Roche, EDTA free) at 1× strength] on ice. Samples were then centrifuged at maximum speed at 4° and supernatant was transferred to a new tube and kept on ice or stored at −80° until preparation for SDS–PAGE.
Microscopy and image analysis
Larval CNS, salivary glands, and imaginal discs were prepared for imaging as previously described with PBS containing 0.1% Triton X-100 (PBT), overnight incubations in primary antibody, and 4-hr incubations in secondary antibody at room temperature (Wu and Luo 2006). F-actin staining in Supporting Information, Figure S2 was carried out by incubating tissues in AlexaFluor 594 conjugated Phalloidin (Invitrogen) at a 1:40 concentration for 20 min in the first PBT wash following secondary antibody incubation. The following primary antibodies were employed for these studies: anti-RpL13 (Abcam), anti-RpS6 (Cell Signaling), and anti-Tyrosine Hydroxylase (ImmunoStar). All images were made on a confocal Leica TCS SP2 laser-scanning microscope. The average pixel intensity of RpL13, RpS6, and NS3 (Figure 5 and Figure S2) was obtained using image J (Schneider et al. 2012). To quantify cytoplasmic RpL13 and RpS6 levels, ellipses were manually drawn proximal, but external to, nuclei as defined by DAPI or lamin–GFP. Ellipses were then converted to curved lines with the “convert region to line” plugin and average pixel intensities for ribosomal proteins under the curved line were measured. The stochastic expression of the c061-gal4 driver in the salivary gland made it possible to compare the outcome of ns3 knockdown to neighboring “wild-type” cells that did not produce Gal4. Each c061-gal4 salivary gland typically had 2–10 cells that did not express Gal4. Gal4/ns3-dsRNA-expressing cells were identified by the presence of lamin–GFP. The average pixel intensity of RpL13 and RpS6 in wild-type cells not expressing ns3-dsRNA (non-lamin–GFP) was normalized by the average pixel intensity of ribosomal protein staining in neighboring ns3-dsRNA (lamin–GFP-positive) cells. Equivalent measurements were carried out in control salivary glands where c061-gal4 drove expression of only uas-lamin-gfp. Quantification of the level of NS3 in DA neurons of wild-type vs. ns3G0431 (Figure S3) was carried out by manually drawing four lines in the cytoplasm of cells carrying the DA marker tyrosine hydroxylase (TH) and then measuring the average pixel intensity of NS3 staining. This value was normalized by dividing it by the average pixel intensity of NS3 in surrounding, TH−, cells. These control values were obtained by manually drawing four boxes in surrounding brain tissue and measuring average pixel intensity.
Figure 5.
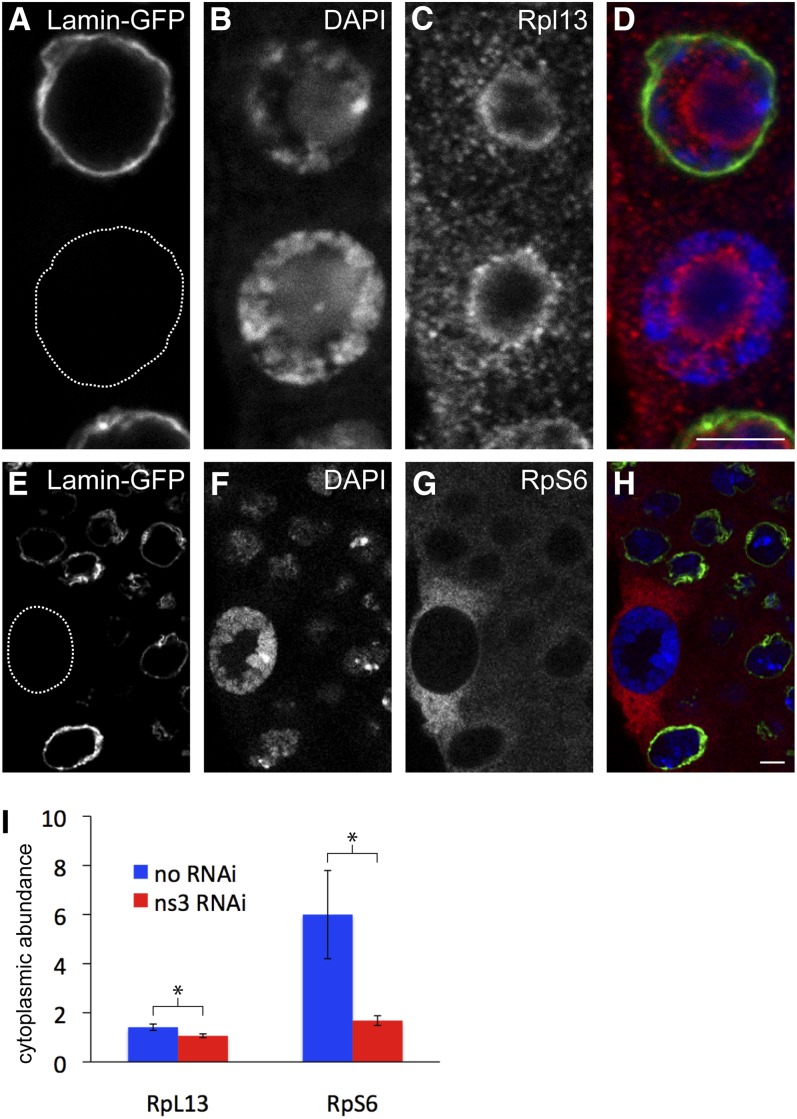
NS3 RNAi reduces the cytoplasmic levels of RpL13 and RpS6 in the salivary glands. (A–D) Anti-RpL13 staining of wild-type and ns3-dsRNA-expressing salivary gland cells. The variably active salivary gland driver c061-gal4 was used to co-express lamin-gfp and ns3-dsRNA. We observed the consequences for RpL13 localization relative to nearby non-lamin-gfp/non-ns3-dsRNA expressing cells. (A) Lamin–GFP marks a nucleus from a cell expressing ns3-dsRNA and the dotted circle highlights a nucleus from a non-ns3-dsRNA-expressing cell. (B) DAPI staining to visualize DNA. (C) Anti-RpL13 staining indicates that RpL13 levels are decreased in the cytoplasm of cells expressing ns3-dsRNA. (D) Merged images of A–C. (E–H) Anti-RpS6 staining of c061 > lamin-gfp, ns3-dsRNA salivary glands. (E) Lamin–GFP staining indicates ns3-dsRNA expressing cells, and the dotted circle highlights a non-ns3-dsRNA expressing cell. (F) DAPI to visualize DNA. (G) Anti-RpS6 staining indicates that RpS6 levels are reduced in the cytoplasm of ns3-dsRNA-expressing cells. (H) Merged images of E–G. (I) Levels of cytoplasmic RpL13 and RpS6 in non-lamin-gfp/non-ns3-dsRNA were quantified with imageJ and normalized to neighboring lamin-gfp/ns3-dsRNA-expressing cells at the same focal plane. Levels of RpL13 and RpS6 are reduced in the cytoplasm after ns3 RNAi. Scale bars, 5 µm.
Results
NS3 is the functional ortholog of yeast Lsg1
In eukaryotic evolution, a clade of GTPases expanded from an ancient prokaryotic protein that resembled modern YlqF, an essential GTPase that promotes ribosome assembly (Leipe et al. 2002; Reynaud et al. 2005; Uicker et al. 2006). Most members of the YlqF-related GTPase (YRG) family have clear roles in ribosome biogenesis, particularly in the maturation and movement of ribosome subunits from nucleoli to the cytoplasm (Table 1) (Reynaud et al. 2005). The expansion of several ribosome biogenesis factors during eukaryotic evolution has evidently facilitated the compartmentalization of ribosome biogenesis steps in the modern eukaryotic cell (Reynaud et al. 2005).
Table 1. YRG family members in yeast, fly and human with documented or likely roles in ribosomal biogenesis.
| YRG member | Implication in growth and/or role in ribosome assembly |
|---|---|
| Nug1 (S.c.) | Nuclear export of 60S ribosomal subunits (Bassler et al. 2001; Bassler et al. 2006) |
| NS1 (D.m.) | Nucleolar export of large ribosomal subunits (Rosby et al. 2009) |
| GNL3L (H.s.) | Nucleolar pre-rRNA processing (Du et al. 2006; Rosby et al. 2009; Yasumoto et al. 2007; Zhu et al. 2009) |
| GNL3/NS (H.s.) | 60S pre-rRNA processing (Cladiere et al. 2006; Tsai and McKay 2002) |
| Nog2/Ngp1 (S.c.) | 60S export from nucleolus to nucleoplasm (Saveanu et al. 2001) |
| NS2 (D.m.) | Decreased function alters ratio of rRNA in nuclear vs. cytoplasm (Matsuo et al. 2011) |
| GNL2/Ngp1 (H.s.) | nucleolar localization (Shin et al. 2004) |
| Lsg1 (S.c.) | 60S nuclear export to cytoplasm (Hedges et al. 2005; Kallstrom et al. 2003) |
| NS3 (D.m.) | Growth, rescues yeast lsg1 null mutant (Kaplan et al. 2008) |
| hLsg1 (H.s.) | Cell growth and viability (Reynaud et al. 2005) |
| NS4 (D.m.) | No phenotype found for putative allele (Kaplan et al. 2008) |
| GNL1 (H.s.) | Shuttles between nucleolus to nucleoplasm; controls growth (Boddapati et al. 2012) |
| Mtg1 (S.c.) | Mitochondrial translation (Barrientos et al. 2003) |
| CG17141 (D.m.) | No known roles |
| Mtg1 (H.s.) | Rescues yeast mtg1 null mutant (Barrientos et al. 2003) |
The YRG family members expanded in eukarya from a single ancestor resembling modern prokaryotic YlqF. All family members carry a circularly permuted GTPase domain and most, if not all, have specific roles in ribosomal biogenesis.
