Abstract
We previously reported on the A. thaliana gene EDM2, which is required for several developmental processes and race-specific immunity. Although EDM2 encodes a nuclear protein with features commonly observed in epigenetic factors, its role in chromatin silencing remains unknown. Here we demonstrate that silencing states of several transposons in edm2 mutants are altered. Levels of their transcripts anti-correlate with those of the repressive epigenetic marks H3K27me1, H3K9me2, and DNA-methylation at CHG sites. In addition, double mutant analysis revealed epistasis between EDM2 and the major histone H3K9-methyltransferase gene KRYPTONITE/SUVH4 in the control of H3K9me2 and CHG methylation. Moreover, we found that the expressivity of several mutant edm2 phenotypes exhibits stochastic variation reminiscent of mutants of known epigenetic modifiers. We propose that EDM2 affects the expression of transposons and developmentally important genes by modulating levels of repressive chromatin marks in a locus dependent manner.
In eukaryotes, activity states of transcription units are often modulated by their chromatin environment. Epigenetic information conditioning the chromatin environment is tightly associated with certain biochemical marks including differential histone modifications, histone variant distribution and DNA methylation1,2. In plants, mutations of several key epigenetic regulators or combination of them can cause pleiotropic phenotypes including delayed or accelerated floral transition, morphological changes and aberrant abiotic or biotic stress responses3,4. This highlights the importance of epigenetic information for orchestrating various biological activities and programs.
Epigenetic mechanisms are critical not only for the regulation of gene expression, but also for the silencing of transposable elements (TEs). While TE transposition is of key significance for the generation of genetic diversity and serves as a major driver in genome evolution5, this process can cause detrimental mutations. To suppress the spread of such invasive DNA elements and to protect host genomes, stringent chromatin-based epigenetic transcriptional silencing mechanisms have evolved6,7. In plants, two types of heterochromatin-associated epigenetic marks have been closely associated with the transcriptional repression of TEs: Methylation of the nucleobase cytosine at carbon 5 (5mC) and post-translational histone modifications, such as di-methylation of lysine 9 of histone H3 (H3K9me2) and mono-methylation of lysine 27 of H3 (H3K27me1)8,9.
H3K9me2 is catalyzed by the Su(var)3-9 family proteins, which were initially identified in Drosophila and are structurally conserved in many organisms10. The model plant Arabidopsis thaliana (Arabidopsis) also has multiple Su(var)3-9 homologs called SUVH proteins. Among these KRYPTONITE/SUVH4 (KYP) is thought to be the major H3K9 methyltransferase11,12,13. However, SUVH5 and SUVH6 act redundantly with KYP to silence transcription at some loci14,15. Mutations in H3K9 methyltransferase genes lead to de-repression of transposons, indicating the importance of H3K9me2 for transposon silencing15. Another histone modification that is associated with constitutive heterochromatin formation is H3K27me1. This histone mark has been shown to be catalyzed by the ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 and 6 (ATXR5 and ATXR6)9. An atxr5 atxr6 double mutant showed re-activation of TEs, indicating that H3K27me1 also plays an important role in TE silencing.
In addition to histone modifications, the role of 5mC in transposon silencing is evident as genome-wide loss of 5mCs in the mutants of DNA methyltransferase 1 (MET1) or a chromatin remodeling factor gene DECREASE IN DNA METHYLATION 1 (DDM1) cause de-repression of many TEs16,17,18. In Arabidopsis, 5mC is observed at Cs in any sequence context (CG, CHG and CHH; H = any base except for G). Each type of DNA methylation is controlled by a distinct class of DNA methyltransferases. 5mC at CG sites (CG methylation) is maintained by the DNA methyltransferase MET1, which is an ortholog of the mammalian Dnmt119. The plant-specific DNA methyltransferase CHROMOMETHYLASE 3 (CMT3) primarily maintains 5mC at CHG sites (CHG methylation)20. DOMAINS REARRANGED METHYLTRANSFERASE 1 (DRM1) and 2 (DRM2), which are plant homologs of the mammalian DNMT3 and serve as the major de novo DNA methyltransferases required for initiation of DNA methylation in all contexts, are also responsible for maintenance of CHH methylation21. These DRM proteins are recruited to their target loci by small interfering RNAs (siRNAs) in the RNA-directed DNA methylation pathway21,22,23. DNA methylation patterns at individual genomic loci are often the outcome of interactive or counteractive interactions of DNA methyltransferases and DNA de-methylases. TEs and other repetitive sequences have been reported to be significantly marked by 5mCs, although 5mC was also found to extend to the euchromatic regions24. CG methylation is common in both genes and TEs, while CHG methylation is almost exclusively present in TEs8.
Importantly, recent genome-wide profiling studies in Arabidopsis using chromatin immunoprecipitation (ChIP) combined with tilling arrays revealed a tight correlation of H3K9me2 with 5mC25. Furthermore, mutations compromising function of MET1 lead to decreased or rearranged H3K9me2 marks26,27, while mutations disrupting the functions of the H3K9 methyltransferase KYP result in decreased CHG methylation11. A mechanism linking CHG methylation and H3K9me2 is based on a self-reinforcing loop28. Methylated CHG sites recruit KYP, which methylates H3K9 predominantly to H3K9me2. H3K9me2 in turn recruits the non-CG cytosine methylase CMT3, which predominantly catalyzes CHG methylation. Thus, H3K9me2 and CHG methylation are intimately coupled. While both CHG methylation and H3K9me2 on the one hand and H3K27me1 on the other hand can co-occur29 and may cooperate in transposon silencing, the latter mark appears to be controlled by mechanisms distinct from those regulating the former two marks.
We previously reported on the Arabidopsis gene EDM2 (enhanced downy mildew 2), which is required for race-specific immunity against the oomycete Hyaloperonospora arabidopsidis (Hpa) mediated by the disease-resistance gene RPP730. It also contributes to several developmental processes, such as floral transition and leaf pavement cell formation30,31,32. EDM2 encodes a nuclear protein with features commonly found in epigenetic factors, including putative nuclear localization signals, PHD-finger like motifs and a region with similarity to the active sites of methyltransferases. EDM2-type PHD fingers are present in many eukaryotic clades, including humans30.
