Abstract
The Burkholderia cepacia complex (Bcc) comprises strains with a virulence potential toward immunocompromised patients as well as plant growth–promoting rhizobacteria (PGPR). Owing to the link between quorum sensing (QS) and virulence, most studies among Bcc species have been directed toward QS of pathogenic bacteria. We have investigated the QS of B. ambifaria, a PGPR only infrequently recovered from patients. The cepI gene, responsible for the synthesis of the main signaling molecule N-octanoylhomoserine lactone (C8-HSL), was inactivated. Phenotypes of the B. ambifaria cepI mutant we observed, such as increased production of siderophores and decreased proteolytic and antifungal activities, are in agreement with those of other Bcc cepI mutants. The cepI mutant was then used as background strain for a whole-genome transposon-insertion mutagenesis strategy, allowing the identification of 20 QS-controlled genes, corresponding to 17 loci. The main functions identified are linked to antifungal and antimicrobial properties, as we have identified QS-controlled genes implicated in the production of pyrrolnitrin, burkholdines (occidiofungin-like molecules), and enacyloxins. This study provides insights in the QS-regulated functions of a PGPR, which could lead to beneficial potential biotechnological applications.
Keywords: Antifungal molecules, Burkholderia, quorum sensing, random mutagenesis, secondary metabolites
Introduction
The Burkholderia cepacia complex (Bcc) encompasses genetically closely related bacteria, currently distributed into 17 validly named species (Vandamme and Dawyndt 2011). Bcc species carry large multireplicon genomes, giving them a remarkable potential to adapt to diverse ecological niches. Indeed, since the first ecological description of B. cepacia as an onion pathogen, Bcc strains have been isolated from soils, waters, rhizospheres, immunocompromised patients, and industrial products (Burkholder 1950; Mahenthiralingam et al. 2005; Vial et al. 2011). Two major reasons of concern for humans are that Bcc species can be efficient plant growth–promoting rhizobacteria (PGPR), but also represent a significant threat to the life of immunocompromised individuals, such as those suffering from cystic fibrosis (CF) (Govan et al. 2000). Tremendous advances in identification techniques and taxonomy have revealed that Bcc members are widely but heterogeneously distributed according to niches, some species displaying particularly high virulence potential toward CF patients, while others acting as efficient PGPR (Mahenthiralingam et al. 2008). Still, almost all Bcc species have been isolated from both environmental and clinical sources, suggesting that adaptation to a specific niche is not strictly linked to the affiliation of a particular species (Mahenthiralingam et al. 2008; Vial et al. 2011). Accordingly, as the environment is apparently a reservoir for life-threatening bacteria, the commercial use of Bcc strains is not recommended, and actually placed under a moratorium imposed by the U.S. Environmental Protection Agency (EPA) (Chiarini et al. 2006).
Quorum sensing (QS) is a bacterial cell-to-cell communication system, based on the release of signaling molecules in the microenvironment; when the bacterial population grows, the concentration of molecules increases, reaching a threshold that triggers regulation of target genes (Williams 2007). The first QS system was discovered in Vibrio fischeri and implicated the LuxI synthase, responsible for the production of an N-acylhomoserine lactone (AHL) as signal molecule, which binds its cognate transcriptional regulator LuxR (Engebrecht and Silverman 1984). Similar LuxRI-type QS systems, relying on AHL molecules, have since been identified in most Gram-negative bacteria. In Bcc species, a LuxRI-type system named CepRI exists (Lewenza et al. 1999). CepRI relies on N-octanoylhomoserine lactone (C8-HSL) and N-hexanoylhomoserine lactone (C6-HSL) as signal molecules, the former being generally the most abundant one (Lutter et al. 2001). Some phenotypes, such as production of siderophores, proteolytic activities, and biofilm formation, have been described to be placed under QS control in Bcc bacteria (Eberl 2006). As QS is typically implicated in the regulation of virulence factors, most studies have been primarily focused on pathogenic bacteria (Sokol et al. 2007). However, QS could be considered more generally as a mean for bacteria to sense and interact with their microenvironments (Williams 2007; Mellbye and Schuster 2011). Recent studies implicating nonpathogenic Burkholderia strains have demonstrated that QS is important for bacterial relationships within the rhizosphere, for interaction with plants as well as in polymicrobial communities (Chan et al. 2011; Suarez-Moreno et al. 2012).
The Bcc Burkholderia ambifaria species is mostly isolated from soils and is especially predominant in the rhizosphere of several crops (Coenye et al. 2001; Coenye and Vandamme 2003). The type strain B. ambifaria LMG19182T (or AMMD) was isolated from the healthy pea rhizosphere and was thereafter used as a biocontrol agent (Coenye et al. 2001; Chiarini et al. 2006). This species is seldom isolated from CF patient, where it causes transient and nontransmissible infections (Chiarini et al. 2006; Mahenthiralingam et al. 2008). Nevertheless, the clinical strain AU0212 was shown to be clonal to AMMD, suggesting that the environment represents a reservoir for clinical strains (Payne et al. 2005). The QS system of B. ambifaria has not been studied in details. It was nevertheless included in a few studies comparing Bcc members, revealing that it also possesses a CepRI system, relying on C6-HSL and C8-HSL signal molecules (Lutter et al. 2001; Zhou et al. 2003). Proteolytic activity, biofilm formation, and virulence toward nematodes are QS-controlled phenotypes, but seem strain-dependent (Wopperer et al. 2006). Interestingly, production of antifungal molecules is also QS-controlled in this Bcc species (Zhou et al. 2003; Schmidt et al. 2009). We have carried out a phenotypic study and a whole-genome transposon mutagenesis screening to identify QS-controlled genes and functions of a clinical B. ambifaria strain we have previously reported (Vial et al. 2008, 2010). This study will contribute to the understanding on the functions regulated by QS in a poorly virulent Bcc species with great biotechnological properties.
Experimental Procedures
Bacterial strains and culture conditions
Burkholderia ambifaria strain HSJ1 was isolated from sputum of a CF patient (Vial et al. 2008). Escherichia coli SM10 λ pir (thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir) was used as a pKNOCK-Cm vector donor for conjugation experiments (Simon et al. 1983; Alexeyev 1999). Escherichia coli SM10pir/pIT2 containing the Tn5-derivative ISlacZ/hah was used as donor for random whole-genome transposon-insertion mutagenesis (Jacobs et al. 2003). Unless otherwise specified, the strains were routinely cultured at 37°C in tryptic soy broth (TSB) (BD) with shaking (240 rpm) in a TC-7 roller drum (New Brunswick, Canada), or on TSB agar plates.
Construction of the cepI and cepR mutants
The HSJ1 cepI and cepR mutants were constructed using a suicide pKNOCK-Cm vector, according to the gene knockout strategy described previously (Alexeyev 1999). Bamb_4118 (cepI) and Bamb_4116 (cepR) from the sequenced B. ambifaria AMMD strain (Winsor et al. 2008) were used as a template to design the primers (Table S1), which carry KpnI and XbaI restriction sites, respectively. The fragment was then cloned as previously described (Vial et al. 2008). Mutants (single cross-over) were selected on TSB agar with 40 μg/mL chloramphenicol, and 25 μg/mL gentamicin to select against the donor (Sigma-Aldrich, Oakville, ON, Canada).
LC/MS-MS analyses for AHL production
Culture samples of HSJ1 wild-type (WT) and cepI strains were retrieved at different time points of growth curve; OD600 was measured and 5 mg/L of methanolic internal liquid chromatography/mass spectroscopy (LC/MS) standard 5,6,7,8-tetradeutero-4-hydroxy-2-heptylquinoline (HHQ-d4) were added to samples (Lépine and Déziel 2011). Culture samples were vortexed, and extracted twice with ethyl acetate (1:1, vol:vol), pooled and evaporated at 35°C under a gentle stream of nitrogen. The residue was then resuspended in acidified acetonitrile (solvent B; details presented in Supplemental Experimental Procedures) at 10× the initial concentration (Lépine and Déziel 2011). Samples were analyzed by high-performance liquid chromatography (HPLC; Waters 2795, Mississauga, ON, Canada) equipped with a C8 reverse-phase column (Eclipse XDB-C8, Agilent Technologies, Mississauga, ON, Canada), and the detector was a mass spectrometer (Quattro Premier XE, Waters). Analyses were carried out in the positive electrospray ionization (ESI+) mode, supplemented by the multiple reactions monitoring (MRM) mode (details presented in Data S1). Samples were prepared in triplicate from three different colonies for each strain, and experiments were carried out at least twice independently.
