Abstract
Vascular wilts are among the most destructive plant diseases that occur in annual crops as well as in woody perennials. These diseases are generally caused by soil-borne bacteria, fungi, and oomycetes that infect through the roots and enter the water-conducting xylem vessels where they proliferate and obstruct the transportation of water and minerals. As a consequence, leaves wilt and die, which may lead to impairment of the whole plant and eventually to death of the plant. Cultural, chemical, and biological measures to control this group of plant pathogens are generally ineffective, and the most effective control strategy is the use of genetic resistance. Owing to the fact that vascular wilt pathogens live deep in the interior of their host plants, studies into the biology of vascular pathogens are complicated. However, to design novel strategies to combat vascular wilt diseases, understanding the (molecular) biology of vascular pathogens and the molecular mechanisms underlying plant defense against these pathogens is crucial. In this review, we discuss the current knowledge on interactions of vascular wilt pathogens with their host plants, with emphasis on host defense responses against this group of pathogens.
Keywords: xylem, defense response, immunity, innate, pathogen, fungi, bacteria
INTRODUCTION
Plants are continuously confronted with potential pests and pathogens that include insects, nematodes, viruses, bacteria, fungi, and oomycetes. While many of these organisms evolved to infect aerial plant parts, such as leaves, stems, flowers, and fruits, others target below-ground organs, such as roots and tubers. Specific pathogens target the vascular system that is composed of xylem vessels, tracheary elements that transport water and minerals that are absorbed by the roots to the photosynthetic organs, and phloem elements, the living tissue that transports organic photosynthesis products. Paradoxically, although the phloem is rich in sugars, most vascular pathogens colonize the nutrient-poor xylem vessels. This may be explained by the accessibility of both types of vessel elements, as the phloem is characterized by living cells with a high osmotic pressure which makes penetration difficult, while the xylem is composed of dead tracheary elements with relatively low osmotic pressure. Consequently, phloem pathogens comprise rickettsias, spiroplasmas, and phytoplasmas that are introduced by vectors such as phloem feeding insects, or by cultural practices like grafting.
Xylem-invading pathogens comprise bacterial, fungal, and oomycete microorganisms that cause vascular wilt diseases. Vascular wilt pathogens are among the most destructive plant pathogens that can wipe out entire crops. Vascular wilt diseases occur worldwide and affect annual crops as well as woody perennials, thus not only impacting food and feed production, but also natural ecosystems. Most of the symptoms caused by vascular wilt pathogens develop in acropetal direction: from bottom to top. Epinasty is the primary disease symptom, followed by flaccidity, chlorosis, vascular browning, and necrostress tolerance when compared with non-stress tolerance when compared with non-sis of the terminal leaflets (Agrios, 2005). A large range of symptoms is caused by vascular wilt pathogens, and the same pathogen may cause different symptoms on different host plants. Depending on the pathogen species and the host, plants may become stunted, wilt partially or completely, and ultimately die. Plant death may occur within days to weeks or, in case of perennials, months to years (Purcell and Hopkins, 1996; Fradin and Thomma, 2006; Niño-Liu et al., 2006; Juzwik et al., 2008; Klosterman et al., 2009, 2011; Michielse and Rep, 2009; Genin, 2010; Janse and Obradovic, 2010; Harwood et al., 2011). Age, fitness, and the nutritional status of the host, environmental conditions, and virulence of the pathogen can all determine the speed and severity at which symptoms develop (Tjamos and Beckman, 1989; Hayward, 1991; Roncero et al., 2003; Niño-Liu et al., 2006; Chatterjee et al., 2008). In all cases where it is observed, wilting symptoms represent a transitory phase of the disease.
Vascular wilt pathogens generally overwinter in soil, plant debris, watercourses, or in insect vectors (Fradin and Thomma, 2006; Niño-Liu et al., 2006; Juzwik et al., 2008; Klosterman et al., 2009,2011; Michielse and Rep, 2009; Genin, 2010; Janse and Obradovic, 2010; Nadarasah and Stavrinides, 2011). While most vascular wilt pathogens are soil-borne and enter their hosts through the roots by penetration via wounds or cracks that appear at the sites of lateral root formation (Vicente et al., 2001; Di Pietro et al., 2003; Fradin and Thomma, 2006; Klosterman et al., 2009; Michielse and Rep, 2009; Genin, 2010), some enter leaves via natural openings such as stomata and hydathodes, such as the bacterial leaf blight pathogen of rice, Xanthomonas oryzae (Niño-Liu et al., 2006). Furthermore, some vascular wilt pathogens are delivered directly into the xylem by insect vectors that feed on xylem sap, such as Xylella fastidiosa bacteria that are transmitted by leafhoppers, or by chewing insects, such as Ophiostoma fungi that are transmitted by bark beetles (Purcell and Hopkins, 1996; Chatterjee et al., 2008; Moser et al., 2010; Nadarasah and Stavrinides, 2011). Regardless of the mechanism used by vascular wilt pathogens to enter their hosts, they subsequently colonize the xylem vessels where they proliferate (Tjamos and Beckman, 1989; Purcell and Hopkins, 1996; Agrios, 2005; Niño-Liu et al., 2006; Klosterman et al., 2009; Genin, 2010).
CONTROL OF VASCULAR WILT DISEASES
Controlling vascular wilt pathogens is difficult for several reasons. First of all, no efficient treatments exist to cure infected plants, and growers generally have to remove them from their crops. Secondly, many vascular wilt pathogens are soil-borne and produce persistent resting structures that are able to survive for long periods of time in the absence of host plants. Thirdly, some of these pathogens can infect a broad range of host plants and as a consequence, cultural control measures such as crop rotation are generally not very effective. Resting structures are desirable targets for control by soil solarization and chemical fumigation. However, limitations in large-scale applicability and ban on chemical fumigants because of public health and environmental issues render these approaches unsuitable. Biological agents and organic soil amendments are used to control vascular wilt diseases (Tsuda et al., 2001; Spadaro and Gullino, 2005; Suárez-Estrella et al., 2007; Ji et al., 2008; Markakis et al., 2008). For instance, injection of the Dutch trig, a bio-control vaccine that contains conidia of a non-pathogenic strain of the vascular wilt fungus Verticillium albo-atrum isolate, into elm trees is used to induce the natural defense against Dutch elm disease caused by the fungi O. ulmi and O. novo-ulmi (Scheffer et al., 2008). However, since biological agents are often affected by biotic and abiotic factors, performance of bio-control microorganisms in the field is often inconsistent (Tsuda et al., 2001).
The most effective strategy to control vascular wilt diseases thus far is the use of genetic resistance in host plants. Due to the fact that vascular wilt pathogens live deep in the interior of their host plants, studies into the biology of vascular pathogens is complicated. However, their high economic impact, combined with the absence of curative treatments, justifies increased attention. The recent availability of a number of genome sequences of vascular pathogens has inspired novel research efforts to unravel the molecular basis of vascular wilt diseases (Table 1). To design novel strategies to combat vascular wilt diseases, understanding the (molecular) biology of vascular pathogens and the molecular mechanisms underlying plant defense against these pathogens is crucial.
Table 1.
Publically available genome sequences of vascular wilt pathogens.
| Organism | Species | Reference |
|---|---|---|
| Fungus | Verticillium dahliae | Klosterman et al. (2011) |
| Verticillium albo-atrum | Klosterman et al. (2011) | |
| Fusarium oxysporum f. sp. lycopersici | Ma et al. (2010) | |
| Bacterium | Ralstonia solanacearum | Salanoubat et al. (2002) |
| Xanthomonas oryzae pv. oryzae | Lee et al. (2005) | |
| Xanthomonas campestris pv.\hb campestris | Qian et al. (2005) | |
| Xylella fastidiosa | Simpson et al. (2000) | |
| Clavibacter michiganensis ssp. michiganensis | Gartemann et al. (2008) | |
| Erwinia amylovora | Sebaihia et al. (2010), Smits et al. (2010), Powney et al. (2011) | |
| Oomycete | Pythium ultimum | Lévesque et al. (2010) |
CAUSAL AGENTS OF VASCULAR WILT DISEASES
FUNGAL VASCULAR WILT PATHOGENS
There are four fungal genera containing the major vascular wilt pathogens: Ceratocystis (vascular wilts of oak, cocoa, and eucalyptus), Ophiostoma (vascular wilts of elm trees), Verticillium (broad host range), and Fusarium (broad host range; Tjamos and Beckman, 1989; Agrios, 2005; Juzwik et al., 2008; Schumann and D’Arcy, 2010; Harwood et al., 2011; López-Escudero and Mercado-Blanco, 2011). In contrast to the other three genera, the vast majority of Fusarium vascular wilt pathogens all belong to a single species, F. oxysporum, which contains morphologically indistinguishable pathogenic and non-pathogenic strains (Lievens et al., 2008). The pathogenic strains cause vascular wilts or root rot in over 100 different host species (Di Pietro et al., 2003; Roncero et al., 2003; Michielse and Rep, 2009). Despite the broad host range of these species, individual strains typically infect only a single or a few hosts and are assigned to formae speciales (Michielse and Rep, 2009). Interestingly, it was experimentally demonstrated that the transfer of two lineage specific (LS) chromosomes from a tomato pathogenic F. oxysporum f. sp. lycopersici strain to a non-pathogenic strain converted the latter into a tomato pathogen, suggesting that host specificity within F. oxysporum may be determined by pathogenicity chromosomes (Ma et al., 2010). Such pathogenicity chromosomes have not been identified in vascular wilt pathogens of the Verticillium genus for which genome sequences have recently been determined as well (Klosterman et al., 2011).
