Abstract
Objective
Within translational research projects in the recent years large biobanks have been established, mostly supported by homegrown, proprietary software solutions. No general requirements for biobanking IT infrastructures have been published yet. This paper presents an exemplary biobanking IT architecture, a requirements specification for a biorepository management tool and exemplary illustrations of three major types of requirements.
Methods
We have pursued a comprehensive literature review for biobanking IT solutions and established an interdisciplinary expert panel for creating the requirements specification. The exemplary illustrations were derived from a requirements analysis within two university hospitals.
Results
The requirements specification comprises a catalog with more than 130 detailed requirements grouped into 3 major categories and 20 subcategories. Special attention is given to multitenancy capabilities in order to support the project-specific definition of varying research and bio-banking contexts, the definition of workflows to track sample processing, sample transportation and sample storage and the automated integration of preanalytic handling and storage robots.
Conclusion
IT support for biobanking projects can be based on a federated architectural framework comprising primary data sources for clinical annotations, a pseudonymization service, a clinical data warehouse with a flexible and user-friendly query interface and a biorepository management system. Flexibility and scalability of all such components are vital since large medical facilities such as university hospitals will have to support biobanking for varying monocentric and multicentric research scenarios and multiple medical clients.
Keywords: Requirement specification, biobanking, translational research information technology infrastructure
Introduction
In translational medicine, research projects in the recent years have aimed at efficiently integrating biological findings, genomic data, new molecular technologies and patient phenotype data in order to improve our knowledge on the human diseases and on the potential and limitations of new diagnostic and therapeutic measures [1]. Such research increasingly relies on high quality collections of human biospecimens leading to a rapid increase in biobanking projects. While traditionally clinical researchers only had to focus on the collection and storage of phenotypic clinical data within clinical research warehouses (e.g. [2–5]), electronic data capture systems (EDC) or clinical trials management systems (e.g. [6–12]), there is an abundant need for also collecting high quality human biological samples in the genomic era (e.g. blood, tissue or DNA).
Biobanking is currently being applied in a wide variety of research initiatives. The collection and storage of tissue samples within hospital-based comprehensive cancer centers (mostly initiated and managed by pathology departments) [13], the creation of large biobanks in multicenter research networks [14, 15] and small monocentric collections within specific clinical trials are just examples of this development. Even though literature shows that much progress has already been made in these areas, it has been stated that the specific development of information systems, especially to support biobanking activities, still is a rate-limiting challenge for even faster high quality translational research [16]. In order to optimize the transformation of biomedical data and genomic data into proactive, predictive, preventive, and participatory health, new concepts and methodologies for sample management, data storage, data linkage, data analysis and data interpretation are required.
The term ‘biobank‘ has been used since about 1996 [17] in order to describe collections of various types of biological samples. In 2006 the OECD [18] has defined a ‘biobank‘ as ‘a collection of biological material and the associated data and information stored in an organized system for a population or a large subset of a population’. Based on this, biobanks can be seen as dedicated research infrastructure components, which enable the linkage of physical samples, their storage locations and their ‘quality assured history since their original time of sampling‘ with associated, well documented clinical data. For managing such a biobank, Yuille et al. have emphasized the need for an efficient information infrastructure as a critical component in life-sciences research and have even created the term ‘biobank informatics‘ as a discipline with the purpose of identifying the complete scope of information structures needed and of analyzing how available nomenclature and coding systems can be used for storing and retrieving biobank information [19].
In this context numerous IT-applications have been developed in the last five years in order to support the daily work of biobanking facilities as well as researchers who need to identify specific patient subsets with respective clinical annotations and samples stored within the biobank freezers. Nevertheless, existing publications are mostly limited to either high level descriptions of the database and communication structure as well as the biobank's data safety concept [14] or very technical implementation details [15]. The latter publication nicely illustrates how the different components of the information processing infrastructure support processes and workflows between the partners, such as sampling, labelling, sample documentation, freezer capacity management and quality control for a three centre clinical study in Greifswald and Berlin.