EDM2 enhances mRNA levels of RPP730 and suppresses those of the floral suppressor gene FLC31. Global transcript profiling showed that the number of genes affected by EDM2 in untreated Arabidopsis plants is relatively small (levels of only 222 transcripts appear significantly affected by EDM2)32. Yeast-two hybrid screens combined with bimolecular fluorescence complementation and reverse genetics revealed the atypical protein kinase WNK8 and members of a small Arabidopsis family of putative EMSY-like (EML) chromatin remodeling factors as EDM2 interactors31,33. PHD-fingers have been reported to mediate docking to the N-terminal tail of histone H3 depending on its methylation or acetylation states and are often present in chromatin-associated epigenetic regulators34. Thus, the domain architecture of EDM2 together with its ability to interact with putative chromatin remodeling factors suggested that EDM2 acts as a chromatin-associated epigenetic factor.
Here we provide data strongly supporting such a role of EDM2. We demonstrate that EDM2 directly or indirectly affects transcript levels of a limited number of TEs, including the DNA transposon Mu1 and the retrotransposon COPIA4. Detailed analyses showed that EDM2 modulates levels of H3K27me1, H3K9me2 and CHG methylation at these two loci. Moreover, double mutant analysis revealed epistatic relationships and genetic interactions between EDM2 and the major H3K9-methylase gene KYP in the control of H3K9me2 and CHG methylation levels of Mu1 and COPIA4. Thus, EDM2 affects the silencing states of these loci at the chromatin level. We also show that, like several mutants of previously described epigenetic regulators, edm2 mutant plants exhibit substantial variation and instability regarding their phenotypes.
Results
Selective regulation of TE transcript levels by EDM2
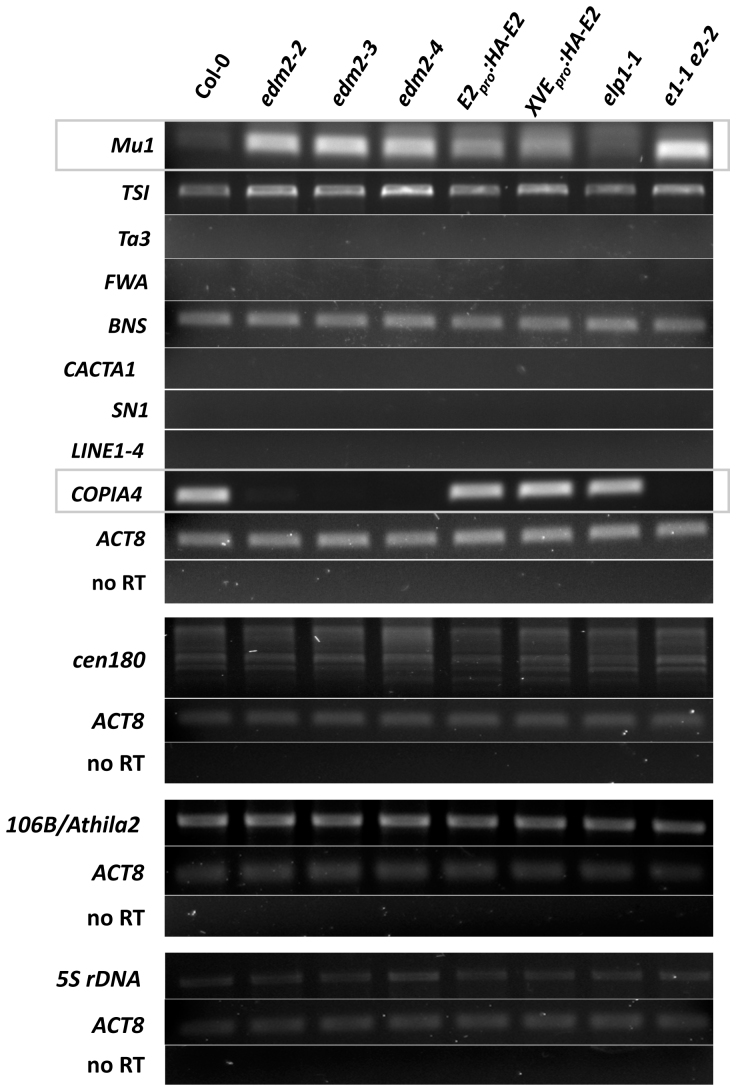
Several well-characterized epigenetic regulators control TEs35. Among EDM2 target genes previously identified by Affymetrix ATH1 microarrays32 is the retrotransposon COPIA4, which is transcriptionally regulated by heterochromatic silencing closely associated with DNA methylation and repressive histone modifications17. The ATH1 microarray does not represent all TEs and other loci known to be epigenetically silenced. Therefore, we determined by RT-PCR transcript levels of various known epigenetically silenced Arabidopsis loci12,17,36,37,38,39,40 in Col-0, edm2 mutants, two independent EDM2 complementation lines as well as a mutant for the EDM2 paralog ELP130 and an elp1-1 edm2-2 double mutant (Fig. 1). Both complementation lines exhibit wild type levels of race-specific immunity and rosette leaf expansion (31,32; not shown). Levels of EDM2 transcripts in these complementation lines are similar to those in Col-0 wildtype (Supplementary Fig. S131). Besides COPIA4, the DNA transposon Mu1 and the retro-TE fragment soloLTR were found to be miss-expressed in edm2 mutants (Fig. 1 and Supplementary Fig. S2). RT-PCRs with three biological replicates reproducibly resulted in the detection of altered transcript levels of Mu1, COPIA4 and soloLTR in all tested edm2 mutants. Consistent with our microarray analysis, we found transcriptional repression of COPIA4 to be tighter in edm2 plants. While transcripts of this TE were clearly detectable in Col-0, they were nearly undetectable in edm2 mutants. In both EDM2 complementation lines, wild type levels of this transcript were clearly restored. ELP1 appears not to affect levels of COPIA4 transcripts.
Figure 1. Effects of EDM2 on transcriptional silencing in constitutively repressed loci.
Transcript levels of the respective loci were determined by RT-PCR. ACTIN8 (ACT8) served as an internal control, while use of template made without reverse transcriptase (no RT) show no genomic DNA contamination. Of the tested transcripts, levels of Mu1 and COPIA4 (framed by grey line) are clearly affected by EDM2. E2pro:HA-E2 and XVEpro:HA-E2: EDM2 complementation lines expressing in the edm2-2 background HA-tagged EDM2 driven by the native EDM2 promoter (E2pro) or the estradiol-inducible XVE promoter, respectively. Expression of EDM2 in the XVEpro:HA-EDM2 line is constitutive and does not require application with estradiol31. elp1-1: apparent mRNA null T-DNA insertion mutant of the EDM2 paralog ELP1; e1-1 e2-2: elp1-1 edm2-2 double mutant. Ta3, LINE1-4 and COPIA4 are retrotransposons. Mu1 and CACTA1 are DNA transposons. TSI, transcriptionally silent information36; FWA, FLOWERING WAGENINGEN37; BNS, BONSAI40; SN1, a short interspersed nuclear retroelement. cen180, centromeric satellite repeats38. 106B/Athila2, centromer repeats of Athila transposon-derived 106B.