The S. cerevisiae YRG member Lsg1p is a cytoplasmic GTPase that is associated with nascent 60S subunits (Figure 1A). An apparent gene duplication of LSG1 in coelomate evolution created one paralog that has the highest similarity to LSG1 and another that diverged in sequence (Reynaud et al. 2005). Based on protein sequence similarity, the likely ortholog of Lsg1p in Drosophila is nucleostemin 3 (53.1% similar), with nucleostemin 4 (40.6% similar) being a divergent paralog (Table 1 and Figure 1A).
Figure 1.
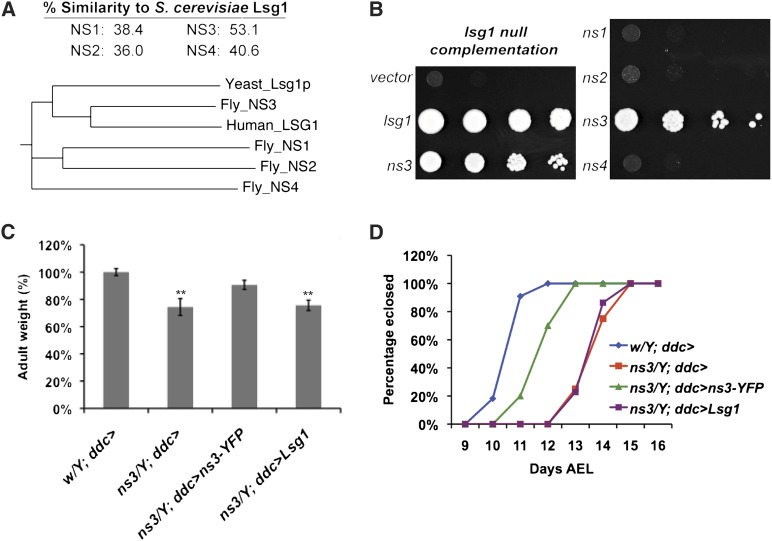
Drosophila NS3 partially rescues yeast lsg1Δ lethality. (A) Percentage similarity and phylogenetic tree of the Drosophila NS family. Of the four Drosophila NS family members, NS3 has the strongest protein sequence similarity to yeast Lsg1p (EMBOSS stretcher). A multiple alignment and phylogram tree of the Drosophila NS family, yeast Lsg1p, and human Lsg1 were generated by ClustalW2 (Larkin et al. 2007). The distance value of each branch shows the number of substitutions as a proportion of the length of the alignment (gaps excluded). Sequence homology of NS3 with yeast and human Lsg1 leads to their clustering within the phylogeny. (B) Yeast lsg1Δ cells were complemented by transformation of LSG1 and Drosophila ns3, but not by transformation with Drosophila ns1, ns2, or ns4. (C) LSG1 driven by the ddc-gal4 driver did not rescue the decreased body weight of the ns3G0431 mutant. The average adult male weight, derived from three groups of 10 flies, is expressed as a percentage of the average weight of negative control flies that carried only ddc-gal4. Expression of uas-ns3-yfp with ddc-gal4 rescued the ns3G0431 mutant phenotype to the size of wild-type flies, but expression of uas-LSG1 did not. (D) LSG1 driven by the ddc-gal4 driver did not rescue the delayed development of the ns3G0431 mutant. The number of eclosed flies was scored daily. The number of ns3G0431 males eclosed per day is plotted as the percentage of the total ns3G0431 males eclosed. Expression of uas-ns3-yfp with ddc-gal4 partially rescued the developmental delay of the ns3G0431 mutant so that eclosion occurred closer to the time of negative-control wild-type flies that carried only ddc-gal4. Expression of uas-LSG1 using ddc-gal4 did not rescue ns3G0431 developmental delay; these flies were as developmentally delayed as ns3G0431/Y; ddc-gal4 flies.
To further validate that NS3 is the functional ortholog of Lsg1p, we transformed LSG1, Drosophila nucleostemins 1-4, or empty vector into the lsg1Δ yeast mutant containing a LSG1 URA3 vector. The LSG1 URA3 vector was eliminated on 5-FOA-containing medium, which is toxic to cells containing a URA3 gene. Strains transformed with ns3 partially rescued lsg1Δ lethality. In contrast, ns1, ns2, and ns4 did not rescue lethality of lsg1Δ (Figure 1B). This suggests that NS3 is a functional ortholog of Lsg1p.
Yeast LSG1 expression was unable to rescue mutant ns3 flies. We previously demonstrated that fly ns3 mutants (ns3G0431) could be rescued by the expression of wild-type ns3-yfp using the ddc-gal4 driver (Kaplan et al. 2008). Expression of LSG1 with this same driver was not capable of rescuing the ns3 decreased fly size (Figure 1C) or the ns3 developmental delay (Figure 1D). We suggest that NS3 acquired unique features during metazoan evolution that preclude Lsg1p from rescuing an NS3 deficiency. This might have entailed alterations that enable unique protein localization properties, as discussed below.
Alleles of l(1)2Ad are strong loss-of-function alleles of ns3 and are larval lethal
The ns3G0431 mutation causes phenotypes common to the Minute phenotypic class: developmental delay, small cell size, and small adults (Lambertsson 1998; Marygold et al. 2007). The ns3G0431 fly is not a typical Minute as its growth defects are non-cell autonomous and caused by defective neuronal signaling (Kaplan et al. 2008). However, a purely non-cell-autonomous growth-control function of NS3 is unlikely based on the following. First, yeast and human orthologs of NS3 have strong effects on cell growth and viability (Hedges et al. 2005; Reynaud et al. 2005). Second, expression of a shRNA targeting ns3 either ubiquitously (tubulin-gal4; 100% lethal) or in neurons (elav-gal4; 100% lethal) causes lethality exceeding that of the ns3G0431 mutant, which has “escapers” that make it to adulthood.
To clarify the roles of ns3, we sought stronger mutant alleles. FlyBase lists several genetic loci that were mapped genetically or cytologically, but not yet to a nucleotide coordinate. Ns3 falls within cytological band 2A4. In a search of genetic aberrations listed on FlyBase that are near or within cytological band 2A4, we acquired 19 candidate lines from the Bloomington Stock Center with lesions possibly falling within the ns3 locus.
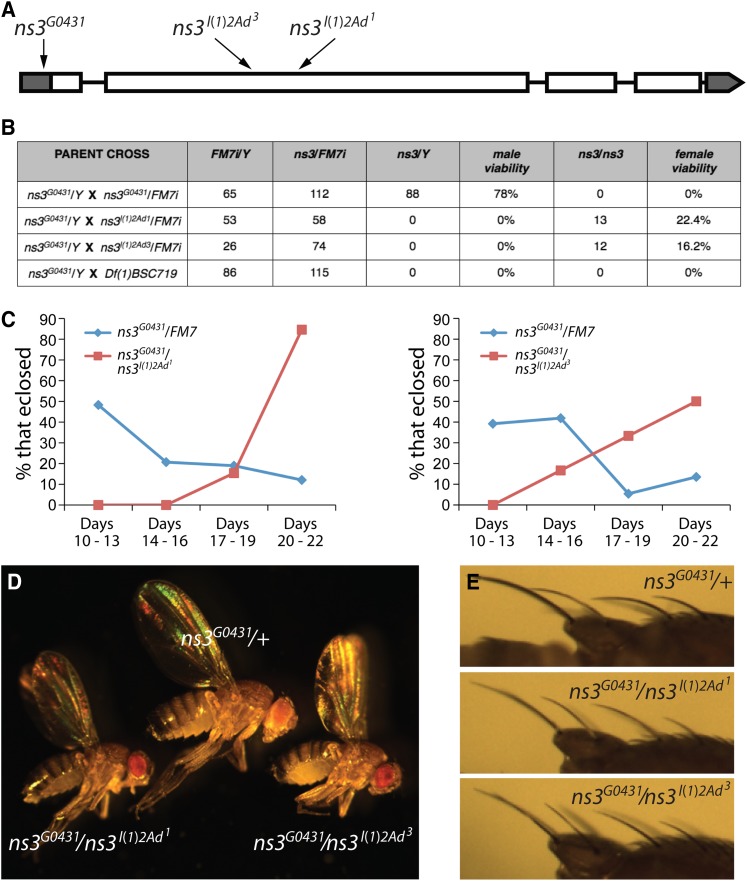
We performed complementation tests with the various candidate alleles. ns3G0431/Y males were crossed to candidate females carrying an X chromosome mutation, often balanced over FM7. Since homozygous female ns3G0431 mutants are lethal and developmentally delayed, we first looked for complementation by virtue of the presence of ns3G0431/X females among early eclosing flies. We then looked for a failure to complement, deduced from the absence of ns3G0431/X females until several days after their siblings had eclosed. These studies revealed that l(1)2Ad1 and l(1)2Ad3 are alleles of ns3. Sequencing the three alleles of ns3 identified the ns3l(1)2Ad1 lesion as an 8-bp deletion in the second exon. This causes a frameshift and the introduction of an early stop codon, possibly creating a protein abnormally shortened by 33%. The ns3l(1)2Ad3 lesion is a 46-bp deletion in the second exon, also causing a frameshift. This allele is expected to make a truncated protein only 22% the length of wild type. Neither ns3l(1)2Ad1 nor ns3l(1)2Ad3 alleles are expected to code for a protein with a functional GTPase. The ns3G0431 allele is a P-element insertion in the 5′-UTR (Figure 2A).