Phenotypic assays
Siderophore production was determined with Chrome Azurol S (CAS) agar plates (Schwyn and Neilands 1987). Bacteria siderophores are visualized by an orange halo around the colonies. Proteolytic activity was determined with 1% skim milk agar plate (Vial et al. 2008). Cholesterol oxidase activity was assessed as described (Doukyu and Aono 2001), with slight modifications as the TSB agar plates contained 0.26% Triton X-100 and 0.68 mmol/L cholesterol (Sigma-Aldrich). Cholesterol oxidase activity is visualized by a turbid precipitate around colonies, corresponding to oxidation of cholesterol into 6β-hydroperoxycholest-4-en-3-one, which is poorly soluble. Hemolytic activity was estimated on 5% sheep blood agar plates (Quelab, Montreal, QC, Canada) and 5% human blood agar plates. Antifungal activity was investigated against Pythium ultimum (Dr Richard Bélanger, Université Laval, Québec, QC, Canada), Rhizoctonia solani Kühn (MUCL number 51654, BCC/MUCL, Louvain-La-Neuve, Belgium), and Candida albicans. Potato dextrose agar (PDA, BD) and malt agar (BD Difco, Mississauga, ON, Canada) plates were inoculated with agar plugs of P. ultimum and R. solani, respectively. An overnight preculture of C. albicans in TSB was homogeneously spread with a sterile swab on a TSB agar plate, and then incubated at 37°C for 24 h. TSB agar plates supplemented with 0.1% Congo Red (Sigma-Aldrich) are used to study the colonial morphology and the ability to bind the red pigment (Vial et al. 2010). For most of phenotypic assays described above, HSJ1 WT and cepI strains were grown overnight in TSB, the cultures were normalized to OD600 = 5 and 3 μL were spotted on different agar plates, incubated at 37°C for 24 h and 48 h. For the tests with P. ultimum and R. solani, plates were incubated at 25°C for 3 and 21 days, respectively. Congo Red agar plates were inoculated with 100 μL of overnight preculture normalized to OD600 = 5 and diluted until 10−7, incubated for 1 day at 37°C, and then allowed to grow at room temperature for 20 days. Each experiment was repeated at least twice independently. For the inhibition test against B. multivorans, an overnight culture of B. multivorans ATCC 17616 was diluted to OD600 = 1 in TSB, and then incorporated in TSB medium containing 7.5 g/L agar (100 μL bacterial suspension/100 mL soft agar medium). The bacterial strains were allowed to grow 24 h at 30°C in liquid Basal Salts Medium (Mahenthiralingam et al. 2011). The supernatants were collected and 100 μL were laid in wells made in plates overlaid with B. multivorans. Burkholderia ambifaria AMMD WT strain was used as a control for the production of enacyloxins (Mahenthiralingam et al. 2011) (data not shown). To estimate if the phenotype could be restored by exogenous addition of signal molecules, control plates were supplemented with 2 mg/L C8-HSL.
Infection of Drosophila
Fruit flies were infected by needle pricking according to the protocol previously described (Castonguay-Vanier et al. 2010). Control solution was composed of 10 mmol/L MgSO4 supplemented with 500 μg/mL ampicillin (Sigma-Aldrich) to avoid infection with nonspecific bacteria. The same solution was used to dilute B. ambifaria HSJ1 and its cepI mutant, precultured in TSB, at a final OD600 of 2. Thirty-six flies distributed in three bottles were pricked with bacterial suspensions containing HSJ1 or its cepI mutant; twelve flies were also pricked with the control solution to assess that the mortality was not due to the injury. Fly survival was scored daily and survival curves were processed with GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA) to perform a statistical log-rank (Mantel–Cox) test.
Transposition mutagenesis
The HSJ1 cepI mutant was used as background for random whole-genome transposon-insertion mutagenesis, by mating with E. coli SM10pir/pIT2 containing Tn5 ISlacZ/hah (Jacobs et al. 2003). Transconjugant cells were selected by plating on TSB agar containing 125 μg/mL tetracycline (Fisher Scientific, Ottawa, ON, Canada), 25 μg/mL gentamicin, 40 μg/mL of 5-bromo-4-chloro-3-indolyl-d-galactoside (X-gal, GoldBio, St. Louis, MO), and 2 mg/mL C8-HSL. Candidate colonies (2496) producing β-galactosidase activity, hence having a transposon inserted with the lacZ gene under the control of an expressed promoter, were then transferred to identical TSB agar plates but without C8-HSL. We found that 275 (11%) colonies produced a blue pigment, from pale to intense coloration, and that displayed a modification of the pigment production according to the absence of C8-HSL. These 275 candidates were then further verified by liquid β-galactosidase activity assay with o-nitrophenyl-β-d-galactopyranoside (ONPG, Thermo Fisher Scientific, Nepean, ON, Canada) (Miller 1972). Results obtained at four time points of the growth curve with C8-HSL were compared to those of the control (without C8-HSL). Finally, 43 transposants that differentially expressed β-galactosidase activity according to the presence of C8-HSL, both in solid and liquid media, were kept for further analyses.
Identification of the transposon insertion sites
Insertion sites of the transposon were successfully determined for 40 mutants, mostly by two-stage semi-degenerate polymerase chain reaction (PCR) (Jacobs et al. 2003). As this approach failed for a few candidates, insertion sites were determined using another protocol (Lewenza et al. 2005), with modifications. Briefly, genomic DNA from mutants was extracted by bacterial DNA extraction kit (EZNA, Omega Bio-tek Norcross, GA). About 500 ng of genomic DNA were then digested for 2 h at 37°C by 10 U of AatII (New England Biolabs Ltd., Whitby, ON, Canada) or 7.5 U of PstI (Amersham Pharmacia Biotech Inc., Piscataway, NJ), in order to generate fragments susceptible to include the left or the right side of the transposon, respectively. Digestions were stopped by heat inactivation, and the DNA then circularized using T4 DNA Ligase (Rapid DNA Ligation Kit, Fermentas, Thermo Fisher Scientific), 30 min at room temperature. After 20 min of inactivation at 65°C, ligation products were used as template for an inverse PCR reaction, using the primers ISLacOut1F and ISLacOut1R, or ISLacOut2F and ISLacOut2R, depending if the template had been initially digested by AatII or PstI, respectively (Table S1). Amplification products were all sequenced at The McGill University and Génome Québec Innovation Centre (Montreal, Canada). Sequencing results were compared to the genomes of B. ambifaria AMMD and MC40-6 strains using BLAST (Winsor et al. 2008).
Beta-galactosidase activity
Selected identified transposants were submitted to an additional β-galactosidase assay (Miller 1972). Conditions of the test were standardized, as differential activity in absence (control) or presence of C8-HSL was monitored for all mutants at two time points of the growth curve, around OD600 = 2 and 5. Each measure was done in triplicate, and all results were standardized as percentage of the control. For β-galactosidase assays performed on solid medium, 100 μL of an overnight culture diluted to OD600 = 0.1 were spread onto a 0.2 μm polycarbonate filter laid on TSB agar plates, supplemented or not with C8-HSL. After incubation for 26 h at 37°C, the filters were collected and the protocol for liquid cultures was followed, except that a total protein extraction followed by a Bradford assay was used instead of the OD600 to estimate bacterial growth.
Quantitative reverse-transcription PCR experiments
Cultures were allowed to grow until around DO600 = 4, corresponding to the end of exponential growth phase. Total RNA was extracted with the RiboPure Bacteria kit (Ambion Life Technologies Inc., Burlington, ON, Canada). Extractions were done at least in triplicate, and twice independently. Concentration and purity of samples were assayed on a ND-1000 Nanodrop, and absence of degradation was confirmed on 1% agarose gel. Quantitative reverse-transcription PCR experiments were performed on a Rotor-Gene 6000 thermocycler (Corbett, Qiagen Inc., Toronto, ON, Canada), using the qScript One-Step qRT-PCR kit (Quanta BioSciences, Inc., Gaithersburg, MD), according to the manufacturer's protocol, with slight modifications (Data S1). The reference gene was ndh (Subsin et al. 2007). Primers used for mRNA amplification are presented in Table S1, and the amplification procedure in Table S2. Gene expression differences between HSJ1 and its cepI mutant were calculated using the 2(−ΔΔ(CT)) formula (Livak and Schmittgen 2001); a threshold of 0.5 was arbitrary chosen as significant.
Bioinformatics search for putative cep boxes
Comparative genomics was used to predict cep boxes. Three different cep box consensus from previous reports (Chambers et al. 2006; Wei et al. 2011) were used in order to write three RNAMotif descriptors (Macke et al. 2001). A search in all the Burkholderia genomes (Winsor et al. 2008) with relaxed parameters provided thousands of hits. These were then compared between the corresponding hits of intergenic regions next to orthologous genes, with 18–24 extra nucleotides on each side. A combination of “cep box” score and conservation across the Burkholderia genus was used to assess significance of the predicted cep box (details in Data S1).
