Abstract
Nuclear actin levels have recently been linked to different cellular fates, suggesting that actin could act as a switch between altered transcriptional states. Here we discuss our latest results on the mechanisms by which nuclear actin levels are regulated and their implications to the functional significance of nuclear actin.
Keywords: actin, nuclear transport, nucleus, transcription
Background
Actin is perhaps the most functionally versatile protein of the whole eukaryotic cell. The key feature of actin is its ability to polymerize into helical filaments, which can then be used to produce force in the cells. The actin polymerization process is tightly regulated by numerous actin-binding proteins and subject to modulation by different signaling pathways. This firm spatial and temporal regulation of actin filament formation allows actin to participate in many cellular events that require dynamic structures, such as cytokinesis, cell migration or establishment of cell shape.1
In addition to being a well studied cytoplasmic protein, actin is also localized to the cell nucleus, where it has been linked to many processes that allow the organization and reading of genetic information. Actin has been described as an actor of most steps of the transcription process from the regulation of transcription initiation to the processing of pre-mRNAs. For example, actin has been connected to transcription by all three RNA polymerases and is a component of many important chromatin remodeling complexes.2
In line with the potentially essential role for actin in various nuclear processes, several reports have linked variable nuclear actin levels to physiologically and pathologically relevant cellular states. Indeed, one of the earliest irrefutable evidence for the presence of actin in the nucleus comes from hand-isolated Xenopus laevis oocyte nucleus, where actin is the most abundant protein in the whole germinal vesicle.3,4 Around the same time, Fukui and Katsumaru observed that cellular stress in the form of dimethyl sulfoxide treatment induces the formation of actin bundles in the cell nuclei from different species, ranging from amoeba to humans.5 Much later, induced nuclear translocation of actin has been described for example in rat mast cells following ATP depletion, or upon treatment with Latrunculin B, which triggers actin depolymerisation.6 Increased nuclear actin levels are also observed after induction of macrophage differentiation of HL-60 cells.7 More recently, Spencer et al. have demonstrated that induction of quiescence, i.e., growth arrest of mammary epithelial cells by growth factor withdrawal or laminin-111 (LN1) addition, results in a severe depletion of the nuclear actin pool. An inverse correlation between LN1 and nuclear actin staining was observed in tissue samples from mammary terminal end buds, indicating that this regulation likely takes place also in vivo.8 Thus, the amount of actin in the cell nucleus can drastically increase or decrease in a wide range of cell types but also in species from different taxonomic phyla. This implies that nuclear localization of actin is tightly regulated, and that this regulation is an evolutionary conserved feature. However, the mechanisms governing the interconnection between nuclear and cytoplasmic actin pools have remained largely unclear. This question, which is central in order to perceive the full range of the potential functions of nuclear actin in cells, has been tackled by our lab in a recent publication in the journal PNAS.9
Molecular Basis and Dynamics of the Nuclear Localization of Actin
The mechanism by which actin actually enters the nucleus to participate in gene expression processes has not been characterized previously. The size of actin, 43 kDa, is at the border of the size exclusion limit for passive diffusion through the nuclear pore complex and this fact has clearly complicated the analysis of nuclear import mechanism for actin. On the contrary, two active nuclear export pathways for actin, dependent on either Crm110 or exportin 6,11 have been described in the literature.