Patel et al. have presented an informatics model for tissue banks as a result of their implementation and experience within the NCI Cooperative Prostate Cancer Tissue Resource. They describe their operating procedures for human subjects’ protection and data collection as well as their data collection application and the methods applied for data transmission [20]. An important issue in their work was to develop a set of common data elements for the annotation of the collected tissues within the four cooperating institutions [21] and to provide a user friendly public query tool supporting researchers in identifying patient subsets from the complete cohort of over 6,000 cases with 38,000 tissue blocks. In another publication Ölund et al. present their development of BIMS (the ‘biobank information management system for the 21st century‘) applied at Karolinska Institute (Stockholm) [22]. They outline the current information management challenges and describe their approach to data integration, their methods for data extraction from the original primary sources, their de-identification concept, the consolidation of data in a shared data model and the abstraction layer which then provides the query environment for the researchers [22].
While all such publications provide informative illustrations of particular IT implementations in dedicated research environments, according to our investigations there is no publication providing a structured catalog of requirements which need to be fulfilled by an IT solution for the different biobanking scenarios. For this purpose in 2009 the German Telematics Platform for Networked Medical Research (TMF) has initiated a strategic project in order to develop a requirements catalog for IT solutions in biobanking and to perform a brief analysis of the respective software market in Germany [23]. Within this publication we aim at presenting the catalog resulting from this project and exemplary illustrating some of the major requirements in various biobanking scenarios.
Methods
As a first step within this project, a comprehensive literature research has been performed to identify publications which have presented concepts, requirements and solutions for information technology implementations for biobanks. As a result of this investigation, numerous publications have been retrieved on various biobanking issues, but real descriptions of functionalities provided by respective IT solutions are sparse. In many publications isolated aspects of information technology are combined with organizational, ethical and data security aspects. Conceptually IT implementations vary between three different levels of biobanking, which are single institution/project biobanks, biobanks of research networks as well as national and international biobanking collaboration scenarios [24–28]. It has then been decided to primarily focus on requirements for the basic management of samples, storage locations and clinical annotations as well as the processes related with sample acquisition, transportation, storage, quality assurance measures and clinical phenotype documentation.
Data security concepts are vital in these IT solutions. They have already been analyzed in an earlier TMF project. In this context a generic data security framework for medical research networks has been defined, based on intense discussions with the German data protection commissioners on national and federal level [29]. This was the basis for the TMF biobanking data protection scheme which relies on the following three key issues:
-
•
separation of informational powers
-
•
pseudonymization
-
•
identity management based on trusted third party (TTP) services, keeping identity data and their associated pseudonyms separate from databases with medical and genetic data.
In this context separation of informational powers implies
-
•
physical separation of identity data (patient list) from medical data (research database)
-
•
physical separation of the data bases storing logistic sample information (sample database), analysis results/genetic data (analysis database) and medical annotation data (research database)
For further details we refer to the comprehensive publications of the TMF data security working group [30, 31].
In special situations (e.g. when not being able to receive a proband’s informed consent for “old” biomaterial or when setting up a cross-institutional biobanking network) pseudonymization of patients may not be sufficient, thus requiring full anonymization. In such scenarios it shall not be neglected, that the combination of experimental (especially genetic) data with detailed patient data poses even further requirements for data protection. Even in the absence of personal data, patient re-identification through the combination of large datasets cannot be excluded [24, 32, 33]. Therefore, particularly when comprehensive patient-related data will be shared with the scientific community outside the hospital environment in which the patient has been treated, the implementation of more far reaching anonymization tools (e.g. based on the concept of k-anonymity [34–36]) is necessary [24].
In order to construct a comprehensive requirements catalog an expert panel has been established with two clinical specialists with comprehensive biobanking experience (MH and MK) and a background in pathology and laboratory medicine, as well as two medical informatics specialists with comprehensive experience in establishing IT infrastructures for medical research networks (US and FÜ). This panel has been moderated by the main author (HUP, a medical informatics specialist and hospital CIO) in order
-
•
to collect a large set of requirements in a first sampling round,
-
•
to create a set of high level categories for such requirements and
-
•
to structure and to categorize the list of requirements into such categories within a second round.
Results
After the first round of consecutive input from the four domain experts about 150 functionalities (process steps which need to be supported) have been identified. From those, three main categories and 20 subcategories have been defined in order to categorize the set of requirements. In the second round wording for the different requirement specifications has been refined and every requirement has been mapped into one of the 20 subcategories. In a final round the complete catalog has been checked again to eliminate redundancies and check for inconsistencies. This finally resulted in a specification catalog with more than 130 requirements grouped into 3 major categories and 20 subcategories (compare ►Table 1)
Table 1.