Results
A cepI mutant of B. ambifaria strain HSJ1 is defective in C6-HSL and C8-HSL production
Previous studies have shown that Bcc members possess a LuxI-type synthase CepI, which produces C6- and C8-HSL signal molecules, and these AHL have also been identified in B. ambifaria strains (Lutter et al. 2001; Zhou et al. 2003). We have used LC/MS-MS to measure AHL production in the clinical WT strain HSJ1; this strain produces mainly C8-HSL, reaching 3 mg/L under our experimental conditions (Fig. 1A) and also traces of C6-HSL, in a range close to 0.06 mg/L (Fig. 1B). As expected, the mutant of the cepI ortholog in the HSJ1 strain produces neither C8-HSL nor C6-HSL (Fig. 1). For following experiments, we focused on C8-HSL, as it is the most abundant and efficient ligand binding the transcriptional regulator CepR (Aguilar et al. 2003).
Figure 1.
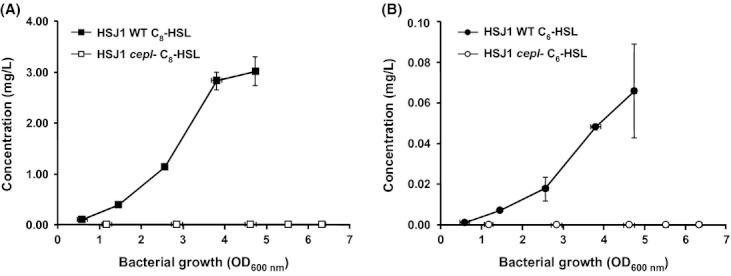
N-acylhomoserine lactone (AHL) production in Burkholderia ambifaria wild-type strain HSJ1 and its cepI mutant. The kinetics of production of (A) C8-HSL and (B) C6-HSL were measured using LC-MS/MS. Results are expressed as means ± standard deviations (SD) for triplicate assays.
Burkholderia ambifaria HSJ1 cepI mutant displays phenotypic modifications
We then looked at phenotypes of B. ambifaria HSJ1 WT strain and its cepI mutant, to compare with other Bcc cepI mutants described in the literature, or to reveal phenotypes associated with B. ambifaria. The inactivation of cepI is associated with the overproduction of siderophores in the Bcc B. cenocepacia K56-2 (Lewenza et al. 1999). This phenotype was thus assessed on CAS agar plates and, after 24 h, the orange-colored halo around HSJ1 cepI mutant colonies was about 27-fold larger (339.05 ± 8.54 mm2) than the one of the WT strain (12.42 ± 4.87 mm2) (Fig. 2A). The WT phenotype was partially restored when C8-HSL was added, as the halo decreased at 191.20 ± 14.21 mm2. Another phenotype generally described in Bcc cepI mutants is the decreased secretion of extracellular proteases (Aguilar et al. 2003; Kooi et al. 2006), which is assessed on milk agar plates. After 24 h, clearing halos around cepI mutant colonies (21.94 ± 6.04 mm2) were roughly 20% of those of HSJ1 WT strain colonies (119.18 ± 8.48 mm2) (Fig. 2B). The difference was reduced by half (49.04 ± 2.45 mm2) when C8-HSL was added to the medium. The cholesterol oxidase activity has been previously reported in HSJ1 WT strain (Vial et al. 2010). We sought if this activity could be modified in the HSJ1 cepI mutant and indeed observed that the characteristic zone of precipitate indicative of cholesterol oxidase activity is present around WT strain colonies, but not around cepI mutant colonies, unless C8-HSL is added to the medium (Fig. 2C). The hemolytic activity was emphasized when B. ambifaria was first described as a Bcc species (Coenye et al. 2001); after 48 h, the beta-hemolytic halo on human blood agar plate around WT strain colonies was 41.33 ± 6.32 mm2, whereas the one of the cepI mutant was about half (18.21 ± 2.44 mm2), except when C8-HSL was added (31.71 ± 3.74 mm2) (Fig. 2D). The hemolytic activity on sheep blood agar plates gives the same kind of results (data not shown).
Figure 2.
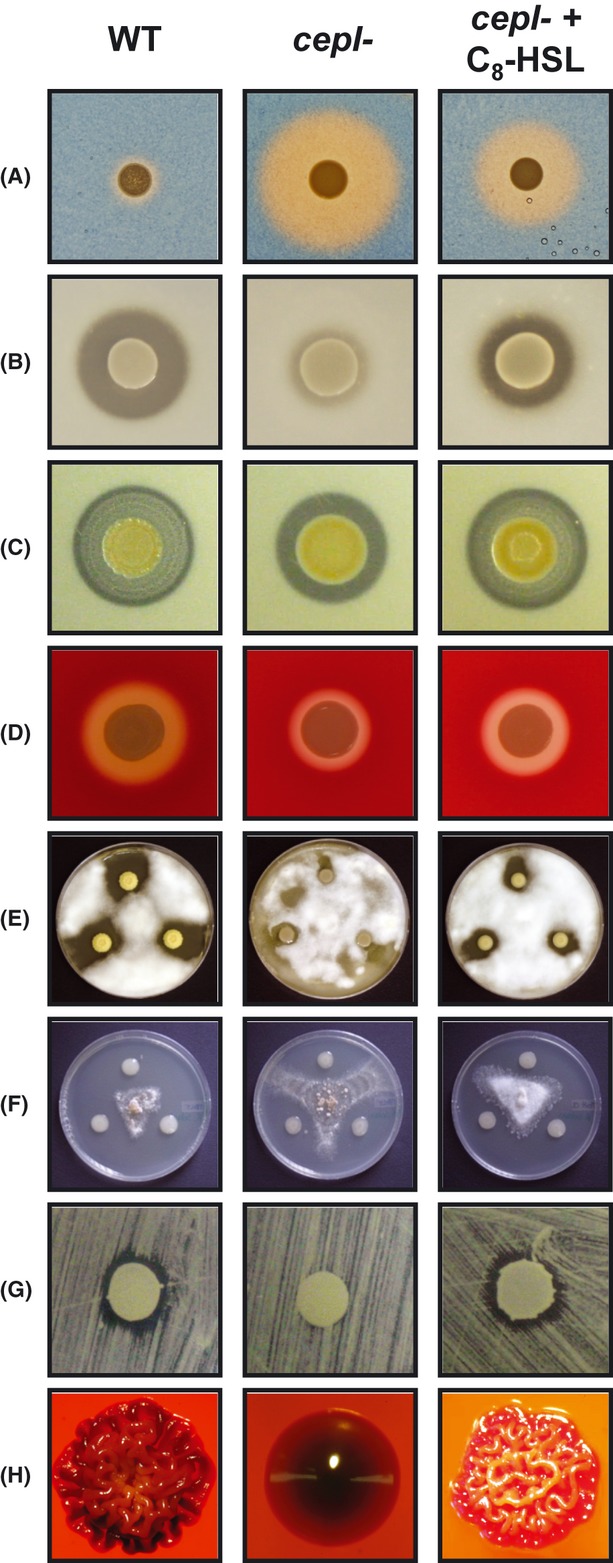
Phenotypic differences between Burkholderia ambifaria wild-type strain HSJ1 and its cepI mutant. (A) Siderophore production after 24 h on Chrome Azurol S (CAS) agar plates, (B) proteolytic activity after 24 h on milk agar plates, (C) cholesterol oxidase activity after 48 h on cholesterol agar plates, (D) hemolytic activity after 24 h on 5% human blood agar plates, (E) antifungal activity against Pythium ultimum after 3 days on potato dextrose agar (PDA) agar plates, (F) antifungal activity against Rhizoctonia solani after 21 days on malt agar plates, (G) antifungal activity against Candida albicans after 24 h on tryptic soy broth (TSB) agar plates, (H) colonial morphology and ability to bind pigment after 21 days on Congo Red agar plates.
Another important phenotype associated with B. ambifaria strains is the inhibitory activity against a broad spectrum of fungi, and some studies reported the influence of QS on such PGPR properties (Zhou et al. 2003; Schmidt et al. 2009). We have thus evaluated the antifungal activity of HSJ1 WT strain and its cepI mutant against P. ultimum (Fig. 2E), R. solani (Fig. 2F), and C. albicans (Fig. 2G), which were previously described to be sensitive to B. ambifaria antifungal properties (Cain et al. 2000; Zhou et al. 2003). In each situation, HSJ1 WT strain displayed an antifungal activity, which was reduced or even absent in the cepI mutant and restored (at least partially) if the medium contained C8-HSL.
We also looked at the colony morphology on Congo Red agar plates, which was previously used in the phenotypic characterization of HSJ1 WT strain (Vial et al. 2010). Although HSJ1 WT strain colonies are wrinkled and able to bind the red pigment, cepI mutant colonies are smooth, but still able to bind the pigment (Fig. 2H).
It is noteworthy that for all these phenotypic tests, the HSJ1 cepR mutant was also investigated; as expected, its phenotypes were similar to those of cepI mutant; therefore, the remaining experiments were only performed with the latter.