We used a wide variety of different imaging techniques to study nuclear actin and our first important conclusion, based on different photobleaching assays, is that actin constantly and rapidly shuttles in and out of the cell nucleus. In fact, one third of nuclear actin is exchanged every 100 sec. This demonstrates that there is extensive and dynamic communication between the cytoplasmic and nuclear actin pools. Interestingly, the availability of actin monomers seems to limit the nuclear transport rate in both directions. This implies that both the extent of actin polymerization or binding to different complexes may modulate nuclear actin levels by restricting the availability of transport-competent actin monomers. By using a combination of imaging approaches and RNA interference (RNAi), we further showed that inhibition of Crm1 does not affect nuclear export of actin. Hence, the earlier study linking Crm1 to actin export could be due to indirect effects on nuclear export of many actin-binding proteins or the actin probes used in this study.10 Instead, we found that exportin 6 mediated the export of actin both in the murine fibroblastic cell line NIH 3T3 and the Drosophila cell line S2R+, demonstrating that this export mechanism is largely shared among eukaryotes.9 This data therefore confirms and extends the previous results implicating exportin 6 as the major nuclear export receptor for actin.11-13
To reveal whether the nuclear import of actin takes place by passive or active mechanism, the nuclear import rates of different sized actin constructs was measured. Even actin tagged with two GFP molecules, giving rise to a construct with a size close to 100 kDa, was imported into the nucleus at the same speed as fluorophore-labeled microinjected actin, which has almost the same size as endogenous actin. This clearly demonstrates that nuclear import of actin is an active process, because passive diffusion would have been sensitive to the size of the construct as demonstrated for GFP and 2GFP. This data was further corroborated by the finding that importin 9, an importin-β family member, is required to maintain nuclear actin levels. As importin 9 interacts both with actin and cofilin in a co-immunoprecipitation experiment, we hypothesize that actin enters the nucleus as a complex with importin 9 and cofilin. This explains why actin, which seems to lack nuclear localization signal (NLS), can enter the nucleus. Combined, this data reveals the regulatory points that the cell can use to modulate its nuclear actin levels. These mechanisms include the concentration or activity of importin 9 and exportin 6 transport proteins, the availability of the transport cofactors cofilin and profilin and the availability of free actin monomers (Fig. 1).9
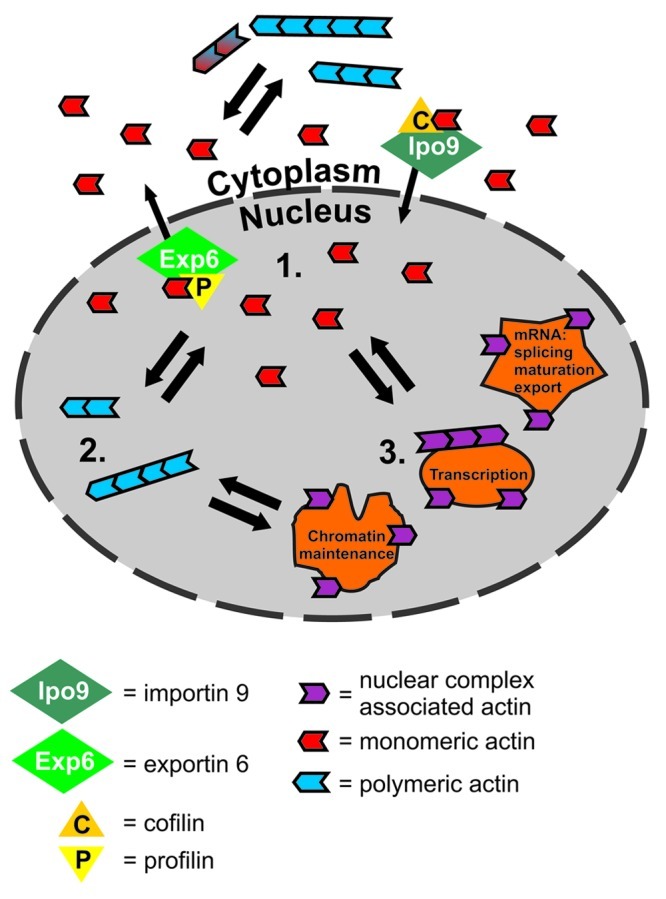
Figure 1. Partitioning of nuclear actin into pools with distinct actin exchange rates and the nuclear import and export mechanisms of actin. Nuclear import of actin is mediated by importin-9 and export by exportin-6. In both cases, a small actin-binding protein acts likely as an adaptor, with profilin operating during export and cofilin during import. Different photobleaching assays demonstrate that nuclear actin is present in three distinct pools, which display different nuclear turnover rates. This rate is highest with actin monomers (1.), which correspond to approximately 20% of total nuclear actin, and which are subject to continuous import and export from the nucleus. The second actin pool, also of about 20%, (2.) in this model consists of some type of polymeric actin, as this pool was absent from an actin mutant that does not polymerize. The exact structural nature of these polymers is currently unknown. The third nuclear actin pool (3.) contains the majority (~60%) of actin in the nucleus and displays slow turnover of actin. We suggest that this represents the numerous interactions described for actin in the nucleus including the monomers in chromatin remodeling complexes, interactions with the transcription machinery, which may or may not require polymeric actin, as well as the specific role described for actin in mRNA processing. Data are based on Dopie et al.9 and Stuven et al.11
Future studies will reveal the exact manner in which these steps are utilized to regulate nuclear actin levels in vivo and which signaling pathways impinge on them. Thus far, the only mechanism elucidated is the regulation in Xenopus oocytes. The large nuclear actin pool observed in these cells is due to low levels of exportin 6 protein, achieved through post-transcriptional downregulation of its expression.12 In addition, the nuclear accumulation of actin upon macrophage differentiation is inhibited by p38 mitogen-activated protein kinase inhibitors7 and, in mammary epithelial cells, treatment with the inhibitor for phosphoinositide 3-kinase reduces the levels of nuclear actin.8 However, the actual regulatory steps that these agents target have not been revealed.