Major categories and subcategories of the requirement specification catalog.
| 1. | Requirements for the organization and management of biobanks | |
| 1.1 | Organizational master data management (projects, institutions, persons, roles) | |
| 1.2 | Biospecimen storage master data management (storage infrastructure, sample metadata) | |
| 1.3 | Sampling, sample tracking, sample delivery, sample reception, sample storage and preanalytics (e.g. sample aliquotation) | |
| 1.4 | Sample administration, management of storage capacities | |
| 1.5 | Management of quality assurance data (e.g. temperature logging) | |
| 1.6 | Providing up to date status and history information on every biobank sample | |
| 1.7 | Providing up to date status information on every storage location and freezer capacities | |
| 1.8 | Management of sample requests | |
| 1.9 | Management of sample shipments | |
| 1.10 | Management of sample analysis results | |
| 1.11 | Support of basic services and routine processes of a biobank | |
| 1.12 | Management of informed consent levels for all samples | |
| 2. | Requirements for the querying of clinical annotations and management of sample/project requests | |
| 2.1 | User-friendly intuitive query module | |
| 2.2 | Linkage of proband data with associated sample storage locations | |
| 2.3 | Creation of research project descriptions and workflow management for receiving project approval by the biobank’s scientific committee | |
| 3. | Requirements for the creation of clinical annotations | |
| 3.1 | Flexible design tool for data assessments (electronic case report forms = eCRFs) | |
| 3.2 | Powerful data validation logic | |
| 3.3 | Inclusion of standardized international classification systems and nomenclatures (e.g. ICD, ICD-O, ICPM, SNOMED, LOINC) |
|
| 3.4 | Graphical annotations | |
| 3.5 | Flexible import and export functions | |
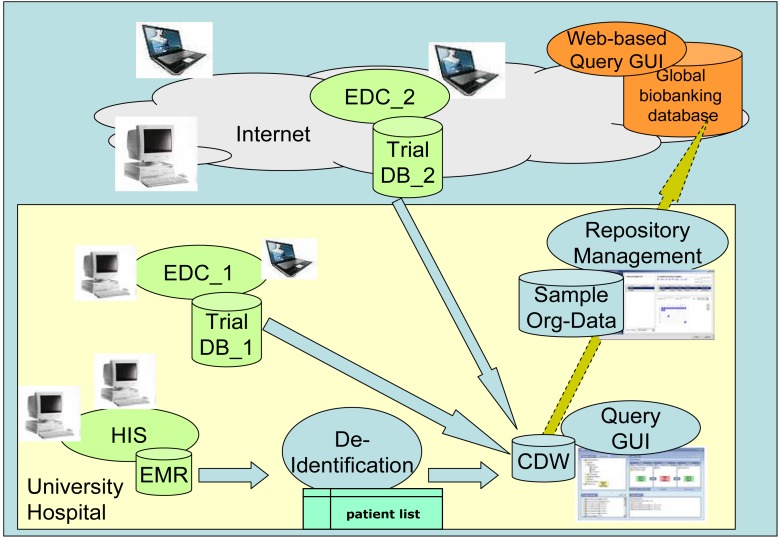
Usually, biobanks compliant with high quality assurance standards will be implemented within either university hospitals or within large non-university research institutions. Especially within a university hospital environment the design of a biobanking information technology infrastructure needs to consider the typically very heterogeneous IT environment which is already established and has grown over decades within hospitals. While the requirements defined in the specification catalog according to the categories presented in ►Table 1 focus on supporting the logistic and medical processes around biobanking, a second, more architectural and technical view needs to contemplate this, considering best practice concepts at the information system level. Within university hospitals biobanking will in the future be pursued by varying clinical specialties and in a variety of research contexts. Nevertheless, it will be impossible to establish and maintain different IT solutions always serving to 100% only one specialized research and biobanking scenario. Instead, a generic architecture (compare ►Fig. 1) is required, which may support varying laboratory environments and storage concepts over time.
Fig. 1.
Architecture and core components of a biobanking IT infrastructure: primary data sources in green (EDC = electronic data capture system, DB = database, HIS = hospital information system, EMR = electronic medical record); biobanking core components in blue (Org-Data = organizational data on samples and storage locations, CDW = clinical data warehouse, GUI = graphical user interface); integration with transnational, European or world-wide biobanking networks in orange.