In contrast, Mu1 was found to be de-repressed in edm2 plants. While low levels of Mu1 transcript were detectable in Col-0, transcript levels of this TE were substantially elevated in all three tested edm2 mutants (Fig. 1). In both EDM2 complementation lines, levels of Mu1 transcripts are intermediate. Interestingly, also the elp1-1 mutant exhibited moderately enhanced Mu1 transcript levels compared to Col-0. Levels of this transcript appear also slightly higher in the elp1-1 edm2-2 double mutant than that in the edm2-2 single mutant. Thus, both EDM2 and ELP1 seem to be involved in the repression of Mu1. However, based on our results, the contribution of EDM2 to Mu1 silencing is much higher than that of ELP1. Real-time quantitative RT-PCRs (qRT-PCRs) further confirmed our observations for COPIA4 and Mu1 (Fig. 2a and d). The amplification specificity for each qRT-PCR was verified by melting curve analysis, which showed in each case that only a single transcript was amplified (Fig. S3).
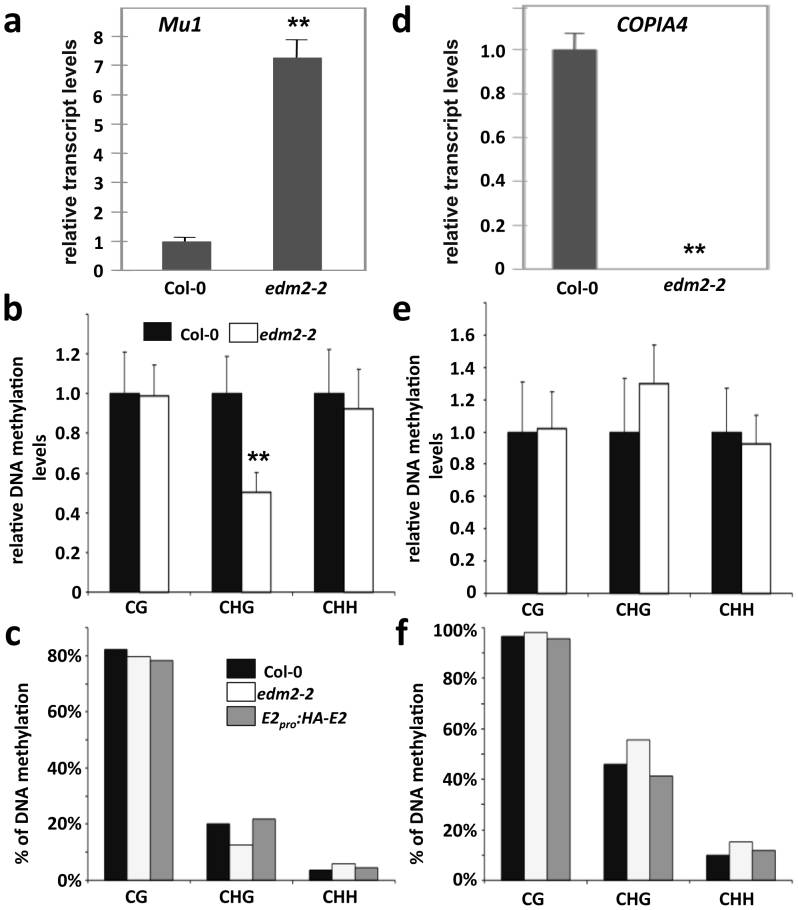
Figure 2. EDM2 affects transcript and CHG methylation levels at Mu1 and COPIA4.
(a) & (d): Mu1 and COPIA4 transcript levels determined by qRT-PCR. (b) & (e): Extent of DNA cytosine methylation at CG, CHG or GHH sites in the bodies of Mu1 (b) and COPIA4 (e) determined by chop-qPCR. Each PCR-amplified genomic region contains 1 recognition site of methylation sensitive restriction enzyme HpyCH4IV (for CG site), BglII (for CHG site), AflII (CHH site in Mu1) or HaeIII (CHH site in COPIA4). Data represent mean plus SD of three biological replicates. Statistical significance was determined using Student's t-test (** P < 0.01 compared with Col-0) as described previously58. (c) & (f): The percentage of methylated cytosine at Mu1 (c) and COPIA4 (f) determined by bisulfite sequencing.
Published genome-wide 5mC data (http://epigenomics.mcdb.ucla.edu/DNAmeth/) suggested 5mC levels to be high in the bodies of both TEs. We performed chop-qPCRs (digestion of genomic DNA with DNA methylation-sensitive restriction enzymes followed by qPCR) to investigate 5mC levels in edm2-2 mutant and wildtype Col-0. 5mC levels of a defined CG and CHH site in each of the two TE loci in edm2-2 mutant were similar to those in wildtype Col-0 (Fig. 2b and e). However we found 5mC levels at a CHG site in Mu1 and COPIA4 to be reduced and enhanced in edm2-2, respectively. Although the magnitude of the enhanced CHG methylation level at COPIA4 is small, this trend was highly reproducible in all three biological replicates we performed. We further analyzed the DNA methylation status at Mu1 and COPIA4 by bisulfite sequencing (Fig. 2c & f), which provides quantitative information on the methylation states of multiple cytosins within a defined genomic area. Consistent with our chop-qPCR data, levels of methylated CHG sites at Mu1 were reduced in edm2-2 plants, while levels of this mark at COPIA4 were enhanced in this mutant. Thus cytosine methylation levels at CHG sites perfectly correlate with the effect of EDM2 on transcript levels of these two TEs.