Functional Consequences of the Variation of Nuclear Actin Levels
In Xenopus oocytes, the high levels of nuclear actin have been suggested to be required for the stabilization of the giant nucleus through the formation of a filamentous actin network.12 In this case, the establishment of a large nuclear actin pool seems to have mainly a structural role, but could also contribute to other functional aspects in the nucleus. Also, the exact structural nature and the rigidity of the polymerized actin network14 is an interesting topic for future research. In mammalian cells, nuclear actin levels have been linked to the transcriptional activity of the cell. Increased nuclear actin in differentiating HL-60 cells has been connected to the transcriptional activation of genes, which are required for macrophage differentiation. Concomitantly, actin is recruited to selected genes upon the differentiation process and its recruitment correlates with RNA polymerase binding to these genes.7 Conversely, decreased nuclear actin has been linked to a quiescent, transcriptionally inactive state in mammary epithelial cells, which also display destabilized binding of RNA polymerases II and III to transcription sites. Importantly, nuclear targeting of actin reversed the effect of LN1 on transcription and RNA polymerase II association, strongly pointing to an important role for nuclear actin in controlling cell quiescence through general effect on transcription.8 Similarly, depletion of importin 9, which decreases nuclear actin levels, significantly impairs transcription. Also this transcription defect was dependent on nuclear actin, because expression of an actin construct fused to an NLS to allow its importin 9-independent nuclear translocation rescued the transcription defect.9 Taken together, these data reveal that efficient transcription requires the maintenance of a sufficient level of nuclear actin and consequently demonstrates how important it is for the cell to master its nuclear actin levels.
In addition to affecting the general transcriptional activity of the cell, modulation of nuclear actin has also been linked to expression of specific sets of genes. One of the best characterized examples of this is the Myocardin-related transcription factor A (MRTF-A) pathway. MRTF-A is a co-activator of the transcription factor serum response factor (SRF), which controls, for instance, the expression of many cytoskeletal genes.15 MRTF-A constantly shuttles in and out of the nucleus via active transport mechanisms. The localization and the activity of MRTF-A depend on the balance between actin monomers and polymers in the cell. If present in high concentration, actin monomers bind to MRTF-A impairing its nuclear import and in parallel stimulating its export from the nucleus, which results in mainly cytoplasmic distribution of MRTF-A. If the rate of actin polymerization increases and the concentration of actin monomers falls below a certain threshold, MRTF-A, free from actin-binding, can accumulate in the nucleus and activate SRF-dependent transcription. In order to function, this regulation requires a coordination of the actin monomer concentration in both the cytoplasm and the nucleus. For example, it has been shown that the MRTF-A-actin complex responds to cellular signaling regardless of the cellular localization.16 It is not clear whether this coordination is accomplished through direct communication of cytoplasmic and nuclear actin pools via nucleo-cytoplasmic shuttling of actin. Another hypothesis is that the balance between monomers and polymers in both compartments is regulated separately in a coordinated manner. Better understanding of the regulatory mechanisms of actin shuttling should provide us precious tools to distinguish between these possibilities. Intriguingly, in the context of MRTF-A-SRF pathway, the effect of actin on transcription is negative,16 whereas the role of actin on “general” transcription appears positive.8,9 It will be interesting to assess in the future how these opposing activities, which seem to take place on the genes, are resolved in the nucleus. A plausible explanation is that the actin-bound and actin-free MRTF-A recruits for example different chromatin remodeling complexes. Future studies will reveal whether different genes are differentially sensitive to nuclear actin, and whether, for example, the three RNA polymerases (I,17 II18 and III),19 all of which have been linked to nuclear actin, utilize it similarly.