Most fungal vascular wilt pathogens overwinter as resting structures in the soil or on dead host tissues. These include microsclerotia, chlamydospores, thick-walled mycelium, and spore-bearing coremia that all can survive for an extended period of time without losing viability. Compounds released from host plants, referred to as exudates, trigger germination of these resting structures. Except for Ophiostoma spp. and the oak wilt pathogen Ceratocystis fagacearum that are transmitted by beetles (Hayslett et al., 2008; Juzwik et al., 2008; Harwood et al., 2011), fungal vascular wilt pathogens enter their host plants through the roots. Following penetration, the fungi colonize the cortical cells from where hyphae migrate intercellularly toward the vascular parenchyma cells and invade the xylem vessels (Di Pietro et al., 2003; Klosterman et al., 2009; Schumann and D’Arcy, 2010; Nadarasah and Stavrinides, 2011). In the xylem, conidiospores are produced which are disseminated acropetally with xylem sap movement. Fungal vascular wilt pathogens are mostly restricted to xylem vessels, but once host tissues become necrotized also these are colonized and the fungus starts to produce resting structures which are released into the soil eventually (Di Pietro et al., 2003; Agrios, 2005; Fradin and Thomma, 2006).
BACTERIAL VASCULAR WILT PATHOGENS
Seven bacterial genera contain vascular wilt pathogens: Clavibacter (causing ring rot of potato and bacterial canker and wilt of tomato), Curtobacterium (bacterial wilt of beans), Erwinia (bacterial wilt of cucurbits), Pantoea (stewart’s wilt of corn), Ralstonia (southern bacterial wilt of Solanaceous crops and Moko disease of banana), Xanthomonas (black rot of crucifers, bacterial blight of rice), and Xylella (Pierce’s disease of grape, citrus variegation chlorosis; Tjamos and Beckman, 1989; Agrios, 2005; Chatterjee et al., 2008; Schumann and D’Arcy, 2010; Nadarasah and Stavrinides, 2011; Roper, 2011). Bacterial vascular wilt pathogens overwinter in plant debris in soil, in seeds, in vegetative propagules, or in their insect vectors as dormant cells (Agrios, 2005). They enter host tissues only passively, via wounds, cracks, or natural openings such as stomata and hydathodes, while others are directly delivered into the xylem by insect vectors, such as Xylella fastidiosa by sharpshooter leafhoppers and spittlebugs, Pantoea stewartii by corn flea beetles and E. tracheiphila by cucumber beetles (Schumann and D’Arcy, 2010; Nadarasah and Stavrinides, 2011; Roper, 2011). After entrance, they rapidly multiply and invade the root cortex and vascular parenchyma intercellularly, from where they spread to the xylem vessels that are used as avenues for passive spread to aerial plant parts. During colonization, bacterial wilt pathogens degrade xylem cell wall components, parenchyma cells, and pit membranes, resulting in slimy masses of bacteria and cellular debris (Agrios, 2005; Schumann and D’Arcy, 2010).
OOMYCETE VASCULAR WILT PATHOGENS
Only one oomycete genus, Pythium, contains vascular wilt pathogens. Pythium mainly infects seeds or seedlings in the soil, causing pre-emergence or post-emergence seedling damping-off disease, and young and juvenile plant tissues (Martin and Loper, 1999; Oliver et al., 2009). The genus Pythium comprises many complex species, most of which are plant pathogens, while others are saprophytes or animal parasites (Martin and Loper, 1999). Pythium species survive in soil or in organic substrates for long periods of time as dormant oospores; thick-walled sexual spores that can withstand harsh environmental conditions (Martin and Loper, 1999). Oospores germinate upon stimulation by exudates released from plants, and often produce a sporangium containing zoospores that are released and encysted after host contact. Alternatively, oospores produce germinating hyphae to penetrate the root epidermis, migrate through the cortex, endodermis, and parenchyma cells, and eventually invade the vascular stele causing typical damping-off symptoms (Rey et al., 1998).
XYLEM STRUCTURE AND DEVELOPMENT
The xylem consists of distinct cells with special wall structures that allow efficient transport of water and solutes from the roots to upper plant parts. The xylem functions not only in long distance transport, but also provides physical strength to the plant. Xylem development occurs in two phases during which primary and secondary xylem is produced (Fukuda, 1997,2004; Ye, 2002; Zhang et al., 2011). Primary development involves the formation of primary xylem from procambium cells which are derived from the apical meristem. Procambium cells give rise to xylem precursor cells that eventually differentiate into treachery elements, xylem parenchyma cells or fiber cells; collectively called the xylem (Ye, 2002; Fukuda, 2004). Treachery elements, which consist of tracheid and vessel elements, are the main conductive tissues. While the xylem parenchyma cells are metabolically active and adapted for storage and transport, the xylem fiber cells together with treachery elements provide physical support (Nieminen et al., 2004). Following xylem differentiation, the treachery elements undergo cell elongation before the initiation of secondary xylem wall development (Ye, 2002; Fukuda, 2004; Nieminen et al., 2004; Zhang et al., 2011). The secondary xylem walls, which are derived from vascular cambium, are deposited onto the primary xylem walls (Fukuda, 1997; De Boer and Volkov, 2003). Secondary xylem is made of cellulose microfibrils, crystalline aggregates of linear polymers of D-glucopyranosyl residues linked in β-(1-4) conformation (Brett, 2000; Emons and Mulder, 2000). The secondary xylem walls are further impregnated with different polysaccharides, such as lignin, hemicellulose, pectin, and structural proteins that add strength and rigidity to the wall (Ye, 2002; Fukuda, 2004; Yokoyama and Nishitani, 2006). Subsequently, the secondary xylem walls are lignified, cross-linked, and eventually waterproofed by polymerization of the aromatic compound monolignol (Fukuda, 1996; De Boer and Volkov, 2003). The patterned secondary xylem walls provide physical strength to the treachery elements to withstand the negative pressure generated during transpiration and by the compressive pressure from surrounding cells (Ye, 2002; Nieminen et al., 2004; Choat and Pittermann, 2009; Zhang et al., 2011).
The final step of xylem development is the induction of programed cell death (PCD) that destroys the cellular contents of treachery elements, leaving behind hollow tube-like vessels through which water and nutrients flow (Fukuda, 1997; Zhang et al., 2011). The PCD is developmentally regulated and is strongly associated with secondary xylem wall formation (Fukuda, 2004). The vessel tubes are dedicated to the unrestricted water and solute movement throughout the plant and individual vessels are interconnected through small openings called pits (De Boer and Volkov, 2003; Choat and Pittermann, 2009). Pits between vessels typically have overarching secondary walls that form a bowl-shaped chamber, referred to as a border pit (De Boer and Volkov, 2003; Jansen et al., 2004). Border pit exists in pairs and contain a pit membrane at the center, which is formed from primary walls and the intervening middle lamella (De Boer and Volkov, 2003). The pit membrane is made of cellulose microfibrils embedded in polysaccharide matrix of hemicellulose and pectin (Tyree and Zimmermann, 2002; Pérez-Donoso et al., 2010). This fine mesh-like and tightly interlocked polysaccharide structure has minute openings through which water and solutes can move with a minimal resistance between vessels or to neighboring parenchyma cells (Choat and Pittermann, 2009). In angiosperm trees, the pit pore diameter varies between 5 and 20 nm (Choat et al., 2003,2004), thus acting as a safety mechanism to limit the spread of embolism within xylem vessels (Tyree and Zimmermann, 2002; De Boer and Volkov, 2003; Choat et al., 2008; Pérez-Donoso et al., 2010).
All vascular wilt pathogens have to breach the highly structured and rigid secondary xylem walls to enter the vessels. Also the pit membranes are a major barriers for vascular pathogens as vascular wilt pathogens are too large to pass pit membrane pores (Choat et al., 2003,2004). For example, the rod-shaped bacterium Xylella fastidiosa has a cell size of 0.25–0.5 μm in diameter (Mollenhauer and Hopkins, 1974), while the conidia of Verticillium species have a diameter of about 2.2 μm (Qin et al., 2008).
THE XYLEM AS A NICHE FOR VASCULAR WILT PATHOGENS
The xylem is a nutritionally poor environment, which could be an important reason why only a limited number of plant pathogens are able to thrive in this environment. Possibly, vascular wilt pathogens exploit this niche to avoid competition with other microbes (McCully, 2001). However, as they reside in the xylem for the major part of their lifecycle, vascular pathogens need to be able to obtain all factors required for growth, reproduction, and survival.