Finally, as several of these factors could be associated with virulence, we used the Drosophila melanogaster host model (Castonguay-Vanier et al. 2010) to assess the virulence of HSJ1 WT and its cepI mutant. As we have previously reported with another B. ambifaria strain, this species is among the less virulent Bcc species toward fruit flies (Castonguay-Vanier et al. 2010). The median survival was 141 h for HSJ1 WT strain (Fig. 3), but 165 h for the cepI mutant, which thus is significantly less virulent (P < 0.05).
Figure 3.

Virulence of Burkholderia ambifaria wild-type strain HSJ1 and its cepI mutant toward Drosophila melanogaster. Mortality was scored daily. *P < 0.05.
For phenotypes for which a comparison with other Bcc cepI mutant is possible, the HSJ1 cepI mutant displays phenotypic modifications in compliance with those already reported.
Twenty QS-controlled genes identified by whole-genome transposon-insertion mutagenesis
To identify genes whose expression is influenced by C8-HSL, we employed the cepI mutant as background for a random whole-genome transposon-insertion mutagenesis. Forty mutants successfully sequenced following the screening revealed 20 QS-regulated genes, corresponding to 17 loci. The identification of these genes was realized by BLAST searches against the genomes of B. ambifaria AMMD and MC40-6 strains (http://www.burkholderia.com) (Winsor et al. 2008). The results were generally identical for the two strains, but e-values were often better with the AMMD strain; furthermore, for several sequences we obtained results only with the AMMD strain. Since this was suggesting that HSJ1 is more closely related to AMMD than to MC40-6, we have only considered the AMMD strain for the remaining of this study.
The identified genes are thus presented in Table 1 according to the AMMD locus codes (Winsor et al. 2008). These genes are homogeneously distributed along the three chromosomes, and some of them were selected several times. Notably, Bamb_4726 (prnA), the first gene of the operon responsible for synthesis of pyrrolnitrin, a potent antifungal compound previously reported to be QS-controlled (el-Banna and Winkelmann 1998; Schmidt et al. 2009), was picked up six times, including four times at the same insertion site. For two mutants, the insertion of the transposon is located in the intergenic region upstream of the corresponding gene. Predicted operons are indicated, including the position of genes identified in the screening (Winsor et al. 2008); for example, Bamb_6469 and Bamb_6472 are predicted to belong to the same operon, as well as Bamb_6476 and Bamb_6477, all four belonging to the gene cluster implicated in the synthesis of occidiofungins, an antifungal compound initially described in B. contaminans MS14 (Lu et al. 2009).
Table 1.
Genes identified in the screening for quorum sensing regulation
| Chromosome | Genes | Strand1 | Transposon position (redundancy)2 | Position of gene in operon | Orthologs3 | C8-HSL-induced regulation4 | Function or predicted function of the genes [compound]5 |
|---|---|---|---|---|---|---|---|
| 1 | Bamb_1141 | − | 27 | 1/2 | 10 | M | Heat shock protein Hsp20 |
| 1 | Bamb_21729 | − | 750 (3) | – | 30 | M | Dihydrolipoamide dehydrogenase |
| 1 | Bamb_22979 | + | 1096 | 2/2 | 29 | M | Sulfate transporter |
| 1158 | |||||||
| 1 | Bamb_2378 | − | 91 | – | 29 | R | Spermidine synthase-like protein (SpeE) |
| 1 | Bamb_2404 | + | 131 | – | 4 | M | Hypothetical protein |
| 1 | Bamb_2520 | − | 235 | 5/6 | 30 | R | Sulfate adenylyltransferase, large subunit (cysN) |
| 1 | Bamb_3128 | + | −170 | – | 22 | I | Hypothetical protein |
| 2 | Bamb_3350 | + | 572 (2) | 6/6 | 30 | R | Tryptophan synthase subunit alpha (trpA) |
| 2 | Bamb_3366 | + | −30 | – | 18 | I | Hypothetical protein |
| 2 | Bamb_45789 | + | 385 | 2/3 | 24 | M | Hypothetical protein |
| 2 | Bamb_47269 | + | 103 | 1/4 | 10 | I | Tryptophan halogenase (prnA) [pyrrolnitrin] |
| 273 | |||||||
| 432 (4) | |||||||
| 2 | Bamb_51099 | + | 113 | – | 3 | I | Hypothetical protein |
| 2 | Bamb_55359 | − | −108 | 1/2 | 29 | M | ElaB (protein of unknown function DUF883) |
| 3 | Bamb_5622 | + | 70 | – | 22 | I | PRC-barrel domain-containing protein |
| 3 | Bamb_5911 | + | 229 | 2/2 | 0 | I | LuxR family transcriptional regulator [enacyloxins] |
| 3 | Bamb_5925 | − | 1463 | 2/10 | 2 | I | Beta-ketoacyl synthase [enacyloxins] |
| 3 | Bamb_6465 | − | 343 | – | 10 | I | FAD linked oxidase domain-containing protein |
| 3 | Bamb_64696 | − | 367 (2) | 4/4 | 1 | I | Cyclic peptide transporter [occidiofungins] |
| 402 | |||||||
| 36 | Bamb_64726,8 | − | 4578 | 1/4 | 70 | I | Amino acid adenylation domain-containing protein [occidiofungins] |
| Bamb_64767,8 | − | 10962 | 3/6 | 38 | Amino acid adenylation domain-containing protein [occidiofungins] | ||
| 3 | Bamb_64777 | − | 204 | 2/6 | 1 | I | Short-chain dehydrogenase/reductase SDR [occidiofungins] |
The sign + or − refers to the DNA strand encoding the gene identified by the BLAST searches (http://www.burkholderia.com).
The position of transposon is indicated in base pairs (bp) since the predicted translational start site; the number between brackets indicates how often the transposon was identified at the same insertion site.
Orthologs indexed in the Burkholderia website (http://www.burkholderia.com).
Effect of C8-HSL on gene expression deduced from LacZ reporter assay; R, repression; I, induction; M, moderate effect.
Function or predicted function listed in the Burkholderia website; if genes are included in clusters, the name of the resulting compound is indicated between square brackets.
Bamb_6469 and Bamb_6472 belong to the same predicted operon.
Bamb_6476 and Bamb_6477 belong to the same predicted operon.
The BLAST search did not allow to discriminate Bamb_6472 from Bamb_6476 for this mutant; thus, both genes are indicated.
Genes for which a putative cep box has been predicted.
We have also verified whether genes identified in the screening have predicted orthologs among the 27 complete and five incomplete genomes present on the Burkholderia website (Winsor et al. 2008). Some genes have about 30 putative orthologs, and appear thus widespread among Burkholderia species, while other seem to be found only in Bcc members. Interestingly, Bamb_5911, which codes for a LuxR-type family transcriptional regulator located upstream of the cluster implicated in the biosynthesis of enacyloxins, an antimicrobial compound recently characterized in B. ambifaria AMMD (Mahenthiralingam et al. 2011), has only one ortholog, in B. ubonensis, which is not included in the Burkholderia website (Winsor et al. 2008).
In order to gain insights in the type of regulation (direct or indirect) exerted by QS on genes identified in our screening, we performed a bioinformatics search for putative cep boxes in promoter regions. cep boxes are sequences recognized by the transcriptional regulator CepR to bind target promoters and modulate the transcription of genes; the presence of these cep boxes upstream of genes pleads in favor of a direct regulation by QS. Concerning the genes identified in the screening, our search predicts the presence of a putative cep box upstream from the transcriptional units of Bamb_2172, Bamb_2297, Bamb_4578, Bamb_4726, Bamb_5109, and Bamb_5535 (Table 1 and Fig. S1). The predictive cep-box sequence, derived from Chambers et al. (2006) and Wei et al. (2011), as well as a cep box derived from these data and the genes identified in our screening, is presented in Figure 4. Putative cep boxes upstream of genes not identified in the screening are presented in Figure S1.
Figure 4.
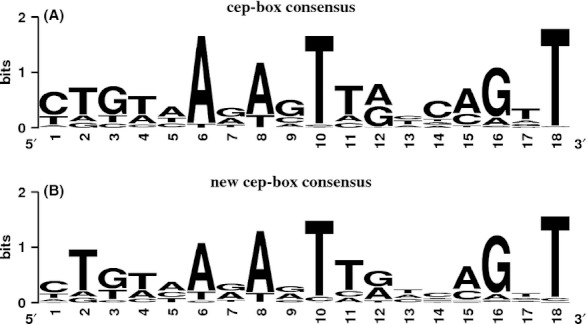
Consensus cep box. (A) The cep box consensus established by Chambers et al. (2006) and by Wei et al. (2011) were combined to obtain this sequence logo with weblogo (Crooks et al. 2004). (B) The putative cep boxes found in this screen (see Supporting Information) were combined with the consensus in (A) to obtain this sequence logo.