Which Form for Actin in the Nucleus?
A burning question that has puzzled nuclear actin researchers since the emergence of the field is the way actin is organized in the nucleus. Phalloidin, which binds to actin polymers of at least seven monomers long, does not normally stain the cell nucleus.20 Similarly, no clear filamentous actin structures can be usually seen in the nuclei of cells transfected with GFP-actin. When observing the nuclear import of actin by fluorescence recovery after photobleaching, we observed three phases in the GFP-actin recovery curve. This indicates that there are at least three populations of nuclear actin with different actin turnover rates.9 The two fastest populations to recover could correspond to free actin monomers and to actin polymers as hypothesized also in a previous work.21 The presence of nuclear actin polymers is also supported by functional studies, which have shown for instance that inhibition of actin polymerization blocks transcription.21,22 Similarly, the nuclear presence of motor protein myosins, which can walk along actin filaments, and their link to the transcription process17,23 advocates the existence of polymerized nuclear actin. However, it is intriguing that, in the context of mammary epithelial cells, an actin mutant that does not polymerize was able to rescue the quiescence phenotype.8 Also, the number of phases with unpolymerizable actin mutant in our FRAP assay was reduced to not one but two, hinting that polymerization may not be required for all nuclear interactions of actin. The third population of nuclear actin, not described previously and which shows relatively slow turnover of actin, could correspond to actin bound to nuclear complexes, for instance as part of chromatin remodeling complexes. Deeper analysis of these profiles and especially the third recovery phase should therefore bring helpful information to understand actin dynamics linked with transcription. It will also be equally important to decipher the biochemical nature of nuclear actin interactions, as this would reveal how monomeric or polymeric actin is linked to different nuclear complexes. At least in the BAF24 and Ino8025 complexes, actin seems to be monomeric, but whether this applies generally to all actin containing chromatin remodelers remains to be shown. Although the RNA polymerase complex seems to be very difficult to isolate without actin,18 it is actually very unclear how actin actually associates with it. This underscores the need to identify direct binding partners for actin in the nucleus.
Concluding Remarks
Recent results have shown that nuclear actin levels have a deep impact on transcription processes and consequently on gene expression. Therefore, nuclear actin levels could be used by the cell as a switch to control its general physiological orientation, for instance between a differentiation state and proliferation. How is actin then able to elicit these very profound changes in the nucleus? One answer to this mechanistic paradox could reside on the link between actin and chromatin modifying complexes, i.e., ATP-dependent remodeling complexes,26 but also epigenetic modifiers such as PCAF.27 Such complexes can affect the accessibility of chromatin to transcription factors, and consequently modulate the global transcriptional activity within these regions by changing chromatin folding. Hence, it is tempting to speculate that instead of just being one of their components, actin could act as a modulator of the activity of these general chromatin modifiers. Increased nuclear actin levels could then trigger a concentration-dependent, controlled unfolding of chromatin, resulting in a global positive effect on transcription. This could be further enforced by the association between actin and the RNA polymerase machinery.