Figure 2.
l(1)2Ad alleles map to the ns3 locus and mimic developmental delay and Minute-like phenotypes. (A) Map indicating positions of ns3 aberrations leading to loss-of-function. Alleles l(1)2Ad1 and l(1)2Ad3 are small deletions mapping to the second exon and the G0431 allele is a P-element insertion into the 5′-UTR. (B) All three ns3 alleles are necessary for viability. ns3G0431/Y males were crossed to heterozygous ns3l(1)2Ad1/FM7i, ns3l(1)2Ad3/FM7i, and ns3Df(1)BSC719/FM7i females. Male and female percentage viability is the number of mutant animals divided by the number of sibling ns3G0431/FM7i that emerged. (C) The surviving ns3l(1)2Ad3/ns3G0431 (N = 12) and ns3l(1)2Ad1/ns3G0431 (N = 13) females eclose later than sibling ns3G0431/FM7i females (N = 58 and 74, respectively) and are thus developmentally delayed. (D) ns3l(1)2Ad3/ns3G0431 and ns3l(1)2Ad1/ns3G0431 females are small in size. (E) ns3l(1)2Ad3/ns3G0431 and ns3l(1)2Ad1/ns3G0431 females have slender bristles mimicking the Minute phenotype.
The alleles l(1)2Ad1 and l(1)2Ad3 were previously characterized as first-instar lethal. In mosaic analyses with these alleles the gene was necessary for female germline viability (Perrimon et al. 1989). Our confirmation that 0% of ns3l(1)2Ad1/Y and ns3l(1)2Ad3/Y males survive to adulthood (Figure 2B) shows that they have less function than ns3G0431, which allows 78% of adult males to survive. In our previous studies we observed some ns3G0431 homozygous females, but we no longer see them. Perhaps the ns3G0431-bearing chromosome has acquired another aberration that affects female viability. If this is the case it would likely be near the ns3G0431 insertion, since ns3G0431 is lethal over the deficiency Df(1)BSC719 (Figure 2B)—the female-specific lethality must be closely linked. We do find ns3l(1)2Ad1/ns3G0431 and ns3l(1)2Ad3/ns3G0431 trans-heterozygotes. These flies are developmentally delayed (Figure 2C) and are small (Figure 2D). Given that our current ns3G0431 stock yields few to no homozygous females (Figure 2B), we performed subsequent analyses with hemizygous males.
In our previous studies, flies carrying ns3G0431 males were developmentally delayed and small due to nonautonomous growth-controlling roles of ns3 (Kaplan et al. 2008). We suspect that this less-lethal phenotype is due to the allele retaining partial function. In contrast, the strong alleles ns3l(1)2Ad1 and ns3l(1)2Ad3 illustrate the expected requirement for NS3 in all cells. These two mutations prevent female germline viability (Perrimon et al. 1989) and are 100% lethal. Given the likely function of NS3 in ribosome biogenesis, strong alleles of ns3 should be considered a class of Minutes. Consistent with this designation, bristles of ns3l(1)2Ad3/ns3G0431 and ns3l(1)2Ad1/ns3G0431 are slender and short in length, a classic Minute phenotype thought to result from suboptimal protein synthesis (Figure 2E).
NS3 is a functioning GTPase and inactivation of this domain confers antimorphic activity
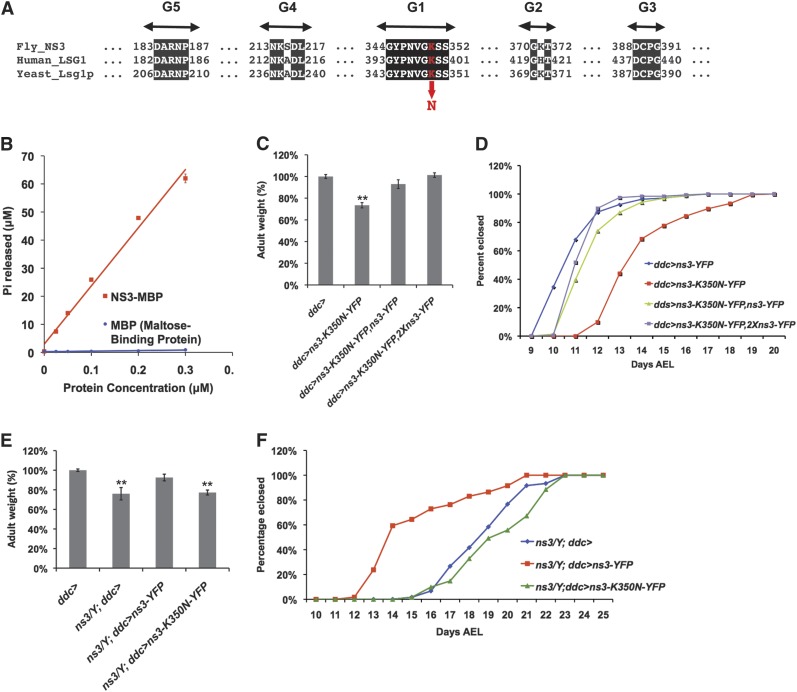
Unlike conventional GTPases that contain five G-domain motifs ordered G1–G2–G3–G4–G5, the YRG family of GTPases (including Lsg1p) carry “circularly permuted” G-motifs in the order G4–G5–G1–G2–G3 (Figure 3A). Mutations within the putative GTP-binding domain of yeast Lsg1 cause a strong antimorphic effect, preventing ribosomal subunit export from the nucleus and stalling growth (Kallstrom et al. 2003; Hedges et al. 2005). This suggests that the GTPase domain is catalytically active and relevant for function, despite the atypical ordering of G-motifs. To directly test the activity of the permuted GTP-binding motifs in NS3, we purified recombinant NS3–MBP (maltose-binding protein) from E. coli and performed in vitro GTPase assays. A linear relationship was observed between the concentration of NS3–MBP and the amount of GTP hydrolyzed within 1 hr at 37° (Figure 3B), while the negative-control MBP protein was incapable of GTP hydrolysis. NS3’s intrinsic GTP hydrolysis rate, Kcat 4.27 ± 0.59 min−1, is on par with that of heterotrimeric G-proteins (Kcat 3–5 min−1) (Kaziro et al. 1991), although relatively high compared to some circularly permuted GTPases (Kcat ∼0.2 min−1) (Daigle and Brown 2004; Himeno et al. 2004; Bassler et al. 2006; Cladiere et al. 2006) and canonical Ras and Rab GTPase members (Kcat <0.01 min−1) (Kaziro et al. 1991). It is unknown whether GTPase activities of Lsg1/NS3 orthologs can be stimulated by GTPase-activating proteins (GAPs) or other factors that can increase catalytic rate >100-fold (Kaziro et al. 1991; Bernards and Settleman 2004). For instance, the Kcat of E. coli YRG member YjeQ is increased ∼500-fold in the presence of small ribosomal subunits (Daigle and Brown 2004).
Figure 3.