C8-HSL addition induces diverse modifications in reporter gene activities
To estimate the impact of C8-HSL addition on the regulation of the identified genes, their expression was measured at two different points of the growth of each mutant, around OD600 = 2 and OD600 = 5. Figure 5 shows the effect of adding 2 mg/L C8-HSL on the activity of the inserted reporters. We considered that mutants were strongly affected by C8-HSL addition if the reporter activity displayed at least a twofold change compared to their respective control (Fig. 5A and B); if the activity displayed only a 1.5-fold change, the mutants were considered moderately affected (Fig. 5C and D). Accordingly, panel A shows mutants which presented a marked decrease in reporter activity after C8-HSL addition (Fig. 5A). For instance, the mutant in which the transposon has interrupted the gene Bamb_2520 (renamed “trBamb_2520” to avoid confusion with the gene itself) displayed an activity nearly four times lower that the one of the control following C8-HSL addition. On the other hand, panel B shows mutants which displayed a strong increase of the reporter expression after C8-HSL addition, displaying activities ranging from two to almost 30 times higher than those of the control (Fig. 5B). The highest score of LacZ reporter activity was obtained with the mutant trBamb_4726, in which the operon coding for pyrrolnitrin is interrupted. The remaining mutants displayed a moderate modification of reporter expression after C8-HSL addition (Fig. 5C and D). Because the mutants were initially screened on agar plates, we reasoned the growth conditions might affect the expression of these genes. We thus submitted them to a β-galactosidase challenge on solid medium. After 26 h, the results obtained for the majority of the mutants were roughly the same as described in panel C (data not shown), except for two mutants presented in panel D, for which the LacZ reporter activity was significantly increased when mutants were grown on solid medium (Fig. 5D). The effects of C8-HSL addition on LacZ reporter activity are summarized in Table 1.
Figure 5.
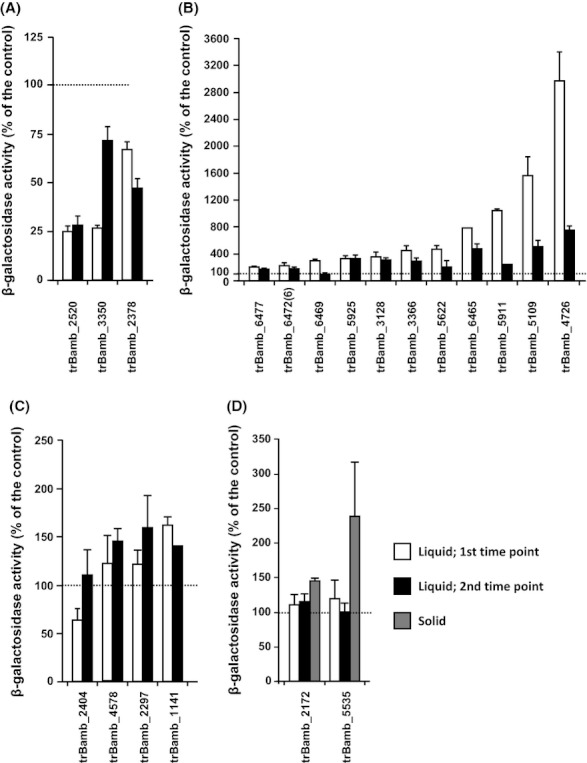
Expression of quorum sensing (QS)-regulated genes in response to C8-HSL. Transposon mutants in putative QS-regulated genes isolated in the cepI mutant background during the screening were tested for their β-galactosidase activity with or without adding of 2 mg/L C8-HSL. Cultures were sampled at two time points of the growth curve, and the activity calculated in Miller units; for (D) cultures were realized in solid medium. Results for transposants supplemented with C8-HSL are expressed in percent of the controls (transposants without C8-HSL), the latter being normalized at 100% and symbolized by the dotted line. Results are expressed as means ± SD for triplicate assays. (A) Highly reduced activity (twofold change of the control or less), (B) highly induced activity (twofold change of the control or more), (C) moderate activity after C8-HSL addition in liquid culture (±1.5-fold change of the control), (D) β-galactosidase activity after C8-HSL addition in solid medium.
It is interesting to note that some mutants displayed a similar reporter gene activity at the two time points (such as trBamb_2520), whereas others were markedly different (such as trBamb_4726) (Fig. 5A and B), reflecting different patterns of regulation.
qRT-PCR experiments confirm LacZ reporter data
To validate β-galactosidase activity results, we then performed real-time quantitative reverse-transcription PCR (qRT-PCR) experiments on a group of genes identified in the screening. We chose genes corresponding to mutants distributed in panels A and B, displaying thus a strong modification of activities. We have also included zmpA and zmpB (Bamb_3836 and Bamb_4475, respectively), previously described to be positively QS-controlled in B. cenocepacia (Kooi et al. 2006; Subsin et al. 2007), as well as Bamb_1196, which was identified in our screening but not confirmed in the β-galactosidase assays. The results presented in Figure S2 show the relative expression obtained in the HSJ1 cepI mutant compared to the HSJ1 WT strain. The level of twofold change was arbitrarily chosen as threshold of significant difference. For Bamb_6469, there is an apparent discrepancy between qRT-PCR experiments and LacZ reporter results. Actually, mRNA were extracted around OD600 = 4, which is closer to the second time point of β-galactosidase assays, for which trBamb_6469 displayed an activity similar to the one of the control (Fig. 5B). For the remaining tested genes, they all confirm data obtained with the LacZ fusions. Under the tested conditions, zmpB was so poorly expressed in the cepI mutant that it did not reach the 0.5 threshold used to calculate the Ct.
Phenotypic confirmation of transposon mutants
As the cepI mutant was used as background strain for the whole-genome transposon-insertion mutagenesis, all the resulting mutants are in fact double mutants, impaired in C8-HSL production and in the function coded by the gene interrupted by the transposon. To explore some phenotypes associated with genes presented in Table 1, the cepI deletion of the transposon mutants was compensated by C8-HSL addition in the media used for the phenotypic challenges. Among the genes identified in the screening, several are implicated in the production of pyrrolnitrin, enacyloxins, and occidiofungins (Table 1), which have antifungal/antibacterial activities. We first tested the corresponding mutants against C. albicans, P. ultimum, and R. solani. Except for Bamb_5925, for which a slight difference against P. ultimum was noted, all the transposon mutants behaved as the cepI mutant (Fig. S3). We therefore sought phenotypic tests providing a better discrimination of these mutants. Hemolytic properties were recently described for genes implicated in occidiofungins biosynthesis (Thomson and Dennis 2012). On sheep blood agar, trBamb_6469 and trBamb_6477 behave as the cepI mutant (data not shown), whereas trBamb_6472/6476 displays no hemolytic activity even with C8-HSL supplementation (Fig. 6A). A specific activity of enacyloxins was reported against B. multivorans (Mahenthiralingam et al. 2011); we thus tested the activity of trBamb_5911 and trBamb_5925 against this bacterium. It is interesting to note that the addition of C8-HSL in the medium allowed the cepI mutant to produce a greater inhibiting zone than the one of the WT; for transposon mutants, trBamb_5911 displayed a slight inhibiting zone when C8-HSL was added, whereas the inhibiting activity of trBamb_5925 was completely abolished (Fig. 6B).
Figure 6.
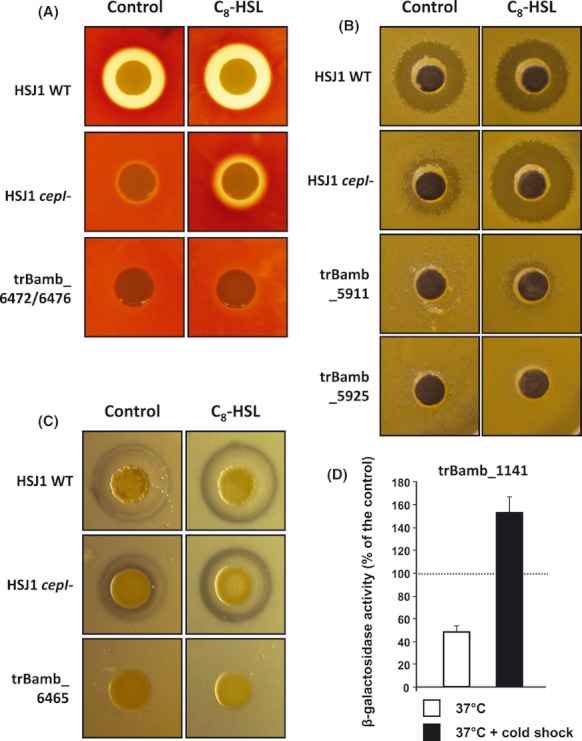
Phenotypic confirmation of transposon mutants. As the transposon mutants are all cepI/transposon mutants, C8-HSL was added in media to compensate the cepI impairment and observe only the effect of transposon mutation on predicted phenotypes. The following phenotypes are presented: (A) hemolytic activity on sheep blood agar, (B) anti-Burkholderia multivorans activity, (C) cholesterol oxidase activity on cholesterol agar plates, (D) effect of temperature on β-galactosidase activity for bacteria grown in tryptic soy broth (TSB) agar plates.