Our own studies bring to light the highly active nucleo-cytoplasmic shuttling of actin that governs the connection between the nuclear and cytoplasmic pools of this multitasking protein.9 These results form the solid base for further investigations of the regulatory steps of actin exchange between cellular compartments that allow the cells to drastically modify this balance when the situation requires it. More investigation is required to unravel the actual biological significance of the variability of nuclear actin levels, considering that some of these roles could be specific to certain cell types or species. Hence, revealing how broad and/or specific is the requirement for nuclear actin in different cell biological processes will be an important research direction in the future. Another important step will be to determine the definite molecular mechanisms by which actin operates during the transcription process. Revealing the forms that actin can adopt when localized in the nucleus, and identification of the direct binding partners of nuclear actin should greatly help in this endeavor.
Acknowledgments
The work in the laboratory of MKV is funded by Academy of Finland and Sigrid Juselius foundation. GH is supported by the Finnish Cultural foundation. KPS is funded by a fellowship from the Viikki Graduate School in Biosciences.
Glossary
Abbreviations:
- LN1
Laminin 111
- MRTF-A
Myocardin-Related Transcription factor-A
- NLS
nuclear localization signal
- PCAF
P300/CBP-associated factor
- RNAi
RNA interference
- SRF
Serum Response Factor
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21062
References
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 2.Skarp KP, Vartiainen MK. Actin on DNA-an ancient and dynamic relationship. Cytoskeleton (Hoboken) 2010;67:487–95. doi: 10.1002/cm.20464. [DOI] [PubMed] [Google Scholar]
- 3.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–91. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 4.Clark TG, Rosenbaum JL. An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell. 1979;18:1101–8. doi: 10.1016/0092-8674(79)90223-X. [DOI] [PubMed] [Google Scholar]
- 5.Fukui Y, Katsumaru H. Nuclear actin bundles in Amoeba, Dictyostelium and human HeLa cells induced by dimethyl sulfoxide. Exp Cell Res. 1979;120:451–5. doi: 10.1016/0014-4827(79)90412-9. [DOI] [PubMed] [Google Scholar]
- 6.Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 7.Xu YZ, Thuraisingam T, Morais DA, Rola-Pleszczynski M, Radzioch D. Nuclear translocation of beta-actin is involved in transcriptional regulation during macrophage differentiation of HL-60 cells. Mol Biol Cell. 2010;21:811–20. doi: 10.1091/mbc.E09-06-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, et al. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci. 2011;124:123–32. doi: 10.1242/jcs.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dopie J, Skarp KP, Rajakylä EK, Tanhuanpää K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A. 2012;109:E544–52. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–41. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–40. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohnsack MT, Stüven T, Kuhn C, Cordes VC, Görlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8:257–63. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- 13.Park SH, Park TJ, Lim IK. Reduction of exportin 6 activity leads to actin accumulation via failure of RanGTP restoration and NTF2 sequestration in the nuclei of senescent cells. Exp Cell Res. 2011;317:941–54. doi: 10.1016/j.yexcr.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Gall JG. Exporting actin. Nat Cell Biol. 2006;8:205–7. doi: 10.1038/ncb0306-205. [DOI] [PubMed] [Google Scholar]
- 15.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–96. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–52. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 17.Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–72. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 19.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–5. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vartiainen MK. Nuclear actin dynamics--from form to function. FEBS Lett. 2008;582:2033–40. doi: 10.1016/j.febslet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 21.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–52. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22:322–30. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann WA, Vargas GM, Ramchandran R, Stojiljkovic L, Goodrich JA, de Lanerolle P. Nuclear myosin I is necessary for the formation of the first phosphodiester bond during transcription initiation by RNA polymerase II. J Cell Biochem. 2006;99:1001–9. doi: 10.1002/jcb.21035. [DOI] [PubMed] [Google Scholar]
- 24.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/S0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 25.Fenn S, Breitsprecher D, Gerhold CB, Witte G, Faix J, Hopfner KP. Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J. 2011;30:2153–66. doi: 10.1038/emboj.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrants AK. Chromatin remodelling and actin organisation. FEBS Lett. 2008;582:2041–50. doi: 10.1016/j.febslet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 27.Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol. 2008;28:6342–57. doi: 10.1128/MCB.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