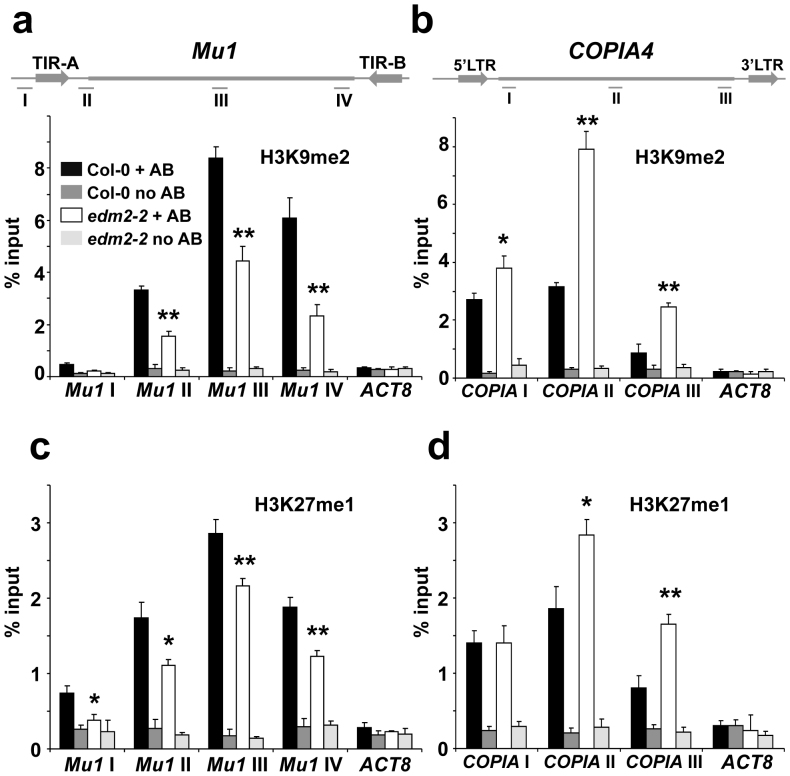
Published genome-wide ChIP-chip data (http://epigenomics.mcdb.ucla.edu/DNAmeth/) also revealed the presence of high levels of the repressive H3K9me2 mark in both Mu1 and COPIA4. ChIP with commercial antibodies against this histone mark combined with qPCR showed that EDM2 affects this type of histone modification at Mu1 and COPIA4. Consistent with the elevated levels of Mu1 transcripts in edm2-2, we found H3K9me2 levels to be reduced at Mu1 in this mutant (Fig. 3a). In addition, the suppression of COPIA4 transcripts in edm2-2 plants is correlated with enhanced H3K9me2 levels at COPIA4 in this mutant (Fig. 3b). Thus, EDM2 affects H3K9me2 levels at Mu1 and COPIA4. The levels of H3K9me2, which is known to be involved in repression of TEs8, perfectly correlate with the effect of EDM2 on transcript levels of these two TEs. Clearly, mutations in EDM2 genetically separate H3K9me2 and CHG methylation from 5mC at CG and CHH sites, which cooperate in transcriptional silencing of plant TEs. Thus, our results suggest involvement of EDM2 at the tested loci in control of the H3K9me2/CHG methylation, which is known to be primarily maintained by the self-reinforcement loop between KYP and CMT3.
Figure 3. EDM2 affects H3K9me2 and H3K27me1 levels at Mu1 and COPIA4.
(a) & (b): ChIP combined with qPCR to measure H3K9me2 levels at Mu1 (a) and COPIA4 (b). (c) & (d): ChIP combined with qPCR to measure H3K27me1 levels at Mu1 (c) and COPIA4 (d). In Mu1 four and in COPIA4, three different regions marked by roman numbers were tested. 3′-part of ACT8 was used as a control region. TIR: terminal inverted repeat, LTR: Long Terminal Repeat. ChIP was performed with antibodies against histone modifications (+AB) or no antibody controls (no AB). Signals from the ChIP assays are represented as percentage of input. Data represent mean plus standard errors of three biological replicates. Statistical significance was determined using Student's t-test (Col0 +AB vs edm2-2 +AB; * P < 0.05, ** P < 0.01).
Besides H3K9me2, we also examined levels of H3K4me3, H3K27me1 and H3K27me3. H3K4me3 is associated with active transcription, while H3K27me1 and H3K27me3 are linked to inactive states of transcription. Levels of H3K27me1 in edm2-2 are reduced or enhanced at Mu1 or COPA4, respectively (Fig. 3c and d). Similar to those of H3K9me2, the levels of the repressive H3K27me1 mark anti-correlate with transcript levels of these two TEs. However, the effect of EDM2 on H3K27me1 levels appears to be smaller than that on H3K9me2 levels. Col-0 exhibited low levels of H3K4me3 and H3K27me3, which were not significantly changed in edm2 mutant plants (Supplementary Fig. S4 and S5). Taken together, we found methylated CHG, H3K9me2 and H3K27me1 to represent EDM2-dependent chromatin marks at the Mu1 and COPIA4 loci that anti-correlate with the transcript levels of these TEs.
Genetic interactions between EDM2 and KYP affecting the Mu1 and COPIA4 loci
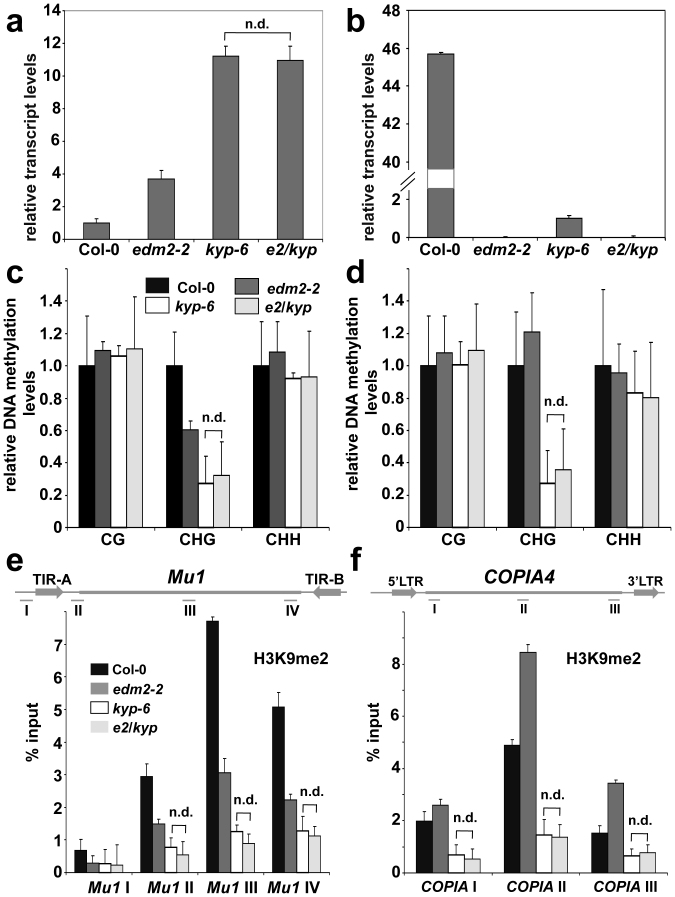
Our data indicate that EDM2-dependent effects on transcript levels of Mu1 and COPIA4 are predominantly associated with H3K9me2 and CHG methylation. KYP is thought to be the major H3K9-specific methyltransferase in Arabidopsis11,12,13. Mutation of KYP leads to loss of CHG methylation but not CG methylation, as seen in edm2-2. Effects of KYP on the regulation of Mu1 and COPIA4 in the Col-0 background of Arabidopsis have not been reported. Interestingly, we found the kyp-6 mutant to exhibit trends of Mu1 and COPIA4 transcript level changes similar to those in edm2-2 (Fig. 4a and b). Levels of Mu1 and COPIA4 transcript levels are enhanced and reduced in kyp-6, respectively.