NUTRIENT COMPOSITION OF XYLEM SAP
Nitrate, sulfate, and phosphate are among the most abundant inorganic anions in the xylem sap, whereas calcium, potassium, magnesium, and manganese are the most predominate inorganic cations present in the xylem sap of oilseed rape (Nakamura et al., 2008). Although only in relatively low amounts, xylem sap also contains various carbohydrates, such as glucose, fructose, saccharose, maltose, raffinose, trehalose, and ribose (Alvarez et al., 2008; Nakamura et al., 2008; Fernandez-Garcia et al., 2011; Krishnan et al., 2011). Of these, glucose, fructose, and saccharose are predominant and are utilized as a carbon source for growth. Xylem sap furthermore contains various proteins, amino acids, and organic acids, which can also act as source of organic and inorganic nutrients (Alvarez et al., 2008; Nakamura et al., 2008; Fernandez-Garcia et al., 2011; Krishnan et al., 2011). For instance, the sulfur-containing amino acids methionine and cysteine can be used as a source of inorganic sulfur (Divon and Fluhr, 2007; Krishnan et al., 2011). Nevertheless, the quantities of the organic and inorganic compounds in the xylem sap are extremely low and fluctuate with day time, growth condition, and plant species (Siebrecht et al., 2003).
NUTRIENT ACQUISITION BY VASCULAR WILT PATHOGENS
Vascular wilt pathogens satisfy their nutritional requirements by efficiently acquiring the scarce nutrients available in the xylem sap, by enzymatic digestion of host cell walls, by invading neighboring cells, or by inducing nutrient leakage from surrounding tissues (Divon et al., 2005; Möbius and Hertweck, 2009; Klosterman et al., 2011).
Nitrogen is one of the limiting nutrients in the xylem sap for vascular wilt pathogens (Divon et al., 2005). The preferred primary nitrogen sources for fungal pathogens, ammonia, glutamine, and glutamate (Marzluf, 1997; Divon et al., 2006), are scarce in xylem sap. In absence of primary nitrogen sources, fungi can utilize secondary nitrogen sources such as nitrate, nitrite, purines, amides, amino acids, and proteins (Marzluf, 1997; Divon et al., 2005,2006). GATA transcription factors, such as the F. oxysporum f. sp. lycopersici global nitrogen regulator (FNR1), are known to regulate utilization of secondary nitrogen sources (Marzluf, 1997; Divon and Fluhr, 2007; Bolton and Thomma, 2008; Donofrio et al., 2009). FNR1 mutants grow normally on primary nitrogen sources, but fail to utilize secondary nitrogen sources such as amino acids, hypoxanthine, and uric acid (Divon et al., 2006). Disruption of FNR1 not only affected virulence of F. oxysporum f. sp. lycopersici on tomato, but also regulation of three nitrogen acquisition genes, Gap1, Mtd1, and Uricase during growth in planta, suggesting that FNR1 regulates the utilization of secondary nitrogen sources in planta (Divon et al., 2006).
Analysis of the whole genome sequences of F. oxysporum f. sp. lycopersici, V. dahliae, and V. albo-atrum showed that these genomes are enriched in genes that encode cell wall-degrading enzymes (CWDEs) that may be used for the enzymatic digestion of xylem walls and pit membranes (Ma et al., 2010; Klosterman et al., 2011). Also other vascular wilt pathogens are known as CWDE producers (Di Pietro et al., 2003; Jha et al., 2005; Sun et al., 2005; Fradin and Thomma, 2006; Michielse and Rep, 2009; Klosterman et al., 2011). While degrading cell wall components, these enzymes liberate sugars that may be used as nutrient sources.
Various vascular wilt pathogens produce high- and low-molecular weight phytotoxins during host colonization that have often been associated with wilt symptom development (Temple and Horgen, 2000; Wang et al., 2004; Palmer et al., 2005; Stipanovic et al., 2011; Zhou et al., 2012; Santhanam et al., 2013). As several phytotoxins disturb plant cell membrane integrity (Möbius and Hertweck, 2009), leakage of nutrients may occur from cells surrounding the xylem vessels that can be utilized by vascular wilt pathogens. For instance, two Verticillium necrosis- and ethylene inducing-like proteins, NLP1 and NLP2, were shown to display cytotoxic activity and differentially contribute to virulence on various host plant species, although the mechanism through which these NLPs contribute to virulence remains unclear (Zhou et al., 2012; Santhanam et al., 2013). Interestingly, compared with other ascomycete plant pathogens that typically contain up to three NLP genes, the NLP gene family is expanded in the V. dahliae genome (Klosterman et al., 2011; Santhanam et al., 2013). A similar expansion has been reported for the F. oxysporum genome, and it has been speculated that this expansion has contributed to their broad host range among dicotyledonous plant hosts (Ma et al., 2010; Klosterman et al., 2011). However, in addition to NLP1 and NLP2, none of the other V. dahliae NLPs were found to display cytotoxic activity, and their potential role in fungal virulence still remains enigmatic (Zhou et al., 2012; Santhanam et al., 2013).
Finally, although most vascular wilt pathogens are confined to xylem vessels, some of them degrade xylem vessel walls to colonize adjacent parenchyma cells (Agrios, 2005). These pathogens may obtain nutrition by parasitizing parenchyma cells.
PLANT DEFENSE AGAINST VASCULAR WILT PATHOGENS
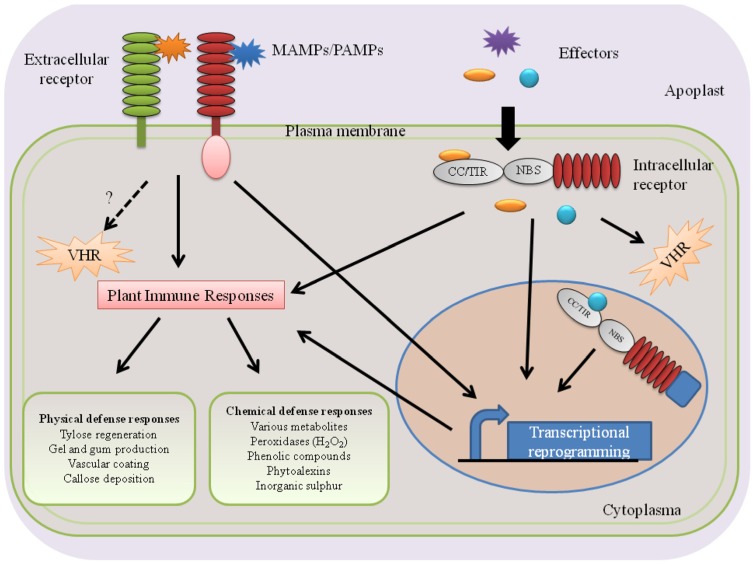
Plants deploy two types of defenses against invading pathogens: pre-existing and inducible plant defense responses. The pre-existing defenses are constitutive and provide physical and chemical barriers against attempted host penetration. Once successful pathogens breach pre-existing defenses, they encounter a spectrum of inducible defense responses with microbe-associated molecular pattern (MAMP)-triggered immunity (MTI) and effector-triggered immunity (ETI) as two extreme ends (Jones and Dangl, 2006; Dodds and Rathjen, 2010). While MTI is activated upon recognition of conserved MAMPs, ETI is activated upon recognition of secreted effector proteins (Figure 1).
FIGURE 1.
Perception of vascular wilt pathogens and activation of subsequent plant immune responses. Plants perceive PAMPs/MAMPs or effector proteins of vascular wilt pathogens using extracellular or intracellular receptors and activate immune responses in the xylem. The tomato receptor-like protein Ve1 and the rice receptor-like kinase Xa21 are examples of extracellular receptors that recognize Verticillium Ave1 and Xanthomonas oryzae pv. oryzae Xa21, respectively. Tomato I-2 and Arabidopsis RRS1-R are examples of intracellular NB–LRR-type receptors that perceive the F. oxysporum f. sp. lycopersici Avr2 effector and the R. solanacearum effector PopP2, respectively. Presumably, these processes take place in the parenchyma cells surrounding the xylem vessels.
PERCEPTION OF VASCULAR WILT PATHOGENS
In general, plants sense invading pathogens by using two types of receptors: extra- and intracellular receptors (Figure 1). While extracellular receptors recognize pathogen molecules on the cellular surface as well as damage-associated host molecules that are released as a consequence of pathogen activity, intracellular receptors recognize pathogen molecules that are delivered inside host cells (Figure 1). This extra- and intracellular receptor-mediated recognition of pathogen molecules [microbe-associated molecular patterns (MAMPs)/pathogen-associated molecular patterns (PAMPs), effectors] leads to the activation of plant innate immunity that wards off invading pathogens. Consequently, failure of a host plant to perceive invading pathogens leads to susceptibility and successful pathogen infections (Figure 2).