Concerning the cholesterol oxidase activity, the cepI mutant is able to produce a clearing halo around the colony, but the characteristic precipitate linked to cholesterol oxidase activity is obtained only if C8-HSL is present (Figs. 2C and 6C). In contrast, the trBamb_6465 mutant’ colony produced neither clearing halo nor precipitate (Table 1 and Fig. 6C). Since Bamb_1141 encodes a heat shock protein HSP20 (Table 1), we ask whether it could exhibit a different LacZ reporter activity according to temperature. Indeed, if the growth of trBamb_1141 occurs in solid medium supplemented by C8-HSL and incubated only at 37°C, the β-galactosidase activity is half the one of the control, whereas it reaches around 150% of the control if the assay is preceded by a 6 h incubation at 4°C (Fig. 6D).
These experiments confirmed the phenotypes of mutants tested and brought some additional clues on the role of C8-HSL in the regulation.
Discussion
Burkholderia ambifaria displays a remarkable potential as a PGPR and biocontrol agent, but like the other Bcc members, its commercial use is placed under a moratorium (Chiarini et al. 2006). The production of commercially interesting molecules in vitro through biotechnological processes requires the understanding of mechanisms that direct and regulate their biosynthesis, such as QS.
Phenotypes of the HSJ1 cepI mutant
Several phenotypes classically associated with QS, such as siderophores production and proteolytic activity, are in fact disparately present and QS-controlled among Bcc members (Gotschlich et al. 2001; Huber et al. 2001; Aguilar et al. 2003). We report here that the cepI mutant of B. ambifaria strain HSJ1 overproduces siderophores, similarly to the cepI mutant of B. cenocepacia K56-2 (Lewenza et al. 1999). The HSJ1 cepI mutant has a decreased protease activity, as most of Bcc strains in absence of C8-HSL (Wopperer et al. 2006). This phenotype has been linked in B. cenocepacia to two QS-regulated metalloproteases, ZmpA and ZmpB (Gingues et al. 2005; Kooi et al. 2006). These proteases were also identified in B. ambifaria strain HSJ1 (Vial et al. 2010); we found here that zmpA and zmpB are strongly downregulated in the HSJ1 cepI mutant (Fig. S2 and data not shown), which probably explains the decreased proteolytic activity.
We have also looked at some phenotypes previously described in B. ambifaria HSJ1 WT strain (Vial et al. 2010), such as the colonial morphology on Congo Red agar plates. The colonial wrinkling is a QS-regulated character in HSJ1 strains; a such colony wrinkling has been linked to the QS-regulation of the exopolysaccharide Pel in Pseudomonas aeruginosa (Gupta and Schuster 2012). The secretion of a FAD-dependent cholesterol oxidase, identified as Bamb_6465 and correlated with a cholesterol-degrading activity, was also previously reported in B. ambifaria HSJ1 (Vial et al. 2010). We found here that this phenotype is positively controlled by QS (Fig. 2C), and confirmed that it is due to Bamb_6465, which is strongly QS-activated in B. ambifaria HSJ1 (Figs. 5B, 6C, and S2).
Another noteworthy phenotype is the beta-hemolytic activity, which was highlighted when B. ambifaria was initially described as a new Bcc species (Coenye et al. 2001). Factors implicated in such effects in Bcc members are poorly identified; however, a hemolytic compound named cepalycins, displaying also antifungal properties, has been previously isolated from the supernatant of B. cepacia JN106 (Abe and Nakazawa 1994). Similar hemolytic properties were recently reported for occidiofungins in B. vietnamiensis DBO1, compounds initially described for their antifungal activities in B. contaminans MS14 (Lu et al. 2009; Thomson and Dennis 2012). These dual activities likely result from the interaction of these extracellular molecules with cholesterol in the membrane; indeed, cepalycins were more inhibited by ergosterol than by cholesterol (Abe and Nakazawa 1994). Moreover, environmental strains of Bcc apparently display more hemolytic activity than clinical strains (Bevivino et al. 2002), which is coherent if hemolytic molecules are in fact antifungal molecules; this hypothesis is also compliant with the natural ecology of B. ambifaria. In our screening, we have identified three genes implicated in occidiofungins biosynthesis (Table 1). The mutant trBamb_6472/trBamb_6476 is impaired in hemolytic function (Fig. 6A), while trBamb_6469 behave as the cepI mutant (data not shown), which is in agreement with recent published data (Thomson and Dennis 2012). On the other hand, trBamb_6477 behaved also like the cepI mutant (data not shown); this discrepancy with the study of Thomson and Dennis (2012) could well be explained by the use of different Bcc species or by the compensation of the disrupted gene by another one with similar function.
Additionally, the antifungal activities of B. ambifaria strains against several fungi have been already described (Cain et al. 2000; Zhou et al. 2003), and implication of the QS-regulation for such antifungal activities has also been reported (Zhou et al. 2003; Schmidt et al. 2009). Accordingly, our phenotypic assay against P. ultimum, R. solani, and C. albicans demonstrated that B. ambifaria HSJ1, although being from clinical origin, exhibits antifungal activity, while its cepI mutant displays reduced (or even abolished) antifungal activities.
High-throughput screening to identify new QS-regulated genes
Global approaches using high-throughput screenings have been developed to identify more rapidly and efficiently a wide range of QS-regulated genes, often with the aim to identify those coding for potential virulence factors. We have identified 20 QS-controlled genes employing a procedure derived from Chambers et al. (2006) that had permitted the identification of seven QS-controlled genes in B. cenocepacia K56-2. This approach does not allow to discriminate genes directly or indirectly controlled by QS (Wei et al. 2011). To partially circumvent this shortcoming, we have performed a bioinformatics search for the presence of putative cep boxes upstream of transcriptional units identified in the screening. cep boxes are short sequences upstream the promoter of target genes that allow CepR to recognize its chromosomal binding site. Although the cep box upstream of cepI is well conserved among Bcc members (Fig. S1), the conservation is less obvious upstream of other QS-controlled genes. We have used the consensus sequence described in B. cenocepacia by Chambers et al. (2006) to predict cep boxes in B. ambifaria HSJ1. We have notably identified a putative cep box upstream of prnA (Bamb_4726), which has been identified six times in our screening and for which the direct regulation by CepR has been experimentally demonstrated in Burkholderia lata 383 (Schmidt et al. 2009). A recent study has reported a different consensus sequence in B. cenocepacia K56-2, as well as the experimental demonstration of the direct regulation for two genes, BCAL0510 and BCAM1869 (Wei et al. 2011). BCAM1869 is an ortholog of Bamb_4117, which is located between cepR and cepI, and for which we have predicted a putative cep box (Fig. S1). On the other hand, BCAL0510 is an ortholog of Bamb_3128, which has been identified in the screening but not in the cep box prediction. We have also used this consensus sequence in our bioinformatics study (Fig. S1). Our predictive method, based on the Chambers’ study, allowed thus to cross data with two others predictive and experimental reports, reinforcing confidence in our results.
Genes identified in the screening
Our screening allowed us to identify 20 genes corresponding to 17 loci (Table 1). According to the LacZ reporter activity challenge, three genes were strongly downregulated after C8-HSL addition (Fig. 5A). These genes are implicated in metabolic functions, such as Bamb_2520, which is the ortholog of B. cenocepacia cysN, part of an operon implicated in sulfur metabolism (Iwanicka-Nowicka et al. 2007). This operon is regulated by two LysR-type regulators, CysB and SsuR, but additional QS-regulation has not been reported (Iwanicka-Nowicka et al. 2007). Bamb_3350 (trpA) is the last of a six-genes operon implicated in the biosynthesis of tryptophan, which can be then catabolized via the tricarboxylic acids (TCA) cycle, or used as the precursor of many metabolites, such as pyrrolnitrin, 4-hydroxy-2-alkylquinolines (HAQ) produced by P. aeruginosa, or their methylated counterparts (HMAQ) discovered in Burkholderia (Déziel et al. 2004; Vial et al. 2008; Schmidt et al. 2009). Some of these metabolites are implicated in, or regulated by QS, but the QS-regulation of tryptophan biosynthesis is not established. However, in a quorum-quenching study realized in Azospirillum lipoferum, TrpA was identified among the QS-repressed proteins (Boyer et al. 2008), which agrees with our observations. The third mutant included in panel A is trBamb_2378, which is interrupted in the gene coding a spermidine synthase (speE)-like protein. Spermidine is a polyamine, implicated in several biological processes, both in eukaryotic and prokaryotic cells (Igarashi and Kashiwagi 2010). As for the other QS-repressed genes, the link with QS is not clear; in B. pseudomallei, inhibition of intracellular spermidine synthesis lead to reduced export of AHL via efflux pumps, which suggested that spermidine had an effect on AHL, but the reciprocal was not suggested (Chan and Chua 2010).