Figure 4. Levels of transcripts, CHG methylation and H3K9me2 at Mu1 or COPIA4 in edm2 and kyp single or double mutants.
(a) & (b): Mu1 (a) and COPIA4 (b) transcript levels determined by qRT-PCR. (c) & (d): Extent of DNA cytosine methylation at CG, CHG or GHH sites in the bodies of Mu1 (c) and COPIA4 (d) determined by chop-qPCR. (e) & (f): ChIP combined with qPCR to measure H3K9me2 levels at Mu1 (e) and COPIA4 (c). Signals from the ChIP assays are represented as percentage of input. (a–d): Data represent mean plus SD of three biological replicates. (e–f): Data represent mean plus standard errors of two biological replicates. (a–f): Using Student's t tests (P > 0.05) no significant differences between data points for kyp-6 and e2/kyp were detected which were, therefore, marked by "n.d. (not different).
In order to examine genetic relationships between EDM2 and KYP, we created an edm2-2 kyp-6 double mutant. The edm2-2 and kyp-6 mutants were previously reported to be strong alleles28,30. We compared H3K9me2, CHG methylation and transcript levels at Mu1 and COPIA4 of edm2-2 or kyp-6 single mutants to those of the edm2-2 kyp-6 double mutant (Fig. 4c–f). At the COPIA4 locus, H3K9me2 and CHG methylation levels were reduced in the kyp-6 single mutant, while opposite trends were observed in edm2-2. The double mutant showed levels of H3K9me2 and CHG methylation similar to those in the kyp-6 single mutant, indicating that KYP is epistatic to EDM2 at COPIA4 in the regulation of these marks. Thus, at COPIA4, KYP is likely to operate downstream from EDM2. At the Mu1 locus, compared to Col-0, H3K9me2 and CHG methylation levels were reduced in the kyp-6 single mutant. Although single mutations in EDM2 and KYP caused similar trends of changes of H3K9me2, CHG methylation and transcript levels, the effects are more pronounced in kyp-6. The double mutant showed levels of H3K9me2 and CHG methylation similar to those in the kyp-6 single mutant.
Interestingly, in the kyp-6 single and edm2-2 kyp-6 double mutants, levels of the repressive H3K9me2 and CHG methylation marks were reduced at COPIA4, although COPIA4 transcripts were suppressed. This may indicate the existence of (an) additional regulatory mechanism(s) activated by KYP dysfunction. This hypothetical mechanism possibly involves post-transcriptional degradation of RNA by RNA interference (RNAi), as reactivated transcription of TEs can induce accumulation of 21 nt siRNA, which is a hallmark of posttranscriptional gene silencing41,42,43,44.
Mutant edm2 phenotypes exhibit phenotypic variability and instability
Besides loss of race-specific immunity against Hpa, edm2 mutants exhibit developmental abnormalities. These phenotypes include late flowering, altered leaf pavement cell shape and reduced rosette leaf expansion31,32 as well as reduced fertility (see Supplementary Fig. S6) and were consistently observed in independent edm2 mutant lines. Intriguingly, expressivity and penetrance of edm2 mutant phenotypes exhibit substantial variability (Fig. 5a–c). Figure 5a represents the range of intensities observed for the reduction in rosette leaf expansion of edm2-2 plants and Figure 5c shows the range of silique sizes typically observed in edm2 mutants. We found other edm2 phenotypes, such as late flowering and race-specific immunity also to exhibit a similar degree of variability (Supplementary Fig. S7; not shown). Due to the combined effects of the delayed transition to reproduction and bolting on the one hand and reduced leaf expansion on the other hand, the size of edm2 mutant plants substantially varied resulting occasionally in extremely stunted individuals (Fig. 5b). The stochastic nature of these edm2 phenotypes is reminiscent to those observed in previously characterized epigenetic Arabidopsis mutants such as ddm1 and met145,46,47,48. Recurrent selection to stabilize particular phenotypic traits of met1 failed due to the stochastic appearance of a broad range of phenotypes in each met1 generation48.
Figure 5. Phenotypic variability and instability in edm2-2.
(a): Morphological variability of edm2-2 plants. Representative 25-day-old WT and edm2-2 plants. (b): Size variation of mature edm2 mutant plants. Representative 38-day-old WT and edm2-2 plants. (c): Fertility variation of edm2 mutant plants documented by reduced size of siliques. (a), (b) & (c): All plants shown in the respective panels are typical examples and were of the same age and grown simultaneously under the same environmental conditions. These phenotypes are also observed in edm2-3 and edm2-4 mutant alleles.
In order to examine if the extent of rosette leaf area reduction is stabilized in later edm2 generations, “small” and “large” edm2-2 siblings were separately selfed and the extent of leaf expansion in their progeny was examined (Fig. 6). There was clearly no correlation between the intensity of the parental and progeny phenotypes. While no progeny exhibited wild-type leaf expansion, the extent of the mutant phenotype varied among each group of progeny. Thus, at least regarding the reduced leaf expansion, variability rather than the extent of the phenotype was inherited.
Figure 6. Inheritance of edm2-2 developmental phenotypes.
Representative progeny from selfing of small and large edm2-2 individuals.
Discussion
Arabidopsis EDM2 was originally isolated as a gene required for race-specific immunity, which is structurally unrelated to any known components of the plant immune system30. Our previous studies30,31 and the detailed analysis of its deduced amino acid sequences indicated that EDM2 encodes a nuclear protein with features commonly observed in epigenetic factors. EDM2 was further shown to interact with putative EMSY-like chromatin remodeling factors33. Epigenetic mutant plants typically show altered states of transcriptional silencing in TEs17. Hence, TEs have become a useful tool in the study of epigenetic regulators as specific subsets of TEs are reactivated in different epigenetic mutants. Therefore, we decided to explore roles of EDM2 in chromatin scilencing by analyzing TEs using edm2 mutants.