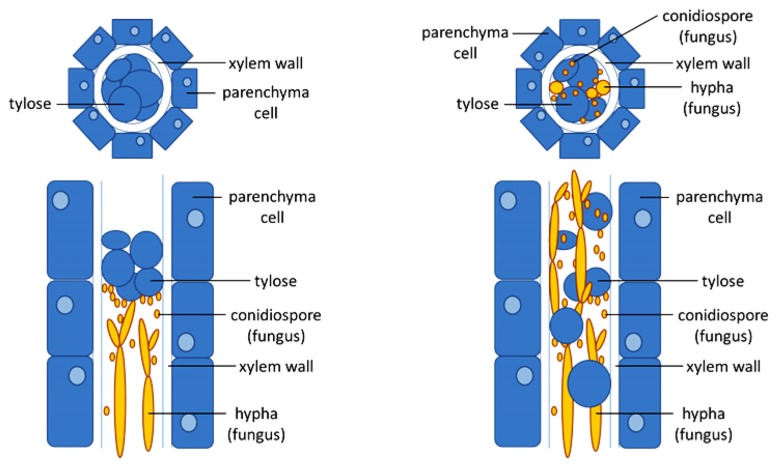
FIGURE 2.
Xylem occlusion limits pathogen growth in resistant plants. Schematic drawing of cross-sections (top) or longitudinal sections (bottom) of a fungal-infected xylem vessel of a resistant (left) and a susceptible (right) plant. In the resistant plant, timely induction of the formation of tyloses, bubble-like outgrowth of the parenchyma contact cells surrounding the xylem vessels that protrude into the lumen of the vessel, are able to trap the fungus after which elimination can occur. As long as the number of vessels that is closed by the tyloses is limited, the host plant will not suffer from droughts stress. In the susceptible plant, tylose formation cannot trap the pathogen which is able to spread and further colonize the xylem.
Extracellular plant receptors
Several MAMP receptors have been characterized, including Arabidopsis FLS2 (flagellin-sensitive 2), EFR (elongation factor Tu receptor), and CERK1 (chitin elicitor receptor kinase 1), and rice CEBiP (chitin elicitor binding protein). While FLS2 and EFR encode receptor-like kinases (RLKs) that recognize the bacterial MAMPs flg22 and EF-Tu, respectively (Gómez-Gómez et al., 2001; Kunze et al., 2004; Zipfel et al., 2006), CERK1 and CEBiP encode LysM domain-containing receptors that recognize chitin, the main constituent of fungal cell walls (Gómez-Gómez et al., 2001; Kunze et al., 2004; Zipfel et al., 2006). These MAMP receptors are considered to display a low degree of specificity and broadly act in pathogen defense.
Extracellular plant receptors that play a role in plant defense against specific vascular wilt pathogens have also been described. Rice Xa21 confers resistance against Xanthomonas oryzae pv. oryzae (Xoo; Song et al., 1995). Xa21 recognizes Ax21 (activator of Xa21), a type I-secreted sulfated protein (Song et al., 1995; Lee et al., 2009). Similar to FLS2 and EFR, Xa21 encodes a RLK (Song et al., 1995; Park et al., 2010b). Xa21 physically interacts with XB24, a protein with a C-terminal ATP synthase (ATPase) motif (Chen et al., 2010). XB24 promotes autophosphorylation of Ser/Thr residues on Xa21 through its ATPase activity, keeping Xa21 in an inactive state (Chen et al., 2010; Chen and Ronald, 2011). Upon Xa21-mediated Ax21 recognition, the Xa21 kinase becomes activated, triggering rice defense responses (Chen et al., 2010; Park et al., 2010b). XB25, plant-specific ankyrin-repeat family protein (Jiang et al., 2012), and BiP3, an endoplasmic reticulum chaperone protein (Park et al., 2010a), are also reported to be involved in Xa21-mediated rice immunity against Xoo.
Tomato Ve1 is another example of extracellular plant receptor that plays a role in xylem defense. Ve1 is an extracellular leucine-rich repeat (LRR) receptor-like protein (RLP; Kawchuk et al., 2001; Wang et al., 2008) that provides resistance against race 1 isolates of V. dahliae and V. albo-atrum (Fradin et al., 2009,2011). Interestingly, Fradin et al. (2011) have recently shown that interfamily transfer of Ve1 gene to Arabidopsis confers resistance against race 1 isolates of V. dahliae and V. albo-atrum. Recently, the pathogen ligand that is perceived by Ve1 was identified as the Ave1 effector, a small (134 aa) effector protein with four cysteines that is required for full virulence on tomato plants lacking Ve1 (De Jonge et al., 2012). Ave1 is homologous to a widespread family of plant natriuretic peptides, mobile signaling molecules that play a role in the regulation of water and ion homeostasis, and it was suggested that Ave1 was acquired by Verticillium through horizontal gene transfer from plants (De Jonge et al., 2012). Ave1 homologs are found in a few other plant pathogens, including the vascular wilt fungus F. oxysporum f. sp. lycopersici, but a role in virulence has not yet been demonstrated for these homologs (De Jonge et al., 2012). Intriguingly, the Ave1 homolog from F. oxysporum f. sp. lycopersici is recognized by Ve1 upon transient co-expression in tobacco, and Ve1 was found to mediate resistance toward F. oxysporum f. sp. lycopersici in tomato (De Jonge et al., 2012).
Recently, Zhang et al. (2013) have reported the presence of a functional Ve1 ortholog in the tobacco species Nicotiana glutinosa, as Ave1 expression in N. glutinosa causes an hypersensitive response (HR), rapid and localized cell death of plant tissue surrounding the site where recognition of pathogen effectors by host immune receptors occurs. Furthermore, N. glutinosa shows resistance against race 1 V. dahliae that is compromised upon inoculation with an Ave1 deletion mutant of a race 1 V. dahliae isolate (Zhang et al., 2013).
Intracellular plant receptors
Also intracellular plant receptors that mediate plant defense against xylem-invading pathogens have been characterized. The tomato I-2 gene is an intracellular receptor that contributes to resistance against race 2 isolates of F. oxysporum f. sp. lycopersici (Huang and Lindhout, 1997; Takken and Rep, 2010). It encodes a cytoplasmic CC (coiled-coil)–NBS (nucleotide-binding site)–LRR receptor protein that recognizes the effector protein Avr2, which was initially identified from the xylem sap of tomato infected by F. oxysporum f. sp. lycopersici and is taken up by tomato cells (Houterman et al., 2007,2009; Takken and Rep, 2010).
The Arabidopsis RRS1-R resistance protein is an intracellular plant receptor that confers resistance against R. solanacearum. RRS1-R encodes a TIR (Toll/interleukin1 receptor)–NBS–LRR R-protein and contains a C-terminal nuclear localization signal (NLS) and a WRKY domain (Deslandes et al., 2002). It recognizes the R. solanacearum type III-secreted effector protein PopP2 (Deslandes et al., 2003). RRS1-R physically interacts with the effector PopP2 (Deslandes et al., 2003). RRS1-R requires RD19, a cysteine protease that also binds to PopP2 (Deslandes et al., 2003; Bernoux et al., 2008). RD19 is localized in the vacuole in absence of PopP2 and re-localizes to the nucleus in the presence of PopP2 (Deslandes et al., 2003; Bernoux et al., 2008). However, no direct interaction between RRS1-R and RD19 has been reported so far. Thus, the current notion is that RRS1-R potentially recognizes the RD19/PopP2 complex in the nucleus and activates the Arabidopsis ETI against R. solanacearum.
PLANT DEFENSE RESPONSES IN THE XYLEM
Comparison of the transcriptional changes in tomato induced by Cladosporium fulvum, a fungal foliar pathogen that causes leaf mold of tomato, with those induced by the vascular wilt pathogen V. dahliae revealed that the Cladosporium fulvum-induced transcriptional changes showed little overlap with those induced by V. dahliae (van Esse et al., 2009). Moreover, within the subset of genes that are regulated by both pathogens, many genes showed inverse regulation (van Esse et al., 2009).
Recognition of vascular wilt pathogens mediated by either extracellular or intracellular receptors leads to the activation of defense responses in the xylem vessels. These comprise physical defense responses which halt or contain the pathogen from further spread in the xylem vessels, and chemical defense responses that kill the pathogen or inhibit its growth (Figure 2).
A common defense mechanism in xylem vessels against vascular wilt pathogens is the formation of tyloses (Beckman, 1964; Talboys, 1972; Rahman et al., 1999; Fradin and Thomma, 2006). Tyloses are outgrowths of vessel-associated parenchyma cells which protrude into the xylem vessel through pits and block the spread of pathogens (Beckman, 1964; Talboys, 1972; Grimault et al., 1994; Agrios, 2005). They are formed during both compatible and incompatible interactions between the host and vascular wilt pathogens, although the time and extent of tylose formation significantly differs (Figure 2). Tyloses form much faster and more extensively in resistant plants when compared to susceptible plants (Grimault et al., 1994; Fradin and Thomma, 2006).