NS3 exhibits GTPase activity and mutating an invariant lysine residue in the GTPase domain confers antimorphic activity. (A) Conserved circularly permuted GTP binding domains among NS3 homologs in human and yeast. The location of the antimorph creating residue (ns3-K350N-yfp) is indicated in red. (B) NS3 exhibits GTPase activity in vitro. (C) Expression of antimorphic ns3-K350N-yfp with ddc-gal4 caused decreased body weight (P < 0.05), which can be rescued by expression of wild-type ns3-yfp. The average adult male weight, derived from three groups of 10 flies, is expressed as a percentage of the average weight of negative-control flies that carried only ddc-gal4. (D) Expression of antimorphic ns3-K350N-yfp via ddc-gal4 caused developmental delay (red line), which can be rescued by expressing wild-type ns3-yfp (green and purple lines). The number of eclosed flies was scored daily. The number of males eclosed per day is plotted as the percentage of the total males eclosed. (E) Decreased body weight of ns3G0431 mutant can be rescued by expressing wild-type ns3-yfp (P < 0.05), but not the antimorphic ns3-K350N-yfp via ddc-gal4. The average ns3G0431 adult male weight, derived from three groups of 10 flies, is expressed as a percentage of the average weight of negative-control flies that carried only ddc-gal4. (F) The ns3G0431 developmental delay phenotype (blue line) can be rescued by expressing wild-type ns3-yfp (red line), but not the antimorphic ns3-K350N-yfp (green line), via ddc-gal4. The number of flies eclosed was scored daily. The number of males eclosed per day is plotted as the percentage of the total males eclosed.
To further study the GTPase function of NS3, we introduced into flies a UAS-controlled ns3 transgene carrying point mutations in the GTPase domains. The “K350N” NS3 mutation we employed falls within the conserved G1 motif (GX4GKS/T) of the GTPase domain (Figure 3A). Previous studies of other GTPases have shown that mutating this lysine residue reduces the GTPase catalytic activity. For example, mutating the lysine to alanine (K220A) in YjeQ, an E. coli circularly permuted GTPase, reduced its Kcat by ∼2.5-fold (Daigle et al. 2002). Hydrogen bonding provided by the K16 residue in H-Ras is thought to coordinate the γ-phosphate of GTP for nucleophilic attack by a water molecule (Pai et al. 1990). A H-Ras K16N mutant, which corresponds to the mutation we created in NS3 (K350N), caused a ∼100-fold reduction in affinity for GDP and GTP, and its expression in yeast suppressed cell growth (Sigal et al. 1986). Thus, these previous studies suggest that replacing the NS3 lysine at position 350 with asparagine will create a mutant protein defective in the binding, coordination, and catalysis of GTP. The corresponding K350N mutation in S. cerevisiae Lsg1 is lethal and antimorphic (Hedges et al. 2005).
When a wild-type ns3-YFP transgene was expressed in ns3G0431 flies, using the ddc-gal4 driver, the delayed development and small body size phenotypes were rescued (Kaplan et al. 2008). To test whether our new ns3-K350N-yfp behaved as an antimorph, we expressed it using the ddc-gal4 driver. The effects of ns3-K350N-yfp were compared to flies expressing the wild-type ns3-yfp or to control flies that had no transgene. Consistent with antimorphic properties, expression of ns3-K350N-yfp using ddc-gal4 phenocopied the ns3G0431 mutant phenotype, causing significantly increased developmental time to eclosion and decreased adult body size (Figures 3, C and D). The co-expression of wild-type ns3-yfp from a single transgene with ns3-K350N-yfp rescued the fly size and developmental timing defects of ns3-K350N-yfp alone, thus validating that the K350N protein variant behaved as an antimorph (Figures 3, C and D). Expression of ns3-yfp from two different transgenes provided an even stronger rescue of the ns3-K350N-yfp phenotypes, although quantitation would be necessary to determine whether more NS3–Yfp is produced when expressed from two vs. a single transgene. Ns3-K350N-yfp expression via ddc-gal4 was unable to rescue the ns3G0431 delayed developmental time to eclosion and decreased adult body size phenotypes (Figures 3, E and F), validating that it does not have full wild type functionality.
NS3 is necessary for cell-autonomous growth control and localizes to cytoplasmic puncta and nuclear and cellular peripheries
We next sought to characterize the tissue expression pattern and subcellular localization of NS3. Using newly made antibodies against the fly protein, we visualized NS3 in the imaginal wing disc and salivary gland (Figures 4, A–K). In parallel with these NS3 localization studies, we expressed a UAS-driven shRNA directed against ns3 in discrete subsets of cells within a tissue. This was performed using a heat-shock-driven FLP recombinase and an actin-gal4 that cannot produce Gal4 unless FLP removes a FRT-flanked transcriptional stop upstream of the gal4 ORF. The stochastic nature of the transcriptional stop removal leads to a mosaic expression pattern of gal4, which subsequently drives expression of cell surface GFP (uas-mCD8-GFP) and ns3-shRNA simultaneously in the same subset of cells. In response to ns3-shRNA, the amount of NS3 protein is clearly reduced in the GFP-marked ns3-shRNA expressing cells in the wing disc (Figures 4, A–D) and salivary gland (Figures 4, E–J). Note that in the salivary gland, decreased ns3 reduces cell size (Figures 4, E–J), presumably because decreased protein production interferes with growth. The largest cross-section of wild-type salivary gland cells had an area of 4096.7 ± 176.0 μm2. In contrast, cells expressing ns3-RNAi, measured in an identical manner, had a much smaller area of 1741.8 ± 367.7 μm2. This difference is significant (P-value = 3 × 10−5, student’s t-test, n = 13 and 10 cells).
Figure 4.
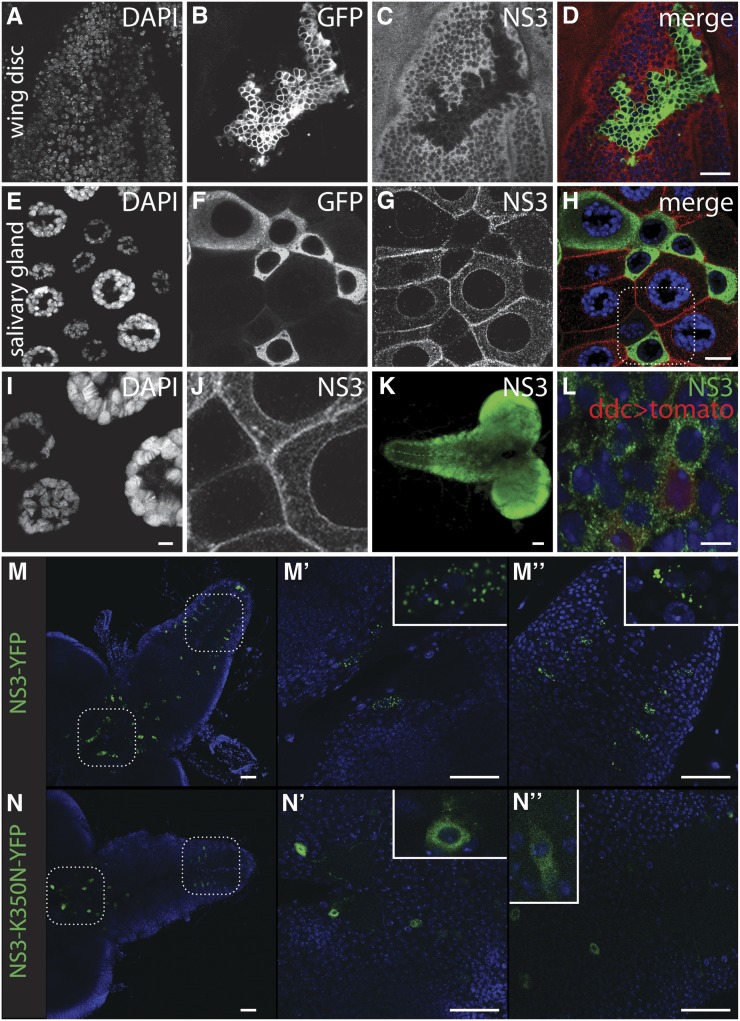
NS3 is localized to novel cytoplasmic punta, and the localization is dependent upon NS3 GTPase activity. (A–D) Image of an imaginal wing disc where an actin-gal4-expressing clone was induced to express mCD8-GFP and ns3-dsRNA and subsequently stained with an anti-NS3 antibody (A) DAPI, (B) mCD8-GFP, (C) anti-NS3, and (D) merged. Note the cytoplasmic NS3 that is substantially reduced in ns3-dsRNA-expressing, mCD8-GFP positive cells, confirming the specificity of the NS3 antibody. Scale bar, 20 µm. (E–H) A salivary gland expressing mCD8-GFP and ns3-dsRNA in a mosaic pattern. (E) DAPI, (F) mCD8-GFP, (G) anti-NS3, and (H) merged. Scale bar, 50 µm. (I–J) Higher magnification image of the salivary gland (see box in H) to show NS3 localization proximal or coincident with the nuclear envelope, cellular membrane, and within cytoplasmic puncta (I), DAPI (J), anti-NS3. Scale bar, 5 µm. (K) Larval CNS stained with anti-NS3 showing the ubiquitous distribution of the protein. Scale bar, 50 µm. (L) NS3 antibody staining in ddc-gal4; uas-TdTomato-labeled ddc neurons. NS3, green; ddc neurons, red; DAPI, blue. Scale bar, 5 µm. (M–M′′) Wild-type ns3-yfp driven by ddc-gal4 demonstrating the punctate localization of NS3 in the cytoplasm. (N–N′′) Ns3-K350N-yfp expressed using ddc-gal4 driver shows a diffuse pattern that extends into neurites. Scale bars, 50 µm.