Consistent with our hypothesis, we found in edm2 mutants the silencing states of transposons including Mu1 and COPIA4 to be altered. The changes of transcript levels of these transposons were tightly associated with levels of the repressive epigenetic modifications H3K9me2, H3K27me1 and CHG methylation. Most pronounced were the effects of EDM2 on H3K9me2 and CHG methylation. It should be noted that EDM2 affected CHG methylation, but not CG or CHH methylation at the tested loci. Each type of DNA methylation is controlled by a distinct maintenance pathway. CHG methylation is predominantly maintained by the KYP/CMT3-dependent self-reinforcement loop28. Thus, EDM2 is likely to impact this mechanism. Indeed, mutation of EDM2 affected levels of H3K9me2 at Mu1 and COPIA4 which are also significantly regulated by KYP at these loci (Fig. 4e and f). Our edm2-2 kyp-6 double mutant analysis revealed epistasis of KYP over EDM2 and suggested that KYP can operate downstream from EDM2. Therefore, EDM2 may serve as an additional component upstream from the KYP/CMT3 regulatory loop in the regulation of H3K9me2 and CHG methylation levels at its target loci. One possibility is that EDM2 enables KYP pathway components to recognize CHG methylation and/or H3K9me2 marks in a locus-specific manner. Alternatively, EDM2 may primarily control chromatin structures, which are accompanied by changes of H3K9me2 and CHG methylation levels catalyzed by KYP/CMT3. Future studies on EDM2-dependent mechanisms affecting chromatin states of Mu1 and COPIA4 are needed to address these hypothetical options.
We also found H3K27me1 levels to be changed in edm2-2 at Mu1 and COPIA4. Although the effects of EDM2 on H3K27me1 appeared to be weaker than those on H3K9me2, their trends are similar. As in the case of H3K9me2 and methylated CHG, levels of the repressive H3K27me1 mark anti-correlate with transcript levels of these two TEs. Thus, EDM2 affects both H3K27me1 and H3K9me2 levels at the tested loci. Unlike H3K9me2, H3K27me1 appears to be independent of DNA methylation9. Decreased H3K27me1 levels in a double mutant of ATXR5 and ATXR6 do not affect H3K9me2 levels. Conversely, mutations in KYP, SUVH5 and SUVH6 or triple mutations of them do not affect H3K27me1 levels. This indicates that H3K9me2 and H3K27me1 are mediated by separate pathways, both of which contribute to heterochromatin formation in parallel. EDM2 affects both of these cooperatively acting but separately regulated histone marks. These observations establish EDM2 as a genetic link between H3K9me2/CHG methylation and H3K27me1 that impacts chromatin silencing at Mu1 and COPIA4.
EDM2 may indirectly affect levels of H3K9me2, H3K27me1 and CHG methylation possibly by controlling transcription of genes encoding histone or DNA-methyltransferases or -demethylases. In order to examine if transcript levels of known H3K9, H3K27 and DNA-methyltransferase genes or -demethylase genes are altered in the edm2 mutant plants, we re-analyzed our previously published genome-wide Affymetrix ATH1 array transcriptome data (32; Supplementary Table S1). Genes not represented on this array were analyzed by RT-PCRs (Supplementary Fig. S8). We did not observe significant transcript level changes of histone or DNA-methyltransferases or -demethylases genes, indicating that the effects of EDM2 on chromatin states at Mu1 and COPIA4 are not mediated by differential expression of known chromatin modifier genes listed in Supplementary Table S1. EDM2 may constitute a novel pathway requited for the regulation of Mu1 and COPIA4 at the chromatin level.
Similar to our observations in edm2-2, transcript levels of Mu1 and COPIA4 are enhanced and reduced in met1-3 plants, respectively49. In addition, mutation of MET1 can have opposite effects on H3K9me2 levels at different TEs. For example, in met1-3 plants H3K9me2 levels at Mu1 are down-regulated, while those at Ta3 are up-regulated49. Thus, opposite effects on the silencing states of distinct TEs by loss of a given type of epigenetic information (e.g. CG methylation in met1-3) are not uncommon. The respective silencing mechanisms in each TE can be significantly different.
It is plausible that neither H3K9me2, H3K27me1 nor methylated CHG are the result of a direct catalytic activity of EDM2, as edm2 mutations affect their levels in opposite directions at both tested TEs. A possible mechanistic explanation is that EDM2 establishes at these TE loci the same epigenetic signal, which is distinct from H3K9me2, H3K27me1 or methylated CHG, but which context-dependently recruits factors either enhancing or suppressing these three marks.
Numerous studies have revealed that PHD-fingers are able to bind to N-terminal tail of histone H334. EDM2 contains a repeat of PHD-finger motifs30. We recently found this PHD-finger repeat to bind in vitro to histone in a histone modification-dependent manner (unpublished data). One intriguing question is why EDM2 affects silencing only of a limited number of TEs. As each TE is targeted by different sets of chromatin modifiers50 and epigenetic factors typically do not directly interact with DNA, those proteins must be directed to their target loci by yet unknown mechanisms. However, the targeting mechanisms of epigenetic factors remain poorly understood. A possibility is that by its PHD finger function, EDM2 mediates targeting specificity for chromatin modifiers. Thus only a subset of TEs among those targeted by KYP or other epigenetic factors are affected by EDM2 mutations.
We demonstrated that edm2 mutant plants show phenotypic variability and instability that resemble the metastable developmental phenotypes caused by dysfunction of several known epigenetic regulators such as MET1 and DDM1. Recurrent selection to stabilize particular phenotypic traits of met1 failed due to the stochastic appearance of a broad range of phenotypes in each met1 generation48. Similarly, in edm2 mutant plants, there was no correlation between the extent or expressivity of defined parental and progeny phenotypes. Thus, the transgenerational phenotypic instability observed in edm2 plants may be caused by stochastic variation of epigenetic states resulting in metastable epialleles, that condition the expression of developmentally important genes, rather than “hard” inheritance51. However, functional EDM2 seems to be able to quickly revert such metastable epialleles, as expression of an epitope-tagged wild type EDM2 protein in the edm2-2 mutant background resulted in phenotypically normal plants (data not shown). One open question is whether, the phenotypic diversity observed in edm2 mutant plants correlates at the molecular level with a differential degree of repression or de-repression of its target genes. Future studies will have to address this. In any case, our observations strongly suggest functions of EDM2 in the epigenetic regulation of developmentally important genes besides TEs.