In order to minimize the effort to gather and document clinical annotations (e.g. phenotype descriptions of study subjects from whom samples are taken) and further comply with the TMF data security framework, one basic biobanking component is a clinical data warehouse (CDW). It may either receive its data input stream from an institution-based electronic medical record (or in more heterogeneous IT environments from several hospital based information systems) or (within a multicenter research project) from a clinical trials database/electronic data capture system or a specialized registry database. While a study subject’s medical data are typically already stored in a de-identified manner within a research project, they are linked with respective identification data within a hospital’s electronic medical record. Therefore a second important component of the biobanking IT environment is a pseudonymization service which shall always be applied before uploading medical data from the hospital’s routine clinical care database into the clinical data warehouse. The third core component is then constituted by a typical LIMS biorepository management submodule. In order to allow sharing of clinical annotations between multiple national and international biobanks the clinical data warehouse component needs to be able to provide anonymized export data sets, which could then be integrated within larger global biobanking networks such as currently established for example within CRIP [24] in Europe or CPCTR in the US [21].
This federated approach implies that the subset of requirements focusing on the creation of clinical annotations typically needs to be fulfilled either by the tools to define clinical assessment forms within the hospital’s clinical information system or by the eCRF generation/definition tool of the clinical trials management system. Secondly, the subsets of requirements associated with the flexible querying of clinical annotations and defining sample/research project requests needs to be fulfilled by the clinical data warehouse. Finally all requirements regarding the efficient sample acquisition, tracking of sample transportation, preanalytical sample annotation, sample storage management and quality assurance measures need to be fulfilled by the biorepository management component.
Listing and describing all the requirements of our specification would go far beyond the scope of this article. Therefore, we have decided to include the complete requirement catalog as additional online material with this publication and in the following only concentrate on few important aspects which describe a) the complexity of such an IT implementation within a university hospital and b) illustrate the flexibility required from the toolbox provided with the biorepository management solution in order to allow the biobanking team to create workflows completely adapted to their specific research scenario.
Project-specific definition of the research and storage context
Before even documenting the acquisition and handling of any biosample, for every research project the particular setup needs to be performed by the biobanking system manager. Roles with associated access rights and permitted functionalities need to be created. All persons involved in the research project need to be entered into the system and linked with their respective role. The storage infrastructure and hierarchy needs to be configured and finally metadata which shall be used to characterize the various sample types to be managed within the project need to be defined. Even when sometimes people or resources within a biobank may overlap between research projects, staff members role definitions can vary across projects and storage sublocations may be specifically reserved for one particular project. Further, sample metadata descriptions may be different from one project to another. Therefore, the setup configuration tool needs to support flexible project-specific parametrization of all such core elements taking into account basic parameters, which may be the same across all projects, but still keeping all project-specific definitions and access rights completely independent from one project to another. Thus, comprehensive multitenancy is most important when such a biorepository management system is considered to be applied in multiple project environments.
Defining Workflows to track sample processing, sample transportation and sample storage
Depending on the particular research project, the requirements for sample tracking may vary extremely. Within a central hospital environment sample acquisition is usually performed in outpatient clinics, on hospital wards, within diagnostic departments such as endoscopy or within the operating rooms. In-house transportation of samples can be standardized and time between sample collection and storage in a freezer is usually short.
Multi-center studies provide a much more complex and less standardized environment. In some projects samples may be collected from study subjects within regionally distributed general medical practices. For liquid samples centrifugation may be carried out within this practice and samples then need to be kept under standardized temperature conditions until they are picked up by a logistic service provider. Transportation then may take several hours and for long distance transports shipping companies sometimes even change transportation means in between. Risk of destroying or simply losing samples on such transports therefore is much higher than within a hospital. To document all transportation steps, pre- and post-centrifugation delays, type of centrifugation, primary container types and types of longterm storage containers as well as transport times and conditions, much more complex workflow steps and metadata input capabilities need to be supported. Tracking every single transportation step needs to be supported and simplified by specific worklists, in which one particular sample may change its status several times until its final storage in a biobank freezer is documented. A workflow and data entry screen generator tool as well as the respective workflow engine need to be flexible enough to individualize those documentation steps for every particular scenario. Data entry must be supported through varying modes of access. Secure remote login over the internet is an absolute must for distinct documentation steps. Tracking transportation and reception data may be pursued either by interactive data entry screens or by interfacing with mobile barcode scanners for samples and storage racks. Interfacing with a laboratory or pathology system may be necessary in order to automatically upload sample orders or pursue batch data import.