Often, the generation of tyloses is associated with the production of gels and gums around the differentiated tylose (Clérivet et al., 2000). Using immuno-gold labeling, strong accumulation of pectin-rich materials around the parenchyma cells, pit membrane, and the newly emerging tylose was observed in the xylem vessels of Platanus acerifolia cultivar infected by Ceratocystis fimbriata f. sp. platani (Clérivet et al., 2000). Plants potentially accumulate these pectin-rich gels and gums around tyloses to completely seal off a xylem vessel to prevent the vascular wilt pathogen from spread to adjacent xylem vessels (Rahman et al., 1999). However, complete sealing of xylem vessels can be disadvantageous for the plant as well. If tylose formation affects too many vessels and no new vessels are formed, tylose formation can result in drought stress (Fradin and Thomma, 2006).
Another typical symptom of Verticillium infection is vein clearing. Based on infections of V. longisporum on Arabidopsis and Brassica napus, it was recently reported that this vein clearing is caused by Verticillium-induced transdifferentiation of chloroplast-containing bundle sheath cells to functional xylem elements (Reusche et al., 2012). In addition, it was shown that infected Arabidopsis wild-type plants display enhanced drought stress tolerance compared with non-infected plants, suggesting that Verticillium infection activates a tissue-specific developmental program that compensates for compromised water transport (Reusche et al., 2012).
Another physical defense response observed during xylem colonization is vascular coating. A quick vascular wall coating around the initially infected and the adjacent xylem vessels, infusing the pit membrane and primary walls was observed in resistant chili pepper inoculated with R. solanacearum, whereas the xylem wall coating was not observed in susceptible chili pepper (Rahman et al., 1999). Similar coating of xylem parenchyma cells induced by V. albo-atrum was reported in tomato (Street et al., 1986) and alfalfa (Newcombe and Robb, 1988), indicating that infusion of pit membranes, primary walls and parenchyma cells with coating materials could prevent lateral and vertical spreading of vascular wilt pathogens in the xylem vessels. Furthermore, callose deposition and swelling of the primary walls of the xylem vessels was reported during the interaction of R. solanacearum with chili pepper (Rahman et al., 1999). Previously, a similar deposition of callose in resistant and susceptible tomato infected with F. oxysporum f. sp. lycopersici was reported (Beckman et al., 1982). However, the resistant cultivar maintains a stronger level of callose deposition during the course of the infection than the susceptible cultivar (Beckman et al., 1982). This high level deposition of callose in the resistant cultivar around the initially infected cells could inhibit pathogens from further spread.
Xylem colonization by Xanthomonas campestris pv. campestris has been reported to activate vascular immunity that triggers an HR, referred to as vascular HR (Xu et al., 2008). Vascular immunity was proposed based on the fact that AvrACXcc8004 (also referred to as XopAC), a type III effector protein of Xanthomonas campestris pv. campestris that confers avirulence in Arabidopsis ecotype Col-0, provides resistance when exclusively targeted to the vascular system (Xu et al., 2008). Infiltration of AvrACXcc8004 into leaf mesophyll tissue of Col-0 did not trigger resistance against Xanthomonas, implying that AvrACXcc8004-mediated activation of defense (vascular immunity) occurs in the xylem. Castañeda et al. (2005) previously reported that the Xanthomonas campestris pv. campestris effector protein AvrXccFM elicits vascular HR on Florida mustard seedlings. It is, however, important to note that unlike the HR occurring in leaf mesophyll cells, vascular HR is difficult to score (Castañeda et al., 2005; Xu et al., 2008).
WAT1 (Walls Are Thin1), which is involved in secondary cell wall deposition, is also implicated in vascular immunity (Denancé et al., 2012). WAT1 mutant Arabidopsis plants are resistant to bacterial and fungal vascular wilt pathogens but not to foliar pathogens (Denancé et al., 2012). In leaf inoculation assays, wat1 provides resistance to R. solanacearum and Xanthomonas campestris pv. campestris only when directly injected into the vascular system, but not when injected into mesophyll tissues (Denancé et al., 2012), demonstrating the tissue-specific immune response. Likely, wat1 resistance involves root-localized metabolic channeling away from indole metabolites to salicylic acid (Denancé et al., 2012).
Arabidopsis AHL19, an AT-hook DNA binding protein, provides enhanced resistance to three plant pathogenic Verticillium spp. but not to the foliar pathogens Botrytis cinerea, Plectosphaerella cucumerina, and Alternaria brassicicola and enhanced susceptibility to Pseudomonas syringae pv. tomato, suggesting a role as positive regulator of xylem-specific plant immunity (Yadeta et al., 2011).
Xylem infection causes drastic metabolic changes in xylem parenchyma cells, which are located adjacent to the infected vessels. These metabolic changes lead to the accumulation of different proteins and secondary metabolites in the xylem sap. Some of the proteins and secondary metabolites that accumulate in the xylem sap during xylem colonization include PR-1, PR-2, PR-3, PR-4, PR-5, peroxidases, proteases, xyloglucan-endotransglycosylase (XET), and xyloglucan-specific endoglucanase inhibitor protein (XEGIP), phenols, phytoalexins, and lignin-like compounds (Cooper et al., 1996; Hilaire et al., 2001; Rep et al., 2002,2003; Williams et al., 2002; Houterman et al., 2007; Basha et al., 2010; Gayoso et al., 2010). These compounds are known to contribute directly or indirectly to plant defense. The PR-1, PR-2, PR-3, and PR-5 were also among the proteins abundantly accumulated in xylem sap during compatible interaction between Fusarium and tomato (Rep et al., 2002; Houterman et al., 2007). For instance, PR-2 (β-1, 3-glucanase) and PR-3 (chitinase) hydrolyze the fungal cell wall component β-1,3-glucan and chitin, respectively (Leubner-Metzger and Meins, 1999; van Loon et al., 2006). In addition, antimicrobial activity of PR-5 proteins has also been demonstrated toward multiple pathogens (van Loon et al., 2006), implying that the presence of these proteins in xylem sap could inhibit or slow down the growth of the fungal vascular wilt pathogens in the xylem vessels.
Peroxidases are among the abundantly accumulated enzymes in xylem sap during host colonization of vascular wilt pathogens. The cationic peroxidase, PO-C1, accumulates in the cytoplasm, the primary and secondary walls of the xylem parenchyma, and lumen cells during incompatible interactions between Xoo and rice (Hilaire et al., 2001). Peroxidases are heme-containing enzymes that catalyze the oxidation of different substrates using hydrogen peroxides as an electron acceptor (Gayoso et al., 2010). Peroxidases are known to be involved in the production of reactive oxygen species through their enzymatic activity and reactive oxygen species are toxic compounds that can eliminate vascular wilt pathogens. Furthermore, peroxidases are implicated in the polymerization of cell wall compounds, lignin and suberin biosynthesis, and regulation of hydrogen peroxide levels, which all can contribute to defense (Hilaire et al., 2001; Passardi et al., 2005).
Plants accumulate different phenolic compounds in the xylem in response to infection. Olive trees accumulate phenols such as rutin, oleuropein, luteolin-7-glucoside, and tyrosol at the site of V. dahliae infection that were shown to have a toxic effect on V. dahliae (Báidez et al., 2007). Interestingly, exogenous treatment of Dutch elm trees with phenolic compounds induces accumulation of suberin-like compounds in the xylem tissue and thereby increases resistance to O. novo-ulmi (Martïn et al., 2008). This indicates that, in addition to direct toxicity, phenolic compounds could also activate other defense responses against vascular wilt pathogens.
Plants employ not only complex organic phytoalexins as defense mechanism against vascular wilt pathogens, but also employ inorganic compounds such as elemental sulfur and sulfur-containing inorganic compounds (Williams et al., 2002; Cooper and Williams, 2004). During an incompatible interaction between V. dahliae and tomato elemental sulfur mainly accumulates in xylem parenchyma cells, xylem vessel walls, and around the vascular occluding gels (Williams et al., 2002). Similar accumulation of elemental sulfur has been observed in an incompatible interaction between V. dahliae and cacao (Theobroma cacao) or cotton (Cooper et al., 1996; Cooper and Williams, 2004). The accumulation of inorganic sulfur specifically in xylem vessel walls and around the vascular occluding gels might suggest its role in eliminating vascular wilt pathogens that are arrested by physical defense responses.
Overall, chemical defense responses play major roles in xylem defense. Some chemical compounds accumulated in xylem sap after infection modulate the morphology of xylem tissue and by doing so inhibit vertical and lateral colonization of the pathogens, whereas other compounds accumulate during xylem infection have antimicrobial activity and can eliminate vascular wilt pathogens contained by the physical defense responses.
CONCLUSION
Vascular wilt pathogens have adapted to thrive in the xylem, which is known as a nutrient-poor niche, causing vascular wilt diseases on hundreds of plant species. Recognition of vascular wilt pathogens by both extra- and intracellular plant receptors triggers plant innate immune responses that comprise physical and chemical defenses. Both types of defense responses occur in the xylem vessels in a coordinated manner, where physical defense responses mainly prevent the pathogens from spreading in the xylem vessels and chemical defense responses kill the pathogen or inhibit its growth.
Currently, little is known about the interaction between vascular wilt pathogens and their hosts. As this interaction takes place in xylem vessels which are located deep in the plant interior, the molecular basis underlying the interaction between vascular wilt pathogens and their hosts remains largely obscure. Genetic resistance is the best strategy for controlling vascular wilt pathogens. To develop genetic resistance, however, a deeper understanding of the host defense mechanisms as well as the biology, evolution and pathogenicity of vascular wilt pathogens is required.