In wild-type cells, NS3 is present in the cytoplasm of diploid imaginal wing disc cells (Figure 4C) and polyploid salivary gland cells (Figure 4G). In the salivary gland, NS3 is not observed in the nucleus but is enriched proximal to, or coincident with, the nuclear envelope and plasma membrane (Figure 4J). NS3 is also present in cytoplasmic punctate structures (Figure 4, J, L, and M). Nuclear envelope proximal localization of NS3 was also observed on the oocyte nucleus of adult egg chambers and larval midgut cells, and cytoplasmic localization was observed in nurse and midgut cells (Figure S1). These patterns using our newly raised antibody represent NS3 localization as Western analysis indicates our antibody most strongly recognizes a protein of NS3’s predicted molecular weight (70 kDa), and NS3–YFP expressed in the salivary gland (c061 > ns3-yfp) has identical localization patterns (cytoplasmic puncta and nucleus/cell membrane proximal) (Figure S1G and Figure S2). Membrane localization of NS3 and NS3–YFP may be unique to salivary gland cells, as it was not observed elsewhere. Yeast Lsg1 has been observed in the cytoplasm, although not in punctate structures or at the cell periphery (Kallstrom et al. 2003; Hedges et al. 2005). Human hLsg1, like NS3, appears to exist in cytoplasmic foci and at the nuclear periphery. However, unlike NS3 localization, hLsg1 was observed in the nucleus colocalized with Cajal bodies as well as coincident with ER (Reynaud et al. 2005).
Our rescue of the ns3G0431 mutant with ns3-yfp expressed in neurons (elav > ns3-yfp) suggests hypersensitivity of neurons to low NS3 dose (Kaplan et al. 2008). Ns3G0431 could be rescued by expressing ns3-yfp using ddc-gal4, a driver that is active in most dopaminergic (DA) and 5-HT neurons (Li et al. 2000). Given that the majority of cells that express both elav and ddc-gal4 drivers are DA and 5-HT neurons, NS3 may have a unique role in those cells. To investigate more closely whether NS3 has a special localization pattern in 5-HT and/or DA neurons, we compared staining levels of NS3 in ddc-gal4-expressing cells to neighboring cells that do not express ddc-gal4 (Figure 4, K and L). To visualize ddc-gal4-expressing neurons, we drove mtd-Tomato with ddc-gal4 and stained the larval CNS with our anti-NS3 antibody. NS3 appears ubiquitous throughout the larval CNS (Figure 4K), with no sign of unique NS3 patterns in ddc-expressing cells (Figure 4L). Thus, the special requirement for NS3 in ddc cells is not reflected in a visibly distinct protein localization pattern.
The GTPase domain is required for NS3 function (Figure 3). We observe an intriguing difference between NS3–YFP and NS3–K350N–YFP protein localization in neurons. NS3–YFP is strongly enriched in punctate structures, while NS3–K350N–YFP is much more diffuse and often spreads from the cell body into axons and dendrites (Figure 4, M and N). GTP coordination or hydrolysis may be necessary for proper NS3 localization.
Lower levels of ribosomal proteins RpL13 and RpS6 in cells with reduced ns3
S. cerevisiae lsg1 loss-of-function mutants cause retention of the 60S subunit protein RpL25 in nucleoli, indicating that Lsg1 is required for 60S export. Lsg1 works by facilitating recycling of the export adapter (Kallstrom et al. 2003). We examined whether NS3 also functions in 60S subunit export by observing the localization of the 60S subunit protein RpL13 in salivary gland cells where ns3 had been knocked down with RNAi. We expressed ns3-dsRNA with c061-gal4 (c061 > ns3-dsRNA), a driver that is active at variable levels in cells of the salivary glands. This convenient tool enabled the assessment of ns3-RNAi knockdown consequence relative to nearby wild-type cells where no ns3-dsRNA was expressed. A third transgene, uas-lamin-gfp, was included to mark cells expressing ns3-dsRNA.
In wild-type cells lacking lamin–GFP and therefore lacking ns3-dsRNA, RpL13 was localized in the nucleolus, especially its outer layer, to the nucleoplasm, and to the cytoplasm. In cells with ns3 knocked down, identified by their lamin–GFP and consequently known to be expressing ns3-dsRNA, cytoplasmic RpL13 levels were lower (Figure 5, A–D). To quantify the results, average pixel intensities of cytoplasmic RpL13 were measured and then normalized by dividing RpL13 levels in wild-type cells by that of ns3-dsRNA-expressing cells. Cytoplasmic RpL13 levels in wild-type cells were 1.4 ± 0.13 times greater than ns3-dsRNA expressing cells. This contrasts with unchanged levels of RpL13 in control salivary gland tissues analyzed in parallel that did not carry ns3-dsRNA and produced only lamin–GFP (1.1 ± 0.08, P = 0.04, t-test) (Figure 5I).
To determine whether the effect of ns3 knockdown was a general influence on ribosomes beyond its specific effect on RpL13, we examined the localization and levels of RpS6, a component of the 40S subunit. RpS6 was observed in the cytoplasm of both wild-type and ns3-dsRNA-expressing cells although, like RpL13, its levels were significantly reduced in cells after ns3 knockdown (Figures 5, E–H). Wild-type cells had 6.0 ± 1.8 times more RpS6 than neighboring cells expressing ns3-dsRNA. This contrasts with control salivary glands that did not express ns3-dsRNA (1.7 ± 0.2, P = 0.005, t-test) (Figure 5I). Reduced cytoplasmic RpL13 and RpS6 after ns3-dsRNA is consistent with a role for NS3 in 60S subunit export.
ns3G0431 developmental defects are rescued with ns3-yfp expression driven by the dopaminergic th-gal4 driver
Previously, a panel of 15 gal4 drivers with expression in cell subpopulations were used to express ns3-yfp in the ns3G0431 mutant background and test for rescue of the developmental delay and small-body-size phenotypes (Kaplan et al. 2008). From this panel, only the elav-gal4 (elav > ns3-yfp) and ddc-gal4-driven ns3-yfp (ddc > ns3-yfp) rescued the mutant. Given that elav-gal4 is neuron specific and that ddc-gal4 is expressed in most larval 5-HT- and DA-positive cells, ns3 mutation likely affects cells in the intersection of the two drivers’ expression patterns: 5-HT and/or DA neurons. Lack of rescue with a th-gal4 driver led to the earlier conclusion that the 5-HT neurons were most important.
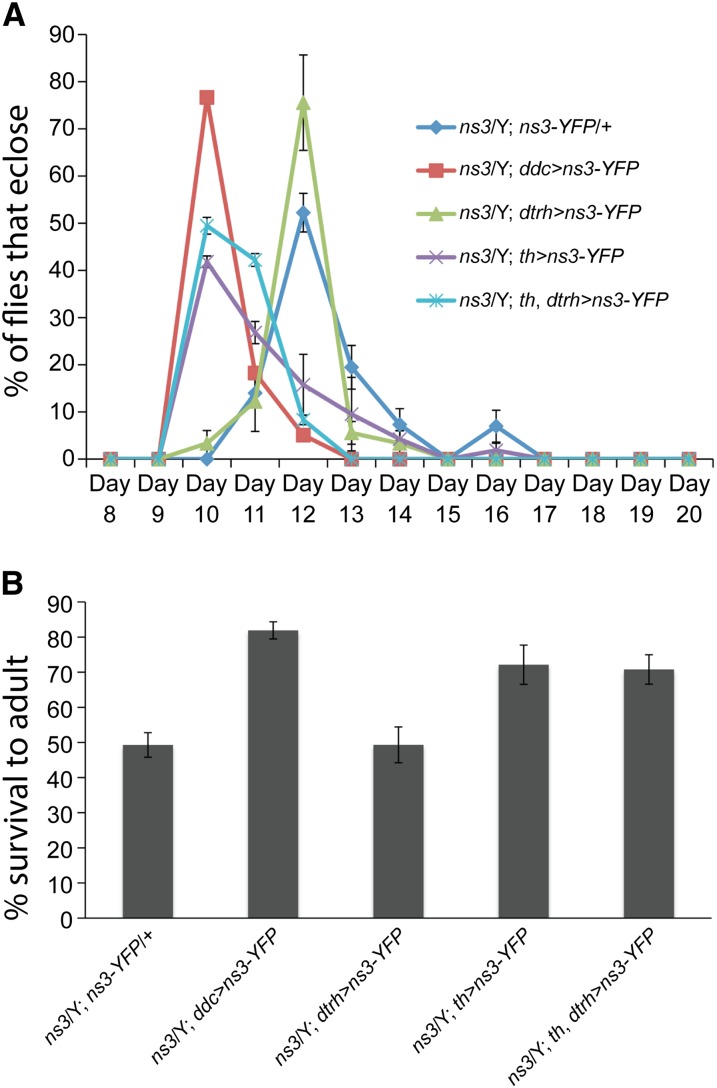
To further define the most highly affected defective neurons in the ns3G0431 mutant, we tested rescue by ns3-yfp expression driven by the newly available dtrh-gal4 (Alekseyenko et al. 2010), which is active in 5-HT neurons. We also retested th-gal4 for DA neurons (Friggi-Grelin et al. 2003). As expected, ddc > ns3-yfp provided strong rescue: 76.7 ± 1.4% of ns3G0431/Y; ddc > ns3-yfp flies eclosed 10 days after embryos were deposited (AEL) (Figure 6A). This contrasts with the negative control ns3G0431/Y; ns3-yfp/+ flies, where no gal4 driver is present and most flies (52.2 ± 4.1%) eclosed 2 days later on day 12 AEL. Driving NS3–YFP production with dtrh > ns3-yfp did not rescue, but ns3G0431/Y flies with th > ns3-yfp were substantially rescued: the largest percentage of flies eclosed on day 10 (41.8 ± 1.2%). Adding the 5-HT driver, dtrh-gal4 to th-gal4 did not improve the rescue: 49.5 ± 1.8% of the ns3G0431/Y; dtrh, th > ns3-yfp flies eclosed on day 10. This trend was matched when looking at viability levels. Only 49.3 ± 3.5% of ns3G0431/Y; ns3-yfp/+ and 49.3 ± 5.1% of ns3G0431/Y; dtrh > ns3-yfp males survived to adulthood, while 81.9 ± 2.4%, 72.1 ± 5.6%, and 70.1 ± 4.2% of the expected ns3G0431/Y males carrying ddc > ns3-yfp, th > ns3-yfp, and dtrh, th > ns3-yfp ns3G0431/Y survived (Figure 6B).