The more precise the recording of processing variables throughout the sample's life span is carried out, the more accurate and extensive will the extraction of explicit information be, when this is required for clinical or research purposes. Recently an international, interdisciplinary group of researchers has argued that that harmonization of methods to trace preanalytical information is a necessary condition for the development of large-scale research involving samples from different settings. Therefore, the International Society for Biological and Environmental Repositories Biospecimen Science Working Group developed a “Standard PREanalytical Code” (SPREC) that identifies the main preanalytical factors of clinical fluid and solid biospecimens and their simple derivatives [37].
Open interfaces to link with high throughput sample preanalytic handling and storage robots
While smaller biobanks may still rely on manual sample storage handling, large biobanks will more and more need to apply liquid handling platforms for aliquotation and storage of liquid samples in order to keep up with the growing number of samples to manage. Therefore, on one hand data entry of storage locations may be simply pursued through interactive screen dialogues, with support for quickly identifying empty rack positions within a freezer. Furthermore however, interfaces to aliquotation robots will be required in order to automatically acquire volume and location data for aliquot plates. Interfaces to storage robots may even go further and need to support automated batch-processing of samples, controlling the positioning of aliquot plates within freezer racks and automatically instantiate robot-based sample retrieval processes when samples are required for experimental analysis.
Furthermore, monitoring temperature conditions within freezers is an important quality assurance measure. Such temperature logging information need to be automatically transferred via electronic interface into the biorepository management system and be linked with the samples stored in the respective freezer.
Discussion
Despite many advances of biobanking described in the literature, significant issues and limitations still remain that are restricting the impact of translational research. The major issues include the need to increase the quality and standardization of biospecimens collected, to enhance accrual capacity in terms of scale and disease representation, and above all, to maintain public trust in these activities [38]. Sung et al. have further stated that inadequate and inefficient information systems for specialized support of all biobanking activities still present a rate-limiting challenge for even faster high quality translational research [16].
In Germany, biobanking has advanced a lot in recent years (e.g. [13–15]). Nevertheless, IT support is still limited and restricted to proprietary self-developed biorepository management solutions. Commercially available LIMS or EDC-systems with biorepository modules have been applied in many countries, but due to the strict data protection regulations in Germany [29–31] they may not be transferred directly to German biobanking environments. Therefore we have defined a federated architectural framework comprising primary data sources for clinical annotations, a pseudonymization service, a clinical data warehouse with a flexible and user-friendly query interface and a biorepository management system. For the latter we have created a comprehensive requirements specification catalog. Some of those requirements have been described in this paper. We have illustrated that flexible information systems are required in order to individualize sample tracking and documentation flows and adapt them to a varying set of biobanking scenarios.
Since results of research using human biological samples often depend on the ‘events’ that samples have undergone during their ‘lifetime’ from sampling through processing, freezing, and thawing prior to usage (the so-called ‘pre-analytical variations’), biobanks and their respective biorepository management systems must guarantee traceability of all such events and ideally perform quality control testing for sample validation/authentication prior to release of materials to researchers [39]. In large high throughput biobanks manual interactive documentation of all such data will not be sufficient. In such environments interfaces with modern automated equipment are essential. Unfortunately application programming interfaces (APIs) for such robots are still proprietary and always need to be specifically adapted to the respective systems. As researchers have already worked on defining a “Standard PREanalytical Code” [37] for annotating samples with metadata, further efforts need to be established in also standardizing robot APIs.
Finally, in the future institutions will probably not only want to establish local institution-based or multicenter biobanks, but need to cooperate on a global transnational, European or even worldwide level [24]. IT-solutions within local institutions therefore need to be adaptable to such future requirements and scalable for their integration in larger biobanking networks. For this purpose, the German ministry of research (BMBF) has just provided grant support for the TMF-initiated P2B2-project, which aims at implementing a web-based national project portal within the newly established German biobanking register and shall also provide links to the upcoming BBMRI network.