It is interesting to note that genes identified in the screening that are QS-repressed or moderately affected are mainly found in the chromosome 1, genes located on chromosome 2 are diversely QS-regulated and genes located on the chromosome 3 are exclusively QS-induced (Table 1). In B. cenocepacia, chromosome 1 carries most of essential (“housekeeping”) genes, while the remaining two chromosomes contain much accessory genes implicated in adaptation to niches; the third chromosome has even been described as a virulence plasmid (Holden et al. 2009; Agnoli et al. 2012; Juhas et al. 2012). As discussed above, the link between QS and metabolism is difficult to decipher, as the genes are also regulated by other factors such as the availability of nutriments, whereas QS-regulation of secreted virulence factors is more obvious. The genes moderately affected by C8-HSL addition, for which a function is predicted, are implicated in metabolic functions and stress adaptation (Fig. 5C and D; Table 1). Four of these six genes are predicted to be preceded by a putative cep box (Table 1; Fig. S1). These genes were at least partially affected by experimental procedures, as two mutants displayed increased β-galactosidase activity if grown in solid rather in liquid medium (Fig. 5D). Another interesting example is trBamb_1141, interrupted in a gene coding a heat shock protein HSP20, which displayed opposite β-galactosidase activities in response to C8-HSL addition, according to the temperature of growth (Fig. 6D). In B. cenocepacia K56-2, the ortholog of this gene is positively regulated by CepR2, an orphan LuxR transcriptional regulator (Malott et al. 2009). We can thus suggest that the genes that appear moderately affected by C8-HSL addition are, additionally to the regulation exerted by C8-HSL, controlled by supplementary factors, such as environmental stresses or other regulation circuitry.
Finally, panel B of Figure 5 contains 11 genes strongly induced by C8-HSL, such as Bamb_6465, responsible for the cholesterol oxidase activity as mentioned above, or Bamb_5109, located upstream of a large nine-genes operon implicated in the biosynthesis and transport of polysaccharide. The most reactive mutant is trBamb_4726, which is interrupted in prnA, the first gene of the operon directing pyrrolnitrin biosynthesis from tryptophan. Pyrrolnitrin is active against a broad spectrum of bacteria and fungi (el-Banna and Winkelmann 1998), and the regulation of its biosynthesis by QS has been demonstrated (Schmidt et al. 2009). The genes Bamb_5911 and Bamb_5925 are implicated in the biosynthesis of enacyloxins, which are antimicrobial compounds produced by B. ambifaria and especially active against B. multivorans (Mahenthiralingam et al. 2011). Bamb_5925 is important in the biosynthesis, whereas Bamb_5911 is involved in the regulation of the cluster, as it is included in a two-gene operon coding LuxR-type transcriptional regulators (Mahenthiralingam et al. 2011). Indeed, the mutant trBamb_5925 is totally impaired in B. multivorans inhibition, while trBamb_5911 is slightly restored when C8-HSL is added, revealing that the second LuxR-type transcriptional regulator could partially activate the biosynthesis of enacyloxins (Fig. 6B). Although several elements indicate that the biosynthesis of enacyloxins is QS-regulated (Mahenthiralingam et al. 2011), our data support this assertion. The remaining genes of panel B are almost all orthologs of those implicated in occidiofungins biosynthesis. As discussed above, these antifungal compounds initially identified in B. contaminans MS14 have recently been described as hemolytic molecules in B. vietnamiensis DBO1 (Lu et al. 2009; Gu et al. 2011; Thomson and Dennis 2012). Another team has identified antifungal molecules named burkholdines in B. ambifaria 2.2N, which have structures similar to occidiofungins, demonstrating that the occidiofungin cluster of B. ambifaria is expressed (Tawfik et al. 2010). In B. contaminans, while the cluster contains two LuxR-type regulators that have C-terminal domains able to bind DNA, neither the autoinducer-binding domain nor the response regulatory domain in N-terminal have been identified, suggesting that the signal molecule is from another nature (Gu et al. 2009). Yet, the results of our screening lead us to conclude that the production of occidiofungins is QS-controlled, at least in B. ambifaria.
In conclusion, we have confirmed in the clinical B. ambifaria HSJ1 strain some genes and phenotypes already known to be QS-regulated in Bcc species, and we have furthermore identified new QS-regulated genes. Predominantly, the production of antifungal/antimicrobial compounds is a very important trait controlled by QS in the HSJ1 WT strain, as our study revealed genes implicated in the biosynthesis of pyrrolnitrin, enacyloxins, and occidiofungins. This arsenal could appear redundant, but each molecule is effective against a different spectrum of microorganisms. Interestingly, a recent study reported that B. cepacia strains produced HMAQs displaying antifungal properties (Kilani-Feki et al. 2011). We have previously reported that at least three species of Burkholderia, including the B. ambifaria strain used in this study, are able to produce HMAQs, and that mutant deficient in the biosynthesis of HMAQs produces increased concentrations of C8-HSL (Vial et al. 2008). Burkholderia ambifaria HSJ1 expresses thus at least four molecules with antifungal/antimicrobial properties, three of them being QS-regulated; studies are currently underway to determine if biosynthesis of the fourth family of molecules, namely HMAQ, is also regulated by QS.
Acknowledgments
We thank Richard Belanger and Caroline Labbé (Department of Phytology, Laval University, Québec) for providing the Pythium ultimum strain. We also thank Fabrice Jean-Pierre for his help in methodological development. This study was supported by a Canadian Institutes of Health Research (CIHR) Operating grant to E. D. Both A. C. and L. V. were recipients of postdoctoral fellowships from the Fondation Armand-Frappier, V. D. of a Ph.D. scholarship from the Fondation Armand-Frappier, and N. L. of an NSERC summer scholarship. E. D. was a Chercheur-boursier Junior 2 of the Fonds de la recherche en santé du Québec (FRSQ) and holds a Canada Research Chair in Sociomicrobiology.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. Supplemental experimental procedures.
Table S1. Primers used in this study.
Table S2. Conditions used in qRT-PCR experiments
Figure S1. Predicted cep-box sequences in Burkholderia species. The detailed method used to determine the putative cep-boxes is described in the explanatory text. All the potential cep-boxes found in the AMMD genome (not only those identified in the screening) are also presented.
Figure S2. Relative expression of candidate quorum sensing-regulated genes by mRNA quantification. The relative expression of the genes was estimated by quantitative reverse transcription PCR (qRT-PCR) experiments on HSJ1 WT and its cepI mutant. The ndh gene was used as reference. The results are expressed as relative quantification of gene expression (log10 scale) in the cepI mutant compared to the WT, normalized to 1. A fold change of two (bottom scale) was chosen as significant threshold. The results are expressed in means ± SD for triplicate assays.
Figure S3. Phenotypic confirmation of transposon mutants for antifungal activities. The antifungal activities of HSJ1 WT, its cepI mutant and three transposon mutants implicated in biosynthesis of pyrrolnitrin, enacyloxins and occidiofungins, were tested against Candida albicans, Pythium ultimum, and Rhizoctonia solani.