Figure 6.
The delayed development and viability of ns3G0431 mutants can be rescued by restoration of ns3 function to th-gal4-positive cells. (A) Developmental time to eclosion of ns3G0431/Y males with and without expression of ns3-yfp in DA, 5-HT, and DA + 5-HT neurons. Five large crosses were set up of males with (1) no Gal4 driver, (2) DA neuron-enriched th-gal4, (3) 5-HT neuron enriched dtrh-gal4, (4) DA and 5-HT-enriched ddc-gal4, and (5) DA and 5-HT enriched th-gal4, dtrh-gal4 to ns3G0431/FM7i, act > gfp; uas-ns3-yfp virgin females. Embryos were collected on yeast/molasses agar plates overnight and aged 24 hr. Forty act > gfp negative, ns3G0431/Y male, and ns3G0431/+ female larvae were transferred to standard molasses food in vials in triplicate. Eclosion of flies after embryos were laid (AEL) was scored daily, and the plot in A indicates the percentage of the final total number of eclosed ns3G0431/Y flies that eclosed on the indicated day AEL. Most ns3G0431/Y flies with no gal4 driver (dark blue) eclosed 12 days AEL (N = 15, 7, and 13 ns3G0431/Y flies for each of 3 replicates). Expression of ns3-yfp in ns3G0431/Y flies with the ddc-gal4 (DA + 5-HT, red, N = 25, 20, and 16 flies) and th-gal4 (DA, purple, N = 14, 18, and 16 flies) provided substantial rescue, with most flies eclosing 10 days AEL. Expression of ns3-yfp with dtrh-gal4 (5-HT, green, N = 10, 7, and 15 flies) did not rescue ns3G0431/Y developmental delay, and dtrh-gal4 in combination with th-gal4 (DA + 5-HT, light blue, N = 17, 11, and 20 flies) did not significantly enhance rescue provided by th-gal4 alone (purple). (B) Survival to adulthood of ns3G0431/Y males with and without expression of ns3-yfp in DA, 5-HT, and DA + 5-HT neurons. The total number of ns3G0431/Y male flies that eclosed over the predicted number (40 − number of ns3G0431/+ females) from three replicates of vials that carried 40 ns3G0431/Y and ns3G0431/+ larvae is plotted. Only 49.3 ± 3.5% of ns3G0431/Y; ns3-yfp/+ and 49.3 ± 5.1% of ns3G0431/Y; dtrh > ns3-yfp males survived to adulthood. In contrast, 81.9 ± 2.4% ns3G0431/Y males carrying ddc > ns3-yfp and 72.1 ± 5.6% carrying th > ns3-yfp eclosed. This indicated that expression of ns3-yfp in ns3G0431/Y ddc/th-gal4+ cells, covering most DA neurons, restored ns3G0431/Y viability. Inclusion of the dtrh-gal4 driver with th-gal4 did not enhance the rescue provided by th-gal4 alone, as 70.1 ± 4.2% ns3G0431/Y; dtrh, th > ns3-yfp males survived.
Given that the cells that activate both elav-gal4 and th-gal4 are mostly DA neurons, we suspect that DA neurons are especially prone to defects when NS3 dose is reduced in the hypomorphic ns3G0431 background. The level of NS3 in ns3G0431 DA neurons relative to surrounding non-DA neurons was unchanged relative to wild type, ruling out an allele-specific effect on NS3 dosage in DA neurons (Figure S3). The role of NS3 in DA neurons may be to support proper translational regulation of transcripts unique and necessary to DA function, or DA neurons may have an especially high demand for efficient protein synthesis. Although a role of DA in wild-type growth has not been directly observed, the rescue of growth defects of ns3G0431 with elav-, ddc-, and th-gal4-driven uas-ns3-yfp implies that DA neurons provide critical functions for proper larval developmental progression.
Discussion
Evolutionary conservation and possible divergence of Lsg1/NS3 functionality
In yeast, Lsg1p is a cytoplasmic GTPase associated with the 60S ribosomal subunit. It is required for releasing the nuclear export adapter Nmd3p from the 60S ribosomal subunit and therefore indirectly promotes 60S nuclear export (Kallstrom et al. 2003; Hedges et al. 2005). In this study, we found that expression of fly NS3 in yeast cells was sufficient to rescue Lsg1p deficiency, confirming their orthology. Thus NS3 appears to ensure proper export of the 60S subunit from the nucleus, like Lsg1p. Consistent with this view, levels of RpL13, a large ribosomal protein, were lower in the cytoplasm of salivary gland cells where ns3 was knocked down with RNAi (Figures 5, A–D and I). Levels of a small ribosomal subunit protein, RpS6, were also reduced after ns3 knockdown (Figure 5, E–H, 5I). Therefore, NS3 likely contributes to one or more steps that culminate in proper protein synthesis.
We previously showed that expression of a uas-ns3-yfp transgene under the control of ddc-gal4 could strongly rescue the ns3G0431 mutant. Expression of uas-lsg1 in this same fashion was not sufficient to provide rescue of the ns3G0431 mutant (Figures 1, C and D). This apparent contradiction may be explained by features of fly NS3 that suggest functional divergence from yeast Lsg1p. NS3 localizes within or proximal to the nuclear envelope and plasma membrane and is found within cytoplasmic puncta. Lsg1p was not described to be in these subcellular regions and exists throughout the cytoplasm (Kallstrom et al. 2003; Hedges et al. 2005). The distinctive localization features of NS3 may reflect special features that were acquired in the metazoan lineage, precluding yeast Lsg1p from rescuing the fly ns3 mutant.
The human protein that is orthologous to Lsg1p/NS3 is hLsg1 (Reynaud et al. 2005). Like yeast and fly Lsg1p/NS3, hLsg1 is necessary for cell viability. It is unclear whether hLsg1 has specific roles in ribosome biogenesis, although this is likely given its protein sequence similarity to the yeast ortholog. NS3 and hLsg1 share localization properties. Both proteins are coincident with the nuclear envelope and located in unidentified cytoplasmic puncta (Figure 4J and Figure S1) (Reynaud et al. 2005). We did not observe NS3 in the nucleus, yet hLsg1 was observed in nuclear Cajal bodies that contain many factors involved in pre-ribosomal RNA and mRNA processing (Reynaud et al. 2005). Just as NS3 and Lsg1p have functional differences, hLsg1 appears to have features and functions different from NS3.
Cell-autonomous role of NS3 revealed by new alleles
In yeast and human cells, Lsg1 is essential for cell viability and growth (Kallstrom et al. 2003; Hedges et al. 2005; Reynaud et al. 2005), yet the Drosophila ns3G0431 allele was only partially lethal. Many ns3G0431 flies survive to adulthood, albeit with developmental delay and decreased overall size (Kaplan et al. 2008). Here we identified two alleles of the previously uncharacterized locus l(1)2Ad as new alleles of ns3. Both ns3l(1)2Ad1 and ns3l(1)2Ad3, which are predicted to have severe truncation of the protein, cause 100% lethality and are therefore likely strong loss-of-function alleles. In contrast, the ns3G0431 lesion is a P-element insertion in the 5′-UTR, which may cause only a subtle loss-of-function. The apparent inconsistency between cell lethality in lsg1 mutant yeast and mammalian cells and semiviability in fly cells was resolved by the lethality of the stronger loss-of-function ns3 alleles. We suspect that most or all Drosophila cells depend upon NS3 for ribosome biogenesis and therefore growth and viability. Flies with the ns3G0431 allele likely survive to adulthood due to a more subtle drop in NS3 levels.