Clinical Relevance Statement
Providing efficient IT-support for biobanking will be essential for clinical and translational research projects. This paper provides an IT-architecture for biobanking, a categorization of functional requirements, the complete requirement specification (as an online appendix) and exemplary illustrations of some major requirements.
Conflict of Interest
The author(s) declare that they have no competing interests.
Human subject research
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects. There were no human subjects involved within this project.
Supplementary Material
Acknowledgments
This work was supported in part by the Deutsche Forschungsgemeinschaft (KFO 179) and by the TMF eV.
References
- 1.Butte AJ. Translational Bioinformatics: Coming of Age. J Am Med Inform Assoc 2008; 15(6):709-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant A, et al. Integrating feedback from a clinical data warehouse into practice organisation. Int J Med Inform 2006; 5(3-4): 232-239 [DOI] [PubMed] [Google Scholar]
- 3.Rubin DL, Desser TS. A data warehouse for integrating radiologic and pathologic data. J Am Coll Radiol 2008; 5:(3): 210-217 [DOI] [PubMed] [Google Scholar]
- 4.Barrett JS, Koprowski SP Jr. The epiphany of data warehousing technologies in the pharmaceutical industry. Int J Clin Pharmacol Ther 2002; 40(3): S3-S13 [PubMed] [Google Scholar]
- 5.Dorda W, Gall W, Duftschmid G. Clinical data retrieval: 25 years of temporal query management at the University of Vienna Medical School. Methods Inf Med 2002; 41(2): 89-97 [PubMed] [Google Scholar]
- 6.Brandt CA, et al. TrialDB: A web-based Clinical Study Data Management System. AMIA Annu Symp Proc 2003: 794. [PMC free article] [PubMed] [Google Scholar]
- 7.Sahoo U, Bhatt A. Electronic data capture (EDC) – a new mantra for clinical trials. Qual Assur 2003; 3-4: 117-121 [DOI] [PubMed] [Google Scholar]
- 8.Cuticchia AJ, Cooley PC, Hall RD, Qin Y. NIDDK data repository: a central collection of clinical trial data. BMC Med Inform Decis Mak 2006; 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welker JA. Implementation of electronic data capture systems: barriers and solutions. Contemp Clin Trials 2007; 3: 329-336 [DOI] [PubMed] [Google Scholar]
- 10.Ene-Iordache B, et al. Developing regulatory-compliant electronic case report forms for clinical trials: experience with the demand trial. J Am Med Inform Assoc 2009; 16(3): 404-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Emam K, et al. The use of electronic data capture tools in clinical trials: Web-survey of 259 Canadian trials. J Med Internet Res 2009; 11(1): e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray E, et al. Methodological Challenges in Online Trials. J Med Internet Res 2009; 11(1): e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stege A, Hummel M. Erfahrungen bei Einrichtung und Betrieb einer Biobank. Pathologe 2008; [Suppl.2] 29: 214-217 [DOI] [PubMed] [Google Scholar]
- 14.Posch MG, et al. The Biomaterialbank of the German Competence Network of Heart Failure (CNHF) is a valuable resource for biomedical and genetic research. Int J Cardiol 2008; 136: 118-111 Doi:10.1016/j.ijcard.2008.03.089 [DOI] [PubMed] [Google Scholar]
- 15.Angelow A, et al. Methods and implementation of a central biosample and data management in a three-centre clinical study. Computer methods and programs in biomedicine 2008; 91: 82-90 [DOI] [PubMed] [Google Scholar]
- 16.Sung NS, et al. Central challenges facing the national clinical research enterprise. JAMA 2003; 289(10): 1278-1287 [DOI] [PubMed] [Google Scholar]
- 17.Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. J Mol Med 1996; 74: 297-312 [DOI] [PubMed] [Google Scholar]
- 18.Creation and Governance of Human Genetic Research Databases. OECD Publishing; 2006. 25 October. ISBN-92-64-02852-8. [Google Scholar]
- 19.Yuille M, et al. Biobanking for Europe. Brief Bioinformatics 2007. 9: 14-24 [DOI] [PubMed] [Google Scholar]
