Abstract
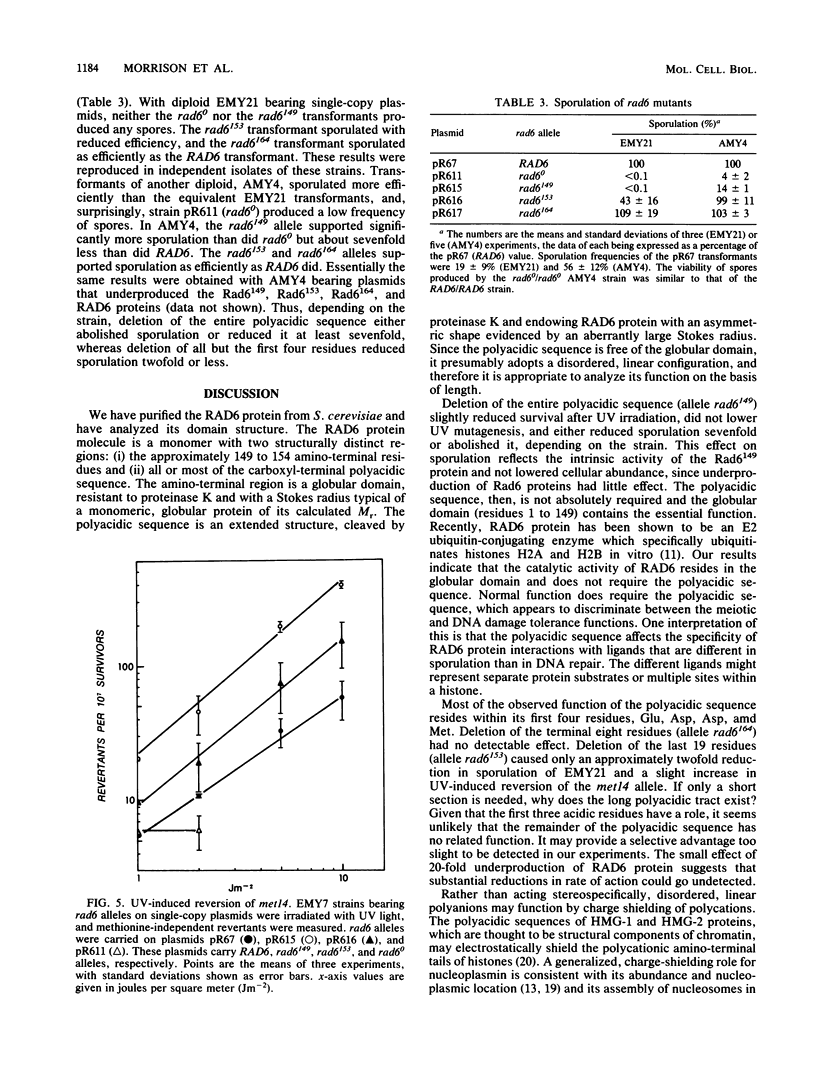
The RAD6 gene of Saccharomyces cerevisiae, which is required for normal tolerance of DNA damage and for sporulation, encodes a 172-residue protein whose 23 carboxyl-terminal residues are almost all acidic. We show that this polyacidic sequence appends to RAD6 protein as a polyanionic tail and that its function in vivo does not require stoichiometry of length. RAD6 protein was purified to near homogeneity from a yeast strain carrying a RAD6 overproducing plasmid. Approximately the first 150 residues of RAD6 protein composed a structural domain that was resistant to proteinase K and had a Stokes radius typical of a globular protein of its calculated mass. The carboxyl-terminal polyacidic sequence was sensitive to proteinase K, and it endowed RAD6 protein with an aberrantly large Stokes radius that indicates an asymmetric shape. We deduce that RAD6 protein is monomeric and comprises a globular domain with a freely extending polyacidic tail. We tested the phenotypic effects of partial or complete deletion of the polyacidic sequence, demonstrating the presence of the shortened proteins in the cell by using antibody to RAD6 protein. Removal of the entire polyacidic sequence severely reduced sporulation but only slightly affected survival after UV irradiation or UV-induced mutagenesis. Strains with deletions of all but the first 4 or 15 residues of the polyacidic sequence were phenotypically almost wild type or wild type, respectively. We conclude that the intrinsic activity of RAD6 protein resides in the globular domain, that the polyacidic sequence has a stimulatory or modifying role evident primarily in sporulation, and that only a short section apparently functions as effectively as the entire polyacidic sequence.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Haber J. E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986 Jul;113(3):551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary P. D., Turner C. H., Leung I., Mayes E., Crane-Robinson C. Conformation and domain structure of the non-histone chromosomal proteins HMG 1 and 2. Domain interactions. Eur J Biochem. 1984 Sep 3;143(2):323–330. doi: 10.1111/j.1432-1033.1984.tb08375.x. [DOI] [PubMed] [Google Scholar]
- Cox B. S., Parry J. M. The isolation, genetics and survival characteristics of ultraviolet light-sensitive mutants in yeast. Mutat Res. 1968 Jul-Aug;6(1):37–55. doi: 10.1016/0027-5107(68)90101-2. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Dilworth S. M., Black S. J., Kearsey S. E., Cox L. S., Laskey R. A. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1987 Jan;6(1):69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Honda B. M., Laskey R. A., Thomas J. O. Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell. 1980 Sep;21(2):373–383. doi: 10.1016/0092-8674(80)90474-2. [DOI] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974 Sep;24(3):281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Game J. C., Zamb T. J., Braun R. J., Resnick M., Roth R. M. The Role of Radiation (rad) Genes in Meiotic Recombination in Yeast. Genetics. 1980 Jan;94(1):51–68. doi: 10.1093/genetics/94.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Kohn J., Wilchek M. A new approach (cyano-transfer) for cyanogen bromide activation of Sepharose at neutral pH, which yields activated resins, free of interfering nitrogen derivatives. Biochem Biophys Res Commun. 1982 Aug;107(3):878–884. doi: 10.1016/0006-291x(82)90604-0. [DOI] [PubMed] [Google Scholar]
- Krohne G., Franke W. W. Immunological identification and localization of the predominant nuclear protein of the amphibian oocyte nucleus. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1034–1038. doi: 10.1073/pnas.77.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Stewart J. W., Sherman F., Christensen R. Specificity and frequency of ultraviolet-induced reversion of an iso-1-cytochrome c ochre mutant in radiation-sensitive strains of yeast. J Mol Biol. 1974 May 5;85(1):137–162. doi: 10.1016/0022-2836(74)90134-x. [DOI] [PubMed] [Google Scholar]
- Mamrack M. D., Olson M. O., Busch H. Amino acid sequence and sites of phosphorylation in a highly acidic region of nucleolar nonhistone protein C23. Biochemistry. 1979 Jul 24;18(15):3381–3386. doi: 10.1021/bi00582a026. [DOI] [PubMed] [Google Scholar]
- McKee R. H., Lawrence C. W. Genetic analysis of gamma-ray mutagenesis in yeast. I. Reversion in radiation-sensitive strains. Genetics. 1979 Oct;93(2):361–373. doi: 10.1093/genetics/93.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A. D., Laskey R. A., Black P., De Robertis E. M. An acidic protein which assembles nucleosomes in vitro is the most abundant protein in Xenopus oocyte nuclei. J Mol Biol. 1980 May 25;139(3):561–568. doi: 10.1016/0022-2836(80)90148-5. [DOI] [PubMed] [Google Scholar]
- Pentecost B. T., Wright J. M., Dixon G. H. Isolation and sequence of cDNA clones coding for a member of the family of high mobility group proteins (HMG-T) in trout and analysis of HMG-T-mRNA's in trout tissues. Nucleic Acids Res. 1985 Jul 11;13(13):4871–4888. doi: 10.1093/nar/13.13.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184(3):471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- Prakash L. Lack of chemically induced mutation in repair-deficient mutants of yeast. Genetics. 1974 Dec;78(4):1101–1118. doi: 10.1093/genetics/78.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Weber S., Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985 Jan;82(1):168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966 Feb 7;112(2):346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]