A family of noncanonical GTPases with unified roles in ribosome biogenesis
The canonical GTPase structure, based on Ras, carries G-domain motifs ordered from the N to C terminus as G1–G2–G3–G4–G5. Lsg1 and NS3 contain circularly permuted G-domain motifs that are instead ordered G4–G5–G1–G2–G3 (Figure 3A). This structure is common for GTPases involved in ribosome biogenesis. Several ribosome biogenesis proteins belong to a family of circularly permuted GTPases that expanded in eukarya from a single circularly permuted GTPase, YlqF (Reynaud et al. 2005) (Table 1). YlqF itself promotes ribosome biogenesis as it is necessary for 70S assembly in Bacillus subtilis (Uicker et al. 2006).
In spite of their atypical structures, several circularly permuted GTPases within the YRG family have been demonstrated to be genuine GTPases using crystal structures or in vitro assays (Daigle and Brown 2004; Levdikov et al. 2004; Shin et al. 2004; Reynaud et al. 2005; Bassler et al. 2006; Cladiere et al. 2006; Matsuo et al. 2007; Moreau et al. 2008; Sudhamsu et al. 2008). Altered G-domain ordering does not change how each motif folds into a tertiary structure. Here we demonstrate that NS3 is proficient in GTP hydrolysis and that proper GTPase activity may confer special protein localization properties in neurons, perhaps by affecting association with other proteins.
Based on previous studies of GTPases, replacement of lysine at position 350 in NS3 with asparagine (K350N) is predicted to disrupt the coordination and catalysis of GTP (Saraste et al. 1990). We found that production of a protein with this mutation conferred antimorphic activities in flies (Figure 3). This mimics what occurs in S. cerevisiae with an analogous mutation (Hedges et al. 2005). We suggest that the antimorphic NS3(K350N) blocks important functions, or sequesters essential cofactors away from endogenous wild-type NS3. GTP hydrolysis may be necessary to remove Nmd3 from the 60S subunit and enable its return to the nucleus for another round of 60S export. In this scenario, NS3(K350N) could prevent GTPase-competent wild-type NS3 from removing Nmd3. On the other hand, NS3(K350N) had altered localization in neurons and appeared to spread from cell bodies into neurites. The altered localization could sequester important cofactors away from endogenous NS3 and thus block its ribosome biogenesis activities.
Identifying neuronal subsets involved in growth control
Previously, we had concluded that ns3 functioned within 5-HT neurons to promote growth (Kaplan et al. 2008). The primary data leading to that conclusion were that expression of uas-ns3-yfp in most 5-HT and DA neurons (ddc > ns3-yfp) rescued the ns3G0431 growth defects, while expression in most DA neurons (th > ns3-yfp) did not provide rescue. We no longer believe that second, negative, result. Since that study, it has become apparent that the penetrance of ns3G0431 phenotypes is highly dependent upon genetic background. For instance, the original background of the 5-HT driver line (dtrh-gal4) was nearly 100% lethal when placed in trans to our ns3G0431 stock. We therefore backcrossed ddc-gal4, dtrh-gal4, and th-gal4 for five generations into an isogenized genetic background (Gohl et al. 2011) that we found to be semiviable in trans to ns3G0431. A second demonstration of the importance of levels of rescuing protein comes from manipulating the temperature of the experiments and therefore the GAL4 activity level. Our previous studies were performed at 25°. The present studies were conducted at 29°, which significantly boosts expression of Gal4-driven transgenes. Under the conditions of boosted gal4 expression in pure genetic backgrounds, expression of uas-ns3-yfp in DA neurons (th > ns3-YFP) provided substantial rescue.
Expression of ns3 with three different Gal4 drivers (elav-gal4, ddc-gal4, and th-gal4) has been sufficient to provide significant rescue of the ns3G0431 mutant. The intersection between these drivers’ expression patterns consists of DA neurons, suggesting that ns3 has an important role in these cells to promote growth. Perhaps factors involved in DA signaling-specific properties have an especially high requirement for protein-translation efficiency. Such growth-promoting properties of DA neurons may include broad-reaching metabolism regulation like that of insulin-like peptides (ILPs), as previously suggested (Kaplan et al. 2008). ILPs are produced from a small cluster of neurosecretory cells known as ILP-producing cells (IPCs) (Rulifson et al. 2002). ILPs accumulate in IPCs in ns3G0431 mutants (Kaplan et al. 2008). Perhaps specific signaling defects from DA neurons in the ns3G0431 mutant lead to failed ILP modification and/or secretion from IPCs. DA neurons may also regulate nutrient acquisition activities at one or more levels such as appetite control, nutrient absorption, or feeding activities.
5-HT signaling could still be altered in the ns3G0431 background. In keeping with that possibility, 5-HT levels were elevated in mutant adult heads. Feeding the 5-HT precursor, 5-HTP, enhanced the ns3G0431 phenotypes (Kaplan et al. 2008). Those results could be a consequence of the primary defect of ns3 loss. Recent studies have implicated 5-HT in promoting some activities of IPCs, which in turn are necessary for growth. Developmental growth roles of 5-HT were not demonstrated, yet the expression of the 5-HT1A receptor gal4 line in IPCs (Luo et al. 2012) suggests that 5-HT may regulate IPC-driven growth in certain circumstances.
The ns3G0431 mutant appears to be a sensitized genetic condition, where important growth control by ddc-gal4 and th-gal4 (ddc/th-gal4)-expressing cells is revealed. Alternatively, growth control via ddc/th-gal4 cells may not happen in wild-type animals, while in ns3G0431 mutants an abnormal circuit of this sort occurs. If some aspect of neuronal signaling is subfunctional in the ns3G0431 mutant, then restoration of ddc/th-gal4 cellular health through expression of uas-ns3-yfp could compensate for neuronal defects elsewhere. Although ddc/th-gal4-expressing cells may convey growth-promoting activities in wild-type flies, these may be DA independent. Both of these drivers are active in a small number of non-dopaminergic neurons and in various other tissues that may be the true source of nonautonomous growth control. In addition, ddc/th-gal4 growth control may be neuronal, yet mediated by a signaling molecule other than DA such as a neuropeptide, hormone, or different neurotransmitter.
A higher demand for NS3 in neurons
Strong alleles of ns3 reveal cell-autonomous growth roles (Figures 1–5), while the weak ns3G0431 allele reveals that ddc/th-gal4-expressing neurons (possibly DA neurons) are sensitive to reduced levels of NS3. Based on high sequence homology, it is likely that NS3 promotes ribosome biogenesis. Thus, the hypomorphic ns3G0431 allele may indicate special ribosome biogenesis requirements of ddc/th-gal4-expressing neurons and/or unique dependency on specific types of translational regulation.
We propose the following alternative working models to explain ddc/th-gal4 hypersensitivity to NS3 dosage. As the NS3 yeast ortholog (Lsg1) promotes export of 60S subunits from nuclei, NS3 in metazoans may have become necessary for positioning ribosomes within the specialized structures of neurons. Most ribosomal subunits appear in the neuronal cell body, but in several instances ribosomes are localized within axons and dendrites (Craig and Banker 1994; Brittis et al. 2002; Kiebler and Bassell 2006; Rolls et al. 2007). This subcellular targeting of ribosomes, causing them to move across long distances, may require specialization of ribosome biogenesis proteins like NS3.
NS3 may instead translationally regulate specific mRNA subsets. Given that DA neurons appear to be especially sensitive to NS3 dose, perhaps one or more mRNAs that code for DA synthesis, degradation, transport, or synaptic dynamics is translationally regulated by NS3. A mRNA-specific translational regulation function would make NS3 analogous to mouse RpL38. This large subunit ribosomal protein binds specifically to a subset of Hox gene mRNAs and controls their association with the 80S ribosome. Mice carrying a mutant copy of RpL38 have specific homeotic transformations due to failed translation of these Hox mRNAs (Kondrashov et al. 2011).
Another unique translational regulation role that NS3 may play is in the proper localization and translational regulation of certain mRNAs to the synapse. Localized translation is not unique to neurons, yet several RNA-binding proteins that transport, tether, activate, and/or repress translation have been identified in screens for molecules involved in neuronal activities (Costa-Mattioli et al. 2009). Perhaps NS3 is necessary for localized translational regulation in neurons by binding certain mRNAs or transporting ribosomal subunits to the relevant site. Further explorations of ribosome biogenesis, and the diversity of ribosomal structures and activities, is likely to reveal specific developmental and physiological functions carried out by presumed “housekeeping” proteins.