References
- Abe M, Nakazawa T. Characterization of hemolytic and antifungal substance, cepalycin, from Pseudomonas cepacia. Microbiol. Immunol. 1994;38:1–9. doi: 10.1111/j.1348-0421.1994.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, et al. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol. Microbiol. 2012;83:362–378. doi: 10.1111/j.1365-2958.2011.07937.x. [DOI] [PubMed] [Google Scholar]
- Aguilar C, Bertani I, Venturi V. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 2003;69:1739–1747. doi: 10.1128/AEM.69.3.1739-1747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev MF. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques. 1999;26:828. doi: 10.2144/99265bm05. [DOI] [PubMed] [Google Scholar]
- el-Banna N, Winkelmann G. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 1998;85:69–78. doi: 10.1046/j.1365-2672.1998.00473.x. [DOI] [PubMed] [Google Scholar]
- Bevivino A, Dalmastri C, Tabacchioni S, Chiarini L, Belli ML, Piana S, et al. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 2002;40:846–851. doi: 10.1128/JCM.40.3.846-851.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Bally R, Perrotto S, Chaintreuil C, Wisniewski-Dye F. A quorum-quenching approach to identify quorum-sensing-regulated functions in Azospirillum lipoferum. Res. Microbiol. 2008;159:699–708. doi: 10.1016/j.resmic.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Burkholder WH. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;40:115–117. [Google Scholar]
- Cain CC, Henry AT, Waldo RH, III, Casida LJ, Jr, Falkinham JO., III Identification and characteristics of a novel Burkholderia strain with broad-spectrum antimicrobial activity. Appl. Environ. Microbiol. 2000;66:4139–4141. doi: 10.1128/aem.66.9.4139-4141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay-Vanier J, Vial L, Tremblay J, Deziel E. Drosophila melanogaster as a model host for the Burkholderia cepacia complex. PLoS ONE. 2010;5:e11467. doi: 10.1371/journal.pone.0011467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers CE, Lutter EI, Visser MB, Law PP, Sokol PA. Identification of potential CepR regulated genes using a cep box motif-based search of the Burkholderia cenocepacia genome. BMC Microbiol. 2006;6:104. doi: 10.1186/1471-2180-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YY, Chua KL. Growth-related changes in intracellular spermidine and its effect on efflux pump expression and quorum sensing in Burkholderia pseudomallei. Microbiology. 2010;156:1144–1154. doi: 10.1099/mic.0.032888-0. [DOI] [PubMed] [Google Scholar]
- Chan KG, Atkinson S, Mathee K, Sam CK, Chhabra SR, Camara M, et al. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:51. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 2006;14:277–286. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003;5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- Coenye T, Mahenthiralingam E, Henry D, LiPuma JJ, Laevens S, Gillis M, et al. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 2001;51:1481–1490. doi: 10.1099/00207713-51-4-1481. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doukyu N, Aono R. Cloning, sequence analysis and expression of a gene encoding an organic solvent- and detergent-tolerant cholesterol oxidase of Burkholderia cepacia strain ST-200. Appl. Microbiol. Biotechnol. 2001;57:146–152. doi: 10.1007/s002530100753. [DOI] [PubMed] [Google Scholar]
- Eberl L. Quorum sensing in the genus Burkholderia. Int. J. Med. Microbiol. 2006;296:103–110. doi: 10.1016/j.ijmm.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingues S, Kooi C, Visser MB, Subsin B, Sokol PA. Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex. J. Bacteriol. 2005;187:8247–8255. doi: 10.1128/JB.187.24.8247-8255.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotschlich A, Huber B, Geisenberger O, Togl A, Steidle A, Riedel K, et al. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 2001;24:1–14. doi: 10.1078/0723-2020-00013. [DOI] [PubMed] [Google Scholar]
- Govan JRW, Balandreau J, Vandamme P. Burkholderia cepacia – friend and foe. ASM News. 2000;66:124–125. [Google Scholar]
- Gu G, Wang N, Chaney N, Smith L, Lu SE. AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol. Lett. 2009;297:54–60. doi: 10.1111/j.1574-6968.2009.01653.x. [DOI] [PubMed] [Google Scholar]
- Gu G, Smith L, Liu A, Lu SE. Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl. Environ. Microbiol. 2011;77:6189–6198. doi: 10.1128/AEM.00377-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Schuster M. Quorum sensing modulates colony morphology through alkyl quinolones in Pseudomonas aeruginosa. BMC Microbiol. 2012;12:30. doi: 10.1186/1471-2180-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 2009;191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B, Riedel K, Hentzer M, Heydorn A, Gotschlich A, Givskov M, et al. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K. Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol. Biochem. 2010;48:506–512. doi: 10.1016/j.plaphy.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Iwanicka-Nowicka R, Zielak A, Cook AM, Thomas MS, Hryniewicz MM. Regulation of sulfur assimilation pathways in Burkholderia cenocepacia: identification of transcription factors CysB and SsuR and their role in control of target genes. J. Bacteriol. 2007;189:1675–1688. doi: 10.1128/JB.00592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Stark M, Lumjiaktase C, von Mering P, Crook DW, Valvano MA, et al. High confidence prediction of essential genes in Burkholderia cenocepacia. PLoS ONE. 2012;7:e40064. doi: 10.1371/journal.pone.0040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilani-Feki O, Culioli G, Ortalo-Magne A, Zouari N, Blache Y, Jaoua S. Environmental Burkholderia cepacia strain Cs5 acting by two analogous alkyl-quinolones and a didecyl-phthalate against a broad spectrum of phytopathogens fungi. Curr. Microbiol. 2011;62:1490–1495. doi: 10.1007/s00284-011-9892-6. [DOI] [PubMed] [Google Scholar]
- Kooi C, Subsin B, Chen R, Pohorelic B, Sokol PA. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect. Immun. 2006;74:4083–4093. doi: 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lépine F, Déziel É. Liquid chromatography/mass spectrometry for the detection and quantification of N-acyl-L-homoserine lactones and 4-hydroxy-2-alkylquinolines. Methods Mol. Biol. 2011;692:61–69. doi: 10.1007/978-1-60761-971-0_5. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Conway B, Greenberg EP, Sokol PA. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, et al. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 2005;15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu SE, Novak J, Austin FW, Gu G, Ellis D, Kirk M, et al. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry. 2009;48:8312–8321. doi: 10.1021/bi900814c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter E, Lewenza S, Dennis JJ, Visser MB, Sokol PA. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 2001;69:4661–4666. doi: 10.1128/IAI.69.7.4661-4666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008;104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Song L, Sass A, White J, Wilmot C, Marchbank A, et al. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria Genomic Island. Chem. Biol. 2011;18:665–677. doi: 10.1016/j.chembiol.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Malott RJ, O'Grady EP, Toller J, Inhulsen S, Eberl L, Sokol PA. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J. Bacteriol. 2009;191:2447–2460. doi: 10.1128/JB.01746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellbye B, Schuster M. More than just a quorum: integration of stress and other environmental cues in acyl-homoserine lactone signaling. In: Storz G, Hengge R, editors. Bacterial stress responses. Washington, DC: ASM Press; 2011. pp. 349–363. [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Payne GW, Vandamme P, Morgan SH, Lipuma JJ, Coenye T, Weightman AJ, et al. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 2005;71:3917–3927. doi: 10.1128/AEM.71.7.3917-3927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, et al. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009;11:1422–1437. doi: 10.1111/j.1462-2920.2009.01870.x. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983;78:4–791. [Google Scholar]
- Sokol PA, Malott RJ, Riedel K, Eberl L. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiol. 2007;2:555–563. doi: 10.2217/17460913.2.5.555. [DOI] [PubMed] [Google Scholar]
- Suarez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonca-Previato L, James EK, Venturi V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012;63:249–266. doi: 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- Subsin B, Chambers CE, Visser MB, Sokol PA. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J. Bacteriol. 2007;189:968–979. doi: 10.1128/JB.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik KA, Jeffs P, Bray B, Dubay G, Falkinham JO, Mesbah M, et al. Burkholdines 1097 and 1229, potent antifungal peptides from Burkholderia ambifaria 2.2N. Org. Lett. 2010;12:664–666. doi: 10.1021/ol9029269. [DOI] [PubMed] [Google Scholar]
- Thomson EL, Dennis JJ. A Burkholderia cepacia complex non-ribosomal peptide-synthesized toxin is hemolytic and required for full virulence. Virulence. 2012;3:286–298. doi: 10.4161/viru.19355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Dawyndt P. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 2011;34:87–95. doi: 10.1016/j.syapm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Vial L, Lepine F, Milot S, Groleau MC, Dekimpe V, Woods DE, et al. Burkholderia pseudomallei B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J. Bacteriol. 2008;190:5339–5352. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial L, Groleau MC, Lamarche MG, Filion G, Castonguay-Vanier J, Dekimpe V, et al. Phase variation has a role in Burkholderia ambifaria niche adaptation. ISME J. 2010;4:49–60. doi: 10.1038/ismej.2009.95. [DOI] [PubMed] [Google Scholar]
- Vial L, Chapalain A, Groleau MC, Deziel E. The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ. Microbiol. 2011;13:1–12. doi: 10.1111/j.1462-2920.2010.02343.x. [DOI] [PubMed] [Google Scholar]
- Wei Y, Ryan GT, Flores-Mireles AL, Costa ED, Schneider DJ, Winans SC. Saturation mutagenesis of a CepR binding site as a means to identify new quorum-regulated promoters in Burkholderia cenocepacia. Mol. Microbiol. 2011;79:616–632. doi: 10.1111/j.1365-2958.2010.07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Khaira B, Lo T, Van Rossum R, Whiteside MD, Brinkman FS. The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopperer J, Cardona ST, Huber B, Jacobi CA, Valvano MA, Eberl L. A quorum-quenching approach to investigate the conservation of quorum-sensing-regulated functions within the Burkholderia cepacia complex. Appl. Environ. Microbiol. 2006;72:1579–1587. doi: 10.1128/AEM.72.2.1579-1587.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yao F, Roberts DP, Lessie TG. AHL-deficient mutants of Burkholderia ambifaria BC-F have decreased antifungal activity. Curr. Microbiol. 2003;47:174–179. doi: 10.1007/s00284-002-3926-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