VASCULAR WILT PATHOGENS INDUCE DROUGHT STRESS
Wilting of plant parts as a consequence of xylem dysfunction is the most conspicuous symptom of vascular wilt disease. Daugherty et al. (2010) have nicely demonstrated using stable carbon isotope labeling that Xylella fastidiosa induces drought stress in alfalfa. Many factors can contribute to xylem occlusion, such as high- and low-molecular weight polysaccharides secreted by vascular wilt pathogens during xylem colonization and the presence of pathogen biomass (bacterial cells and fungal and oomycete mycelium and spores) in the xylem vessels. However, also plant defense responses can contribute to xylem occlusion, such as tyloses that are formed by the parenchyma cells and gum and gels that are secreted (Fradin and Thomma, 2006; Klosterman et al., 2009; Beattie, 2011). Embolism (the formation of air bubbles) in xylem vessels is another factor that can reduce the hydraulic conductivity of the xylem. Pérez-Donoso et al. (2007) have demonstrated using magnetic resonance imaging that Xylella fastidiosa-infected grape displayed early occurrence of embolism, which correlated with decreased xylem conductivity and drought stress. Although several research reports identify a correlation between xylem infection and drought stress, a recent report revealed enhanced drought tolerance in Arabidopsis upon Verticillium infection (Reusche et al., 2012). Arabidopsis plants infected with V. longisporum exhibited increased de novo xylem formation with newly transdifferentiated xylem vessels that were able to compensate for the occluded ones. Consequently, the plants showed higher drought stress tolerance when compared with non-infected plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Agrios G. N. (2005). Plant Pathology. Burlington, MA: Elsevier Academic Press [Google Scholar]
- Alvarez S., Marsh E. L., Schroeder S. G., Schachtman D. P. (2008). Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ. 31 325–340 [DOI] [PubMed] [Google Scholar]
- Báidez A. G., Gómez P., Del Rïo J. A, Ortuño A. (2007). Dysfunctionality of the xylem in Olea europaea L. plants associated with the infection process by Verticillium dahliae Kleb. role of phenolic compounds in plant defense mechanism. J. Agric. Food Chem. 55 3373–3377 [DOI] [PubMed] [Google Scholar]
- Basha S., Mazhar H., Vasanthaiah H. (2010). Proteomics approach to identify unique xylem sap proteins in Pierce’s disease-tolerant Vitis species. Appl. Biochem. Biotechnol. 160 932–944 [DOI] [PubMed] [Google Scholar]
- Beattie G. A. (2011). Water relations in the interaction of foliar bacterial pathogens with plants. Annu. Rev. Phytopathol. 49 533–555 [DOI] [PubMed] [Google Scholar]
- Beckman C. H. (1964). Host responses to vascular infection. Annu. Rev. Phytopathol. 2 231–252 [Google Scholar]
- Beckman C. H., Mueller W. C., Tessier B. J., Harrison N. A. (1982). Recognition and callose deposition in response to vascular infection in Fusarium wilt-resistant or susceptible tomato plants. Physiol. Plant Pathol. 20 1–10 [Google Scholar]
- Bernoux M., Timmers T., Jauneau A., Brière C., de Wit P. J. G. M., Marco Y., et al. (2008). RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M. D, Thomma B. P. H. J. (2008). The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 72 104–110 [Google Scholar]
- Brett C. T. (2000). Cellulose microfibrils in plants: biosynthesis, deposition, and integration into the cell wall. Int. Rev. Cytol. 199 161–199 [DOI] [PubMed] [Google Scholar]
- Castañeda A., Reddy J. D., El-Yacoubi B., Gabriel D. W. (2005). Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant Microbe Interact. 18 1306–1317 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Almeida R. P. P., Lindow S. (2008). Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 46 243–271 [DOI] [PubMed] [Google Scholar]
- Chen X., Chern M., Canlas P. E., Ruan D., Jiang C., Ronald P. C. (2010). An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc. Natl. Acad. Sci. U.S.A. 107 8029–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ronald P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choat B., Ball M., Luly J., Holtum J. (2003). Pit membrane porosity and water stress-induced cavitation in four co-existing dry rainforest tree species. Plant Physiol. 131 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choat B., Cobb A. R., Jansen S. (2008). Structure and function of bordered pits: new discoveries and impacts on whole-plant hydraulic function. New Phytol. 177 608–626 [DOI] [PubMed] [Google Scholar]
- Choat B., Jansen S., Zwieniecki M. A., Smets E., Holbrook N. M. (2004). Changes in pit membrane porosity due to deflection and stretching: the role of vestured pits. J. Exp. Bot. 55 1569–1575 [DOI] [PubMed] [Google Scholar]
- Choat B., Pittermann J. (2009). New insights into bordered pit structure and cavitation resistance in angiosperms and conifers. New Phytol. 182 557–560 [DOI] [PubMed] [Google Scholar]
- Clérivet A. Déon V., Alami I., Lopez F., Geiger J. P., Nicole M. (2000). Tyloses and gels associated with cellulose accumulation in vessels are responses of plane tree seedlings (Platanus × acerifolia) to the vascular fungus Ceratocystis fimbriata f. sp platani. Trees Struct. Funct. 15 25–31 [Google Scholar]
- Cooper R. M., Resende M. L. V., Flood J., Rowan M. G., Beale M. H., Potter U. (1996). Detection and cellular localization of elemental sulphur in disease-resistant genotypes of Theobroma cacao. Nature 379 159–162 [Google Scholar]
- Cooper R. M., Williams J. S. (2004). Elemental sulphur as an induced antifungal substance in plant defence. J. Exp. Bot. 55 1947–1953 [DOI] [PubMed] [Google Scholar]
- Daugherty M., Lopes J. S., Almeida R. P. (2010). Strain-specific alfalfa water stress induced by Xylella fastidiosa. Eur. J. Plant Pathol. 127 333–340 [Google Scholar]
- De Boer A. H., Volkov V. (2003). Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant Cell Environ. 26 87–101 [Google Scholar]
- De Jonge R., Van Esse H. P., Maruthachalam K., Bolton M. D., Santhanam P., Saber M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109 5110–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denancé N., Ranocha P., Oria N., Barlet X., Rivière M.-P., Yadeta K. A., et al. (2012). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 10.1111/tpj.12027 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D. X., Khounlotham M., Boucher C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Theulières F., Hirsch J., Feng D. X., Bittner-Eddy P., et al. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. U.S.A. 99 2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A., Madrid M. P., Caracuel Z., Delgado-Jarana J, Roncero M. I. G. (2003). Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4 315–325 [DOI] [PubMed] [Google Scholar]
- Divon H. H., Fluhr R. (2007). Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiol. Lett. 266 65–74 [DOI] [PubMed] [Google Scholar]
- Divon H. H., Rothan-Denoyes B., Davydov O., Di Pietro A., Fluhr R. (2005). Nitrogen-responsive genes are differentially regulated in planta during Fusarium oxysporum f. sp. lycopersici infection. Mol. Plant Pathol. 6 459–470 [DOI] [PubMed] [Google Scholar]
- Divon H. H., Ziv C., Davydov O., Yarden O., Fluhr R. (2006). The global nitrogen regulator, FNR1, regulates fungal nutrition-genes and fitness during Fusarium oxysporum pathogenesis. Mol. Plant Pathol. 7 485–497 [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11 539–548 [DOI] [PubMed] [Google Scholar]
- Donofrio N. M., Mitchell T. K., Dean R. A., Wang G.-L., Valent B. (2009). “The significance of nitrogen regulation, source and availability on the interaction between rice and rice blast,” in Advances in Genetics, Genomics and Control of Rice Blast Disease, edsWang G.-L., Valent B. (Dordrecht: Springer; ) 59–72 [Google Scholar]
- Emons A. M. C., Mulder B. M. (2000). How the deposition of cellulose microfibrils builds cell wall architecture. Trends Plant Sci. 5 35–40 [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia N., Hernandez M., Casado-Vela J., Bru R., Elortza F., Hedden P., et al. (2011). Changes to the proteome and targeted metabolites of xylem sap inBrassica oleracea in response to salt stress. Plant Cell Environ. 34 821–836 [DOI] [PubMed] [Google Scholar]
- Fradin E. F., Abd-El-Haliem A., Masini L., van den Berg G. C. M., Joosten M. H. A. J., Thomma B. P. H. J. (2011). Interfamily transfer of tomato ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin E. F, Thomma B. P. H. J. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7 71–86 [DOI] [PubMed] [Google Scholar]
- Fradin E. F., Zhang Z., Ayala J. C. J., Castroverde C. D. M., Nazar R. N., Robb J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato ve1. Plant Physiol. 150 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H. (1996). Xylogenesis: initiation, progression, and cell death. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 299–325 [DOI] [PubMed] [Google Scholar]