Supplementary Material
Acknowledgments
We thank Michael Dellinger for critiquing the manuscript. We are grateful to Kevin Cook and the Bloomington Drosophila Stock Center for help identifying stocks carrying putative ns3 alleles. We thank Kathleen Beckingham, Sangbin Park, Seung Kim, Wendy Neckameyer, Jay Hirsh, Eric Rulifson, Ron Alfa, and members of Tom Clandinin’s laboratory for helpful discussions. The research was supported by National Institutes of Health grant GM53655 to A.J. and by the Howard Hughes Medical Institute. T.H. is a Robert Black fellow of the Damon Runyon Cancer Research Foundation (DRG-2045-10). J.N. is a postdoctoral fellow, and M.P.S. an Investigator, of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: P. K. Geyer
Literature Cited
- Alekseyenko O. V., Lee C., Kravitz E. A., 2010. Targeted manipulation of serotonergic neurotransmission affects the escalation of aggression in adult male Drosophila melanogaster. PLoS ONE 5: e10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A., Korr D., Barwell K. J., Sjulsen C., Gajewski C. D., et al. , 2003. MTG1 codes for a conserved protein required for mitochondrial translation. Mol. Biol. Cell 14: 2292–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., et al. , 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8: 517–529. [DOI] [PubMed] [Google Scholar]
- Bassler J., Kallas M., Hurt E., 2006. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 281: 24737–24744. [DOI] [PubMed] [Google Scholar]
- Bernards A., Settleman J., 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14: 377–385. [DOI] [PubMed] [Google Scholar]
- Boddapati N., Anbarasu K., Suryaraja R., Tendulkar A. V., Mahalingam S., 2012. Subcellular distribution of the human putative nucleolar GTPase GNL1 is regulated by a novel arginine/lysine-rich domain and a GTP binding domain in a cell cycle-dependent manner. J. Mol. Biol. 416: 346–366. [DOI] [PubMed] [Google Scholar]
- Bridges C. B., Morgan T. H., 1923. The Third Chromosome Group of Mutant Characters of Drosophila melanogaster. Carnegie Institution, Washington, DC. [Google Scholar]
- Brittis P. A., Lu Q., Flanagan J. G., 2002. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell 110: 223–235. [DOI] [PubMed] [Google Scholar]
- Cladiere L., Hamze K., Madec E., Levdikov V. M., Wilkinson A. J., et al. , 2006. The GTPase, CpgA(YloQ), a putative translation factor, is implicated in morphogenesis in Bacillus subtilis. Mol. Genet. Genomics. 275: 409–420. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N., 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron 61: 10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A. M., Banker G., 1994. Neuronal polarity. Annu. Rev. Neurosci. 17: 267–310. [DOI] [PubMed] [Google Scholar]
- Daigle D. M., Brown E. D., 2004. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J. Bacteriol. 186: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle D. M., Rossi L., Berghuis A. M., Aravind L., Koonin E. V., et al. , 2002. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41: 11109–11117. [DOI] [PubMed] [Google Scholar]
- Du X., Rao M. R., Chen X. Q., Wu W., Mahalingam S., et al. , 2006. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol. Biol. Cell 17: 460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F., Coulom H., Meller M., Gomez D., Hirsh J., et al. , 2003. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 54: 618–627. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., et al. , 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21: 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl D. M., Silies M. A., Gao X. J., Bhalerao S., Luongo F. J., et al. , 2011. A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods 8: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A. W., 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24: 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A. K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., et al. , 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cellular and molecular life sciences. Cell. Mol. Life Sci. 65: 2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Hanawa-Suetsugu K., Kimura T., Takagi K., Sugiyama W., et al. , 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 32: 5303–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J. H., Kallstrom G., Johnson A. W., 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151: 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Hannus S., Schmelzl B., Lau D., Tollervey D., et al. , 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallstrom G., Hedges J., Johnson A., 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23: 4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. D., Zimmermann G., Suyama K., Meyer T., Scott M. P., 2008. A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size. Genes Dev. 22: 1877–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaziro Y., Itoh H., Kozasa T., Nakafuku M., Satoh T., 1991. Structure and function of signal-transducing GTP-binding proteins. Annu. Rev. Biochem. 60: 349–400. [DOI] [PubMed] [Google Scholar]
- Kiebler M. A., Bassell G. J., 2006. Neuronal RNA granules: movers and makers. Neuron 51: 685–690. [DOI] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C. R., Shimizu K., Hsieh A. C., et al. , 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson A., 1998. The minute genes in Drosophila and their molecular functions. Adv. Genet. 38: 69–134. [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L., 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317: 41–72. [DOI] [PubMed] [Google Scholar]
- Levdikov V. M., Blagova E. V., Brannigan J. A., Cladiere L., Antson A. A., et al. , 2004. The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis viability. J. Mol. Biol. 340: 767–782. [DOI] [PubMed] [Google Scholar]
- Li H., Chaney S., Roberts I. J., Forte M., Hirsh J., 2000. Ectopic G-protein expression in dopamine and serotonin neurons blocks cocaine sensitization in Drosophila melanogaster. Curr. Biol. 10: 211–214. [DOI] [PubMed] [Google Scholar]
- Lin J. I., Mitchell N. C., Kalcina M., Tchoubrieva E., Stewart M. J., et al. , 2011. Drosophila ribosomal protein mutants control tissue growth non-autonomously via effects on the prothoracic gland and ecdysone. PLoS Genet. 7: e1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K. Y., Li Z., Bussiere C., Bresson S., Marcotte E. M., et al. , 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 39: 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Becnel J., Nichols C. D., Nassel D. R., 2012. Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5–HT1A receptor: cellular and molecular life sciences. Cell. Mol. Life Sci. 69: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold S. J., Roote J., Reuter G., Lambertsson A., Ashburner M., et al. , 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8: R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Oshima T., Loh P. C., Morimoto T., Ogasawara N., 2007. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J. Biol. Chem. 282: 25270–25277. [DOI] [PubMed] [Google Scholar]
- Matsuo E., Nagamine T., Matsumoto S., Tsuneizumi K., 2011. Drosophila GTPase nucleostemin 2 changes cellular distribution during larval development and the GTP-binding motif is essential to nucleoplasmic localization. Biosci. Biotechnol. Biochem. 75: 1511–1515. [DOI] [PubMed] [Google Scholar]
- Moreau M., Lee G. I., Wang Y., Crane B. R., Klessig D. F., 2008. AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283: 32957–32967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy T. I., Silver P. A., 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13: 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., et al. , 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9: 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V. G., Johnson A. W., 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., Engstrom L., Mahowald A. P., 1989. Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics 121: 333–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud E. G., Andrade M. A., Bonneau F., Ly T. B., Knop M., et al. , 2005. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M. M., Satoh D., Clyne P. J., Henner A. L., Uemura T., et al. , 2007. Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev. 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosby R., Cui Z., Rogers E., deLivron M. A., Robinson V. L., et al. , 2009. Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol. Biol. Cell 20: 4424–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., Nusse R., 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A., 1990. The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15: 430–434. [DOI] [PubMed] [Google Scholar]
- Saveanu C., Bienvenu D., Namane A., Gleizes P. E., Gas N., et al. , 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20: 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiser R. M., Sundberg A. E., Wollam B. J., Zobel-Thropp P., Baldwin K., et al. , 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174: 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. H., Lou Y., Jancarik J., Yokota H., Kim R., et al. , 2004. Crystal structure of YjeQ from Thermotoga maritima contains a circularly permuted GTPase domain. Proc. Natl. Acad. Sci. USA 101: 13198–13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., Gibbs J. B., D’Alonzo J. S., Temeles G. L., Wolanski B. S., et al. , 1986. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc. Natl. Acad. Sci. USA 83: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt U., Silver P. A., 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11: 3777–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. P., Woolford J. L., Jr, 2009. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 21: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk B. S., Karbstein K., 2009. Powering through ribosome assembly. RNA 15: 2083–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhamsu J., Lee G. I., Klessig D. F., Crane B. R., 2008. The structure of YqeH: an AtNOS1/AtNOA1 ortholog that couples GTP hydrolysis to molecular recognition. J. Biol. Chem. 283: 32968–32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. Y., McKay R. D., 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker W. C., Schaefer L., Britton R. A., 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59: 528–540. [DOI] [PubMed] [Google Scholar]
- Wu J. S., Luo L., 2006. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 1: 2110–2115. [DOI] [PubMed] [Google Scholar]
- Xue S., Barna M., 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto H., Meng L., Lin T., Zhu Q., Tsai R. Y., 2007. GNL3L inhibits activity of estrogen-related receptor gamma by competing for coactivator binding. J. Cell Sci. 120: 2532–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Kutay U., 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581: 2783–2793. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Meng L., Hsu J. K., Lin T., Teishima J., et al. , 2009. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 185: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.