- 20.Patel AA, et al. An informatics model for tissue banks – lessons learned from the Cooperative Prostate Cancer Tissue Resource. BMC Cancer 2006; 6: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel AA, et al. The development of common data elements for a multi-institute prostate cancer tissue bank: the Cooperative Prostate Cancer Tissue Resource (CPCTR) experience. BMC Cancer 2005; 5: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ölund G, Lindqvist P, Litton JE. BIMS: An information management system for biobanking in the 21st century. IBM Systems Journal 2007; 46: 171-182 [Google Scholar]
- 23.Prokosch HU, et al. TMF IT Strategie, Teilprojekt 3: Erstellung eines Anforderungskatalogs zur IT-Unterstützung von Biomaterialbanken und Analyse der derzeit in Deutschland verfügbaren IT-Werkzeuge zur Unterstützung des Managements von Biomaterialbanken. TMF Abschlussbericht, 18.1.2010 [Google Scholar]
- 24.Asslaber M, Zatloukal K. Biobanks: transnational, European and global networks. Brief Funct Genomic Proteomic 2007; 3: 193-201 [DOI] [PubMed] [Google Scholar]
- 25.Yuille M, et al. The UK DNA banking network: a "fair access" biobank. Cell Tissue Bank 2009; 11: 241-251 Doi: 10.1007/s10561-009-9150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asslaber M, et al. The Genome Austria Tissue Bank (GATiB). Pathobiology 2007; 74(4): 251-258 [DOI] [PubMed] [Google Scholar]
- 27.Viertler C, Zatloukal K. Biobanking and Biomolecular Resources Research Infrastructure (BBMRI). Implications for pathology. Pathologe 2008; 29 (Suppl 2): 210-213 [DOI] [PubMed] [Google Scholar]
- 28.Rudan I, et al. “10001 Dalmatians:” Croatia launches its national biobank. Croat Med J 2009; 50(1): 4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reng CM, Debold P, Specker CH, Pommerening K. Generische Lösungen zum Datenschutz für die Forschungsnetze in der Medizin. Schriftenreihe des TMF e.V., Band 1, 2006 [Google Scholar]
- 30.Becker R, Ihle P, Pommerening K, Harnischmacher U. Ein generisches Datenschutzkonzept für Biomaterialbanken (Version 1.0), TMF Bericht, April2006 [Google Scholar]
- 31.Pommerening K, Becker R, Sellge E, Semler SC. Datenschutz in Biomaterialbanken. In: Steyer G, Tolxdorff T. TELEMED 2006: Gesundheitsversorgung im Netz. Tagungsband zur 11. Fortbildungsveranstaltung und Arbeitstagung – Nationales Forum zur Telematik für die Gesundheit. Berlin: Aka GmbH, 2006: 89-99 [Google Scholar]
- 32.Godard B, et al. Data storage and DNA banking for biomedical research: informed consent, confidentiality, quality issues, ownership, return of benefits. A professional perspective. Eur J Hum Genet 2003; 11(Suppl.2): S88-S122 Doi: 10.1038/sj.ejhg.5201114 [DOI] [PubMed] [Google Scholar]
- 33.van Veen EB, et al. TuBaFrost 3: regulatory and ethical issues on the exchange of residual tissue for research across Europe. Eur J Cancer 2006; 42: 2914-2923 [DOI] [PubMed] [Google Scholar]
- 34.Sweeney L. k-Anonymity: A Model for Protecting Privacy. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems 2002; 10(5): 557-570 [Google Scholar]
- 35.Sweeney L. Achieving k-Anonymity privacy protection using generalization and suppression. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems 2002; 10(5): 571-588 [Google Scholar]
- 36.Stark K, Eder H, Zatloukal K. Priority-based k-anonymity accomplished by weighted generalisation structures. Computer Sci 2006; LNCS4081: 394-404 [Google Scholar]
- 37.Betsou F, et al. Standard preanalytical coding for biospecimens: Defining the sample PREanalytical Code. Cancer Epidemiol Biomarkers Prev; 19(4): OF1-8 [DOI] [PubMed] [Google Scholar]
- 38.Watson PH, et al. Evolutionary concepts in biobanking – the BC BioLibrary. J Transl Med 2009; 7: 95 doi: 10.1186/1479-5876-7-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betsou F. Biobanks in immunoanalysis and specialised biology. Immunoanalyse et Biologie Spécialisée 2006; 21(6): 327-332 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.