- Fukuda H. (1997). Programmed cell death during vascular system formation. Cell Death Differ. 4 684–688 [DOI] [PubMed] [Google Scholar]
- Fukuda H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391 [DOI] [PubMed] [Google Scholar]
- Gartemann K.-H., Abt B., Bekel T., Burger A., Engemann J., Flugel M., et al. (2008). The genome sequence of the tomato-pathogenic Actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayoso C., Pomar F., Novo-Uzal E., Merino F, Martinez de Ilarduya O. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10:232 10.1186/1471-2229-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187 920–928 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Bauer Z., Boller T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Grimault V., Gélie B., Lemattre M., Prior P., Schmit J. (1994). Comparative histology of resistant and susceptible tomato cultivars infected by Pseudomonas solanacearum. Physiol. Mol. Plant Pathol. 44 105–123 [Google Scholar]
- Harwood T. D., Tomlinson I., Potter C. A., Knight J. D. (2011). Dutch elm disease revisited: past, present and future management in Great Britain. Plant Pathol. 60 545–555 [Google Scholar]
- Hayslett M., Juzwik J., Moltzan B. (2008). Three Colopterus beetle species carry the oak wilt fungus to fresh wounds on red oak in Missouri. Plant Dis. 92 270–275 [DOI] [PubMed] [Google Scholar]
- Hayward A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29 65–87 [DOI] [PubMed] [Google Scholar]
- Hilaire E., Young S. A., Willard L. H., McGee J. D., Sweat T., Chittoor J. M., et al. (2001). Vascular defense responses in rice: peroxidase accumulation in xylem parenchyma cells and xylem wall thickening. Mol. Plant Microbe Interact. 14 1411–1419 [DOI] [PubMed] [Google Scholar]
- Houterman P. M., Ma L., Van Ooijen G., De Vroomen M. J., Cornelissen B. J. C., Takken F. L. W., et al. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58 970–978 [DOI] [PubMed] [Google Scholar]
- Houterman P. M., Speijer D., Dekker H. L., De Koster C. G., Cornelissen B. J. C., Rep M. (2007). The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants: short communication. Mol. Plant Pathol. 8 215–221 [DOI] [PubMed] [Google Scholar]
- Huang C.-C., Lindhout P. (1997). Screening for resistance in wild Lycopersicon species to Fusarium oxysporum f. sp. lycopersici race 1 and race 2. Euphytica 93 145–153 [Google Scholar]
- Janse J. D., Obradovic A. (2010). Xylella fastidiosa: its biology, diagnosis, control and risks. J. Plant Pathol. 92 S1.35–S31.47 [Google Scholar]
- Jansen S., Baas P., Gasson P., Lens F., Smets E. (2004). Variation in xylem structure from tropics to tundra: evidence from vestured pits. Proc. Natl. Acad. Sci. U.S.A. 101 8833–8837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha G., Rajeshwari R., Sonti R. V. (2005). Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol. Plant Microbe Interact. 18 891–898 [DOI] [PubMed] [Google Scholar]
- Ji X., Lu G., Gai Y., Zheng C., Mu Z. (2008). Biological control against bacterial wilt and colonization of mulberry by an endophytic Bacillus subtilis strain. FEMS Microbiol. Ecol. 65 565–573 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Chen X., Ding X., Wang Y., Chen Q., Song W.-Y. (2012). The XA21 binding protein XB25 is required for maintaining XA21-mediated disease resistance. Plant J. 73 814–823 [DOI] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Juzwik J., Harrington T. C., MacDonald W. L., Appel D. N. (2008). The origin of Ceratocystis fagacearum, the oak wilt fungus. Annu. Rev. Phytopathol. 46 13–26 [DOI] [PubMed] [Google Scholar]
- Kawchuk L. M., Hachey J., Lynch D. R., Kulcsar F., Van Rooijen G., Waterer D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U.S.A. 98 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman S. J., Atallah Z. K., Vallad G. E., Subbarao K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47 39–61 [DOI] [PubMed] [Google Scholar]
- Klosterman S. J., Subbarao K. V., Kang S., Veronese P., Gold S. E., Thomma B. P. H. J., et al. (2011). Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathol. 7:e1002137 10.1371/journal.ppat.1002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H. B., Natarajan S. S., Bennett J. O., Sicher R. C. (2011). Protein and metabolite composition of xylem sap from field-grown soybeans (Glycine max). Planta 233 921–931 [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-W., Han S.-W., Sririyanum M., Park C.-J., Seo Y.-S., Ronald P. C. (2009). A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326 850–853 [DOI] [PubMed] [Google Scholar]
- Lee B. M., Park Y. J., Park D. S., Kang H. W., Kim J. G., Song E. S., et al. (2005). The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G., Meins F. (1999). “Functions and regulation of plant beta-1,3-glucanases (PR-2),” in Pathogenesis-Related Proteins in Plants, eds Datta S. K., Muthukrishnan S. (Boca Raton: CRC Press; ) 49–76 [Google Scholar]
- Lévesque C. A., Brouwer H., Cano L., Hamilton J., Holt C., Huitema E. (2010). Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11 R73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens B., Rep M., Thomma B. P. (2008). Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manag. Sci. 64 781–788 [DOI] [PubMed] [Google Scholar]
- López-Escudero F., Mercado-Blanco J. (2011). Verticillium wilt of olive: a case study to implement an integrated strategy to control a soil-borne pathogen. Plant Soil 344 1–50 [Google Scholar]
- Ma L.-J., van der Does H. C., Borkovich K. A., Coleman J. J., Daboussi M.-J., Di Pietro A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis E. A., Tjamos S. E., Chatzipavlidis I., Antoniou P. P., Paplomatas E. J. (2008). Evaluation of compost amendments for control of vascular wilt diseases. J. Phytopathol. 156 622–627 [Google Scholar]
- Martin F. N., Loper J. E. (1999). Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. Crit. Rev. Plant Sci. 18 111–181 [Google Scholar]
- Martïn J. A., Solla A., Domingues M. R., Coimbra M. A., Gil L. (2008). Exogenous phenol increase resistance of Ulmus minor to Dutch elm disease through formation of suberin-like compounds on xylem tissues. Environ. Exp. Bot. 64 97–104 [Google Scholar]
- Marzluf G. (1997). Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully M. E. (2001). Niches for bacterial endophytes in crop plants: a plant biologist’s view. Funct. Plant Biol. 28 983–990 [Google Scholar]
- Michielse C. B., Rep M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius N., Hertweck C. (2009). Fungal phytotoxins as mediators of virulence. Curr. Opin. Plant Biol. 12 390–398 [DOI] [PubMed] [Google Scholar]
- Mollenhauer H. H., Hopkins D. L. (1974). Ultrastructural study of Pierce’s disease bacterium in grape xylem tissue. J. Bacteriol. 119 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J. C., Konrad H., Blomquist S. R., Kirisits T. (2010). Do mites phoretic on elm bark beetles contribute to the transmission of Dutch elm disease? Naturwissenschaften 97 219–227 [DOI] [PubMed] [Google Scholar]
- Nadarasah G., Stavrinides J. (2011). Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol. Rev. 35 555–575 [DOI] [PubMed] [Google Scholar]
- Nakamura S.-I., Akiyama C., Sasaki T., Hattori H., Chino M. (2008). Effect of cadmium on the chemical composition of xylem exudate from oilseed rape plants (Brassica napus L.). Soil Sci. Plant Nutr. 54 118–127 [Google Scholar]
- Newcombe G., Robb J. (1988). The function and relative importance of the vascular coating response in highly resistant, moderately resistant and susceptible alfalfa infected by Verticillium albo-atrum. Physiol. Mol. Plant Pathol. 33 47–58 [Google Scholar]
- Nieminen K. M., Kauppinen L., Helariutta Y. (2004). A weed for wood? Arabidopsis as a genetic model for xylem development. Plant Physiol. 135 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño-Liu D. O., Ronald P. C., Bogdanove A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7 303–324 [DOI] [PubMed] [Google Scholar]
- Oliver J. P., Castro A., Gaggero C., Cascón T., Schmelz E. A., Castresana C., et al. (2009). Pythium infection activates conserved plant defense responses in mosses. Planta 230 569–579 [DOI] [PubMed] [Google Scholar]
- Palmer C. S., Saleeba J. A., Lyon B. R. (2005). Phytotoxicity on cotton ex-plants of an 18.5 kDa protein from culture filtrates of Verticillium dahliae. Physiol. Mol. Plant Pathol. 67 308–318 [Google Scholar]
- Park C.-J., Bart R., Chern M., Canlas P. E., Bai W., Ronald P. C. (2010a). Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS ONE 5:e9262 10.1371/journal.pone.0009262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. J., Han S. W., Chen X., Ronald P. C. (2010b). Elucidation of XA21-mediated innate immunity. Cell. Microbiol. 12 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F., Cosio C., Penel C., Dunand C. (2005). Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 24 255–265 [DOI] [PubMed] [Google Scholar]