In summary, our results clearly indicated the selective effects of EDM2 on the regulation of TEs at the chromatin level. In addition, we revealed a relationship between EDM2 and the major histone H3K9-methyltransferase gene KYP with their genetic interaction and influence on histone modifications and DNA methylation in a locus dependent manner. This sheds new light on the modes-of-action of EDM2, impacting regulatory mechanisms of plant development and immunity as well as the epigenetic mechanisms in which this gene participates.
Methods
Mutant plants and phenotypic analysis
Wild-type plants of the Arabidopsis accession Col-0 were obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio State University, Columbus, OH). All mutants used are exclusively in the Col-0 background. We obtained a EDM2-LIKE PROTEIN 1 (ELP1) mutant elp1-1 (SALK_145992) and a KRYPTONITE/SUVH4 null mutant kyp-6 (SALK_04147428;) from the SALK T-DNA insertion collection from the ABRC52. The elp1-1 SALK T-DNA line contains an insertion in the 14th of 17 introns in ELP1 gene. The edm2-2, edm2-3 and edm2-4 alleles have been described previously30. The edm2-2 kyp-6 or edm2-2 elp1-1 double mutant was created by crossing edm2-2 to kyp-6 or elp1-1 respectively and genotyping their F2 progeny by PCR amplification. Primers used for genotyping are listed in Supplementary Table S2. For phenotypic analysis, plants were grown simultaneously under the same environmental conditions (22°C, long day condition of 16 h light/8 h dark).
Transgenic lines
To make EDM2pro:HA-EDM2 lines, a full-length coding region of EDM2 cDNA was amplified by PCR using HA-tag sequence containing primers HA:EDM2GW-F1/F2 and EDM2GW-R. The HA-EDM2 cDNA clone was introduced into pDONR-Zeo plasmid (Invitrogen, Carlsbad, CA, USA) to generate a Gateway entry clone. To construct a Gateway destination expression vector containg EDM2 native promoter and nopaline synthase terminator (NOSterm), NOSterm DNA fragment of pMDC32 53 was PCR-amplified using primers, NOSterm-F and NOSterm-R. The PCR product was ligated into between SpeI and SacI sites of pMDC123 53. The EDM2 promoter region was PCR-amplified using primers, EDM2pro-F and EDM2pro-R, and ligated into between HindIII and AscI sites of the NOSterm containing pMDC123. The HA-EDM2 cDNA was then transferred into the destination vector. The resulted binary vector was mobilized into the Agrobacterium tumefaciens strain GV3101 (pMP90) and was used to transform the edm2-2 mutant plants by the floral dipping method54. T3 plants homozygous for a single insertion locus were used for experiments. The primers used are listed in Supplementary Table S2. The XVEpro:HA-EDM2-a plants have been described previously31.
RNA isolation and RT-PCR analysis
Plants were grown in a controlled culture room at 22°C under long-day conditions (16 h light/8 h dark) with white light illumination. RNAs are isolated from 2-week-old plants using TRIzol Reagent (Invitrogen). RT-PCRs are performed as described previously30. Oligo(dT)18 primer was used for reverse-transcription otherwise commented in Supplementary Table S2. Templates prepared with no reverse-transcriptase were analyzed to detect contaminating DNA. Amplification of Actin8 cDNA was used as an internal control. For each reaction of qRT-PCRs, the threshold cycle value (Ct) was determined with MyiQ qPCR system (Bio-rad). Serial dilutions of total cDNA were used to calculate the amplification efficiency (E) of each primer set according to the equation E = 10−1/slope. Relative gene expression data were calculated by a comparative Ct method55 using Actin8 as a internal control. Data were taken in three independent biological replicates with three technical replicates. The primers used for RT-PCRs are listed in Supplementary Table S2.
DNA methylation analysis
For chop-PCR, genomic DNA was isolated from 2-week-old seedlings using the DNeasy Plant Mini kit (Qiagen, Valencia, CA, USA). 150 ng of genomic DNA was digested with 10 units of methylation sensitive restriction enzymes in total of 20 ul at 37°C for 8 hours. The restriction enzymes used were HpyCH4IV (for CG sites), BglII (for CHG sites), AflII (CHH sites in Mu1) and HaeIII (CHH sites in COPIA4). After the restriction enzyme digestions, 1 μl of the digested DNA was applied for quantitative real-time PCR with the Bio-rad iQ5 system (Bio-Rad, Hercules, CA, USA). Genomic regions that do not contain the restriction enzyme recognition sites were also measured and used as internal controls to normalize between samples. Efficient digestions of genomic DNA with restriction enzymes were assessed by PCR amplifications of a part of Actin2 or Tubulin8, where DNA methylation levels are expected to be low. The primers used for real-time PCRs are listed in Supplementary Table S2.
For bisulfite sequencing, 1 μg of genomic DNA was used for bisulfite modification using the EpiTect Bisulfite Kit (Qiagen). After the chemical bisulfite reaction, PCR fragments were amplified using primers shown in Supplementary Table S2. The amplified DNA fragments were gel-purified with the QIAquick Gel Extraction Kit (Qiagen) and cloned into pGEM-T easy vector (Promega, Madison, WI, USA). At least 18 independent clones were sequenced and analyzed using the Kismeth program56.
Chromatin immunoprecipitation
Aerial parts of 3-week-old plants were harvested, and ChIP was performed as described57 with some modifications. Reverse cross-linked chromatin samples were purified with the QIAquick PCR Purification Kit columns (QIAGEN, Valencia, CA, USA), and DNA was eluted twice with 50 μl of buffer EB. 2 μl of eluted DNA was applied for qPCR. Samples were analyzed using SYBR green dye on MyiQ machine (Bio-rad). We used commercially available antibodies specific to dimethyl H3K9 (Wako 308-32361), monomethyl and trimethyl H3K27 (Millipore 07-448, lot # 2019561; Abcam ab6002) or trimethyl H3K4 (Millipore 05-1339) (Wako Pure Chemical Industries, Osaka, Japan; Millipore, Billerica, MA, USA; Abcam, Cambridge, UK). Enrichments of histone modifications were normalized to total input and expressed as % input using the equation; %(ChIP/Total input) = 2∧((Ctinput − log2[input dilution factor]) − (CtChIP)) × 100%. The primers used for real-time PCRs are listed in Supplementary Table S2.