- Pérez-Donoso A. G., Greve L. C., Walton J. H., Shackel K. A., Labavitch J. M. (2007). Xylella fastidiosa infection and ethylene exposure result in xylem and water movement disruption in grapevine shoots. Plant Physiol. 143 1024–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Donoso A. G., Sun Q., Roper M. C., Greve L. C., Kirkpatrick B., Labavitch J. M. (2010). Cell wall-degrading enzymes enlarge the pore size of intervessel pit membranes in healthy and Xylella fastidiosa-infected grapevines. Plant Physiol. 152 1748–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powney R., Smits T. H. M., Sawbridge T., Frey B., Blom J., Frey J. E., et al. (2011). Genome sequence of an Erwinia amylovora strain with pathogenicity restricted to Rubus plants. J. Bacteriol. 193 785–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell A. H., Hopkins D. L. (1996). Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34 131–151 [DOI] [PubMed] [Google Scholar]
- Qian W., Jia Y., Ren S.-X., He Y.-Q., Feng J.-X., Lu L.-F., et al. (2005). Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G. M., Vallad G. E., Subbarao K. V. (2008). Characterization of Verticillium dahliae and V. tricorpus isolates from lettuce and artichoke. Plant Disease 92 69–77 [DOI] [PubMed] [Google Scholar]
- Rahman M. A., Abdullah H., Vanhaecke M. (1999). Histopathology of susceptible and resistant Capsicum annuum cultivars infected with Ralstonia solanacearum. J. Phytopathol. 147 129–140 [Google Scholar]
- Rep M., Dekker H. L., Vossen J. H., De Boer A. D., Houterman P. M., De Koster C. G., et al. (2003). A tomato xylem sap protein represents a new family of small cysteine-rich proteins with structural similarity to lipid transfer proteins. FEBS Lett. 534 82–86 [DOI] [PubMed] [Google Scholar]
- Rep M., Dekker H. L., Vossen J. H., De Boer A. D., Houterman P. M., Speijer D., et al. (2002). Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 130 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche M., Thole K., Janz D., Truskina J., Rindfleisch S., Drübert C., et al. (2012). Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell 24 3823–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P., Benhamou N., Tirilly Y. (1998). Ultrastructural and cytochemical investigation of asymptomatic infection by Pythium spp. Phytopathology 88 234–244 [DOI] [PubMed] [Google Scholar]
- Roncero M. I. G., Hera C., Ruiz-Rubio M., Garcïa Maceira F. I., Madrid M. P., Caracuel Z., et al. (2003). Fusarium as a model for studying virulence in soilborne plant pathogens. Physiol. Mol. Plant Pathol. 62 87–98 [Google Scholar]
- Roper M. C. (2011). Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol. Plant Pathol. 12 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415 497–502 [DOI] [PubMed] [Google Scholar]
- Santhanam P., van Esse H. P., Albert I., Faino L., Nürnberger T, Thomma B. P. H. J. (2013). Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 26 278–286 [DOI] [PubMed] [Google Scholar]
- Scheffer R. J., Voeten J. G. W. F. , Guries R. P. (2008). Biological control of Dutch elm disease. Plant Dis. 92 192–200 [DOI] [PubMed] [Google Scholar]
- Schumann G. L, D’Arcy C. J. (2010). Essential Plant Pathology. St. Paul, MN: APS Press [Google Scholar]
- Sebaihia M., Bocsanczy A. M., Biehl B. S., Quail M. A., Perna N. T., Glasner J. D., et al. (2010). Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J. Bacteriol. 192 2020–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebrecht S., Herdel K., Schurr U., Tischner R. (2003). Nutrient translocation in the xylem of poplar – diurnal variations and spatial distribution along the shoot axis. Planta 217 783–793 [DOI] [PubMed] [Google Scholar]
- Simpson A. J., Reinach F. C., Arruda P., Abreu F. A., Acencio M., Alvarenga R., et al. (2000). The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406 151–159 [DOI] [PubMed] [Google Scholar]
- Smits T. H., Rezzonico F., Kamber T., Blom J., Goesmann A., Frey J. E., et al. (2010). Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol. Plant Microbe Interact. 23 384–393 [DOI] [PubMed] [Google Scholar]
- Song W. Y., Wang G. L., Chen L. L., Kim H. S., Pi L. Y., Holsten T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806 [DOI] [PubMed] [Google Scholar]
- Spadaro D., Gullino M. L. (2005). Improving the efficacy of biocontrol agents against soilborne pathogens. Crop Prot. 24 601–613 [Google Scholar]
- Stipanovic R. D., Puckhaber L. S., Liu J., Bell A. A. (2011). Phytotoxicity of fusaric acid and analogs to cotton. Toxicon 57 176–178 [DOI] [PubMed] [Google Scholar]
- Street P. F. S., Robb J., Ellis B. E. (1986). Secretion of vascular coating components by xylem parenchyma cells of tomatoes infected with Verticillium albo-atrum. Protoplasma 132 1–11 [Google Scholar]
- Suárez-Estrella F., Vargas-Garcïa C., López M. J., Capel C., Moreno J. (2007). Antagonistic activity of bacteria and fungi from horticultural compost against Fusarium oxysporum f. sp. melonis. Crop Prot. 26 46–53 [Google Scholar]
- Sun Q. H., Hu J., Huang G. X., Ge C., Fang R. X., He C. Z. (2005). Type-II secretion pathway structural gene xpsE, xylanase- and cellulase secretion and virulence in Xanthomonas oryzae pv. oryzae. Plant Pathol. 54 15–21 [Google Scholar]
- Takken F., Rep M. (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 11 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talboys P. W. (1972). Resistance to vascular wilt fungi. Proc. R. Soc. Lond. B Biol. Sci. 181 319–332 [Google Scholar]
- Temple B., Horgen P. A. (2000). Biological roles for cerato-ulmin, a hydrophobin secreted by the elm pathogens, Ophiostoma ulmi and O. novo-ulmi. Mycologia 92 1–9 [Google Scholar]
- Tjamos E. C., Beckman C. H. (1989). Vascular Wilt Diseases of Plants: Basic Studies and Control (NATO ASI Series H: Cell Biology). Berlin: Springer-Verlag [Google Scholar]
- Tsuda K., Kosaka Y., Tsuge S., Kubo Y., Horino O. (2001). Evaluation of the endophyte Enterobacter cloacae SM10 isolated from spinach roots for biological control against Fusarium wilt of spinach. J. Gen. Plant Pathol. 67 78–84 [Google Scholar]
- Tyree M. T., Zimmermann M. H. (2002). Xylem Structure and the Ascent of Sap. Berlin: Springer [Google Scholar]
- van Esse H. P., Fradin E. F., de Groot P. J., de Wit P. J. G. M., Thomma B. P. H. J. (2009). Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol. Plant Microbe Interact. 22 245–258 [DOI] [PubMed] [Google Scholar]
- van Loon L. C., Rep M, Pieterse C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162 [DOI] [PubMed] [Google Scholar]
- Vicente J. G., Conway J., Roberts S. J., Taylor J. D. (2001). Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars. Phytopathology 91 492–499 [DOI] [PubMed] [Google Scholar]
- Wang G., Ellendorff U., Kemp B., Mansfield J. W., Forsyth A., Mitchell K., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Y., Cai Y., Gou J. Y., Mao Y. B., Xu Y. H., Jiang W. H., et al. (2004). VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl. Environ. Microbiol. 70 4989–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. S., Hall S. A., Hawkesford M. J., Beale M. H., Cooper R. M. (2002). Elemental sulfur and Thiol accumulation in tomato and defense against a fungal vascular pathogen. Plant Physiol. 128 150–159 [PMC free article] [PubMed] [Google Scholar]
- Xu R.-Q., Blanvillain S., Feng J.-X., Jiang B.-L., Li X.-Z., Wei H.-Y., et al. (2008). AvrACXcc8004, a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J. Bacteriol. 190 343–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta K. A., Hanemian M., Smit P., Hiemstra J. A., Pereira A., Marco Y., et al. (2011). The Arabidopsis thaliana DNA-binding protein AHL19 mediates Verticillium wilt resistance. Mol. Plant Microbe Interact. 24 1582–1591 [DOI] [PubMed] [Google Scholar]
- Ye Z. H. (2002). Vascular tissue differentiation and pattern formation in plants. Annu. Rev. Plant Biol. 53 183–202 [DOI] [PubMed] [Google Scholar]
- Yokoyama R., Nishitani K. (2006). Identification and characterization of Arabidopsis thaliana genes involved in xylem secondary cell walls. J. Plant Res. 119 189–194 [DOI] [PubMed] [Google Scholar]
- Zhang J., Elo A., Helariutta Y. (2011). Arabidopsis as a model for wood formation. Curr. Opin. Biotechnol. 22 293–299 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fradin E., Jonge R., van Esse H. P., Smit P., Liu C.-M., et al. (2013). Optimized agroinfiltration and virus-induced gene silencing to study Ve1-mediated Verticillium resistance in tobacco. Mol. Plant Microbe Interact. 26 182–190 [DOI] [PubMed] [Google Scholar]
- Zhou B.-J., Jia P.-S., Gao F., Guo H.-S. (2012). Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 25 964–975 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D. G., Boller T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760 [DOI] [PubMed] [Google Scholar]