Author Contributions
T.T. conceived, planned and performed all experiments, analyzed the data and wrote parts of the manuscript. T.E. conceived experiments, supervised the project and wrote parts of the manuscript.
Supplementary Material
Supplementary data items
Acknowledgments
We thank M.D. Curtis (University of Zürich) for pMDC32 and pMDC123 vectors. We also thank the ABRC for providing Arabidopsis T-DNA mutants. This work was supported by the United States National Science Foundation [grants IOS-0724838 and IOS-1052556 to TE].
References
- Rando O. J. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev 22, 148–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Tsugane K., Kanno T. & Nishimura T. DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol 53, 766–84 (2012). [DOI] [PubMed] [Google Scholar]
- Huff J. T. & Zilberman D. Regulation of biological accuracy, precision, and memory by plant chromatin organization. Curr Opin Genet Dev 22, 132–8 (2012). [DOI] [PubMed] [Google Scholar]
- Mirouze M. & Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14, 267–74 (2011). [DOI] [PubMed] [Google Scholar]
- Dooner H. K. & Weil C. F. Give-and-take: interactions between DNA transposons and their host plant genomes. Curr Opin Genet Dev 17, 486–92 (2007). [DOI] [PubMed] [Google Scholar]
- Bestor T. H. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond B Biol Sci 326, 179–87 (1990). [DOI] [PubMed] [Google Scholar]
- Flavell A. J. Retroelements, reverse transcriptase and evolution. Comp Biochem Physiol B Biochem Mol Biol 110, 3–15 (1995). [DOI] [PubMed] [Google Scholar]
- Saze H. & Kakutani T. Differentiation of epigenetic modifications between transposons and genes. Curr Opin Plant Biol 14, 81–7 (2011). [DOI] [PubMed] [Google Scholar]
- Jacob Y. et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16, 763–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S. R. & Wallrath L. L. Gene regulation by chromatin structure: paradigms established in Drosophila melanogaster. Annu Rev Entomol 52, 171–92 (2007). [DOI] [PubMed] [Google Scholar]
- Jackson J. P., Lindroth A. M., Cao X. & Jacobsen S. E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–60 (2002). [DOI] [PubMed] [Google Scholar]
- Johnson L., Cao X. & Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12, 1360–7 (2002). [DOI] [PubMed] [Google Scholar]
- Jasencakova Z. et al. Histone modifications in Arabidopsis- high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33, 471–80 (2003). [DOI] [PubMed] [Google Scholar]
- Ebbs M. L., Bartee L. & Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol 25, 10507–15 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M. L. & Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18, 1166–76 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A. et al. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–4 (2001). [DOI] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T. & Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1, E67 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S. et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature 461, 423–6 (2009). [DOI] [PubMed] [Google Scholar]
- Finnegan E. J. & Kovac K. A. Plant DNA methyltransferases. Plant Mol Biol 43, 189–201 (2000). [DOI] [PubMed] [Google Scholar]
- Cao X. & Jacobsen S. E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci U S A 99 Suppl 4, 16491–8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. & Jacobsen S. E. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12, 1138–44 (2002). [DOI] [PubMed] [Google Scholar]
- Cao X. et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13, 2212–7 (2003). [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B. & Matzke A. J. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21, 367–76 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–201 (2006). [DOI] [PubMed] [Google Scholar]
- Bernatavichute Y. V., Zhang X., Cokus S., Pellegrini M. & Jacobsen S. E. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3, e3156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W. J. et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21, 6549–59 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M. et al. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A 100, 8823–7 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M. et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17, 379–84 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F. et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30, 1928–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. et al. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J 49, 829–39 (2007). [DOI] [PubMed] [Google Scholar]
- Tsuchiya T. & Eulgem T. The Arabidopsis defense component EDM2 affects the floral transition in an FLC-dependent manner. Plant J 62, 518–28 (2010). [DOI] [PubMed] [Google Scholar]
- Tsuchiya T. & Eulgem T. Co-option of EDM2 to distinct regulatory modules in Arabidopsis thaliana development. BMC Plant Biol 10, 203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T. & Eulgem T. EMSY-like genes are required for full RPP7-mediated race-specific immunity and basal defense in Arabidopsis. Mol Plant Microbe Interact 24, 1573–81 (2011). [DOI] [PubMed] [Google Scholar]
- Sanchez R. & Zhou M. M. The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36, 364–72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60, 43–66 (2009). [DOI] [PubMed] [Google Scholar]
- Steimer A. et al. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12, 1165–78 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W. J. et al. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell 6, 791–802 (2000). [DOI] [PubMed] [Google Scholar]
- May B. P., Lippman Z. B., Fang Y., Spector D. L. & Martienssen R. A. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1, e79 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I., Schubert I., Tourmente S. & Mathieu O. MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep 7, 1273–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H. & Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26, 3641–52 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanurdzic M. et al. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol 6, 2880–95 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K. et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H. et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–9 (2011). [DOI] [PubMed] [Google Scholar]
- McCue A. D., Nuthikattu S., Reeder S. H. & Slotkin R. K. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet 8, e1002474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J. A., Flowers S. K., Munakata K. & Richards E. J. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci U S A 93, 12406–11 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T. Genetic characterization of late-flowering traits induced by DNA hypomethylation mutation in Arabidopsis thaliana. Plant J 12, 1447–51 (1997). [DOI] [PubMed] [Google Scholar]
- Kankel M. W. et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163, 1109–22 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C. & Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130, 851–62 (2007). [DOI] [PubMed] [Google Scholar]
- Rigal M., Kevei Z., Pélissier T. & Mathieu O. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J 31, 2981–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P. DNA methylation systems and targets in plants. FEBS Lett 585, 2008–15 (2011). [DOI] [PubMed] [Google Scholar]
- Richards E. J. Inherited epigenetic variation--revisiting soft inheritance. Nat Rev Genet 7, 395–401 (2006). [DOI] [PubMed] [Google Scholar]
- Alonso J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–7 (2003). [DOI] [PubMed] [Google Scholar]
- Curtis M. D. & Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133, 462–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–43 (1998). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E. et al. Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9, 371 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R. & Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3, 1018–25 (2008). [DOI] [PubMed] [Google Scholar]
- Yuan J. S., Reed A., Chen F. & Stewart C. N. Statistical analysis of real-time PCR data. BMC Bioinformatics 7, 85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data items