Abstract
Insect fungiculture is practiced by ants, termites, beetles, and gall midges and it has been suggested to be widespread among plant–ants. Some of the insects engaged in fungiculture, including attine ants and bark beetles, are known to use symbiotic antibiotic-producing actinobacteria to protect themselves and their fungal cultivars against infection. In this study, we analyze the bacterial communities on the cuticles of the plant–ant genera Allomerus and Tetraponera using deep sequencing of 16S rRNA. Allomerus ants cultivate fungus as a building material to strengthen traps for prey, while Tetraponera ants cultivate fungus as a food source. We report that Allomerus and Tetraponera microbiomes contain >75% Proteobacteria and remarkably the bacterial phyla that dominate their cuticular microbiomes are very similar despite their geographic separation (South America and Africa, respectively). Notably, antibiotic-producing actinomycete bacteria represent a tiny fraction of the cuticular microbiomes of both Allomerus and Tetraponera spp. and instead they are dominated by γ-proteobacteria Erwinia and Serratia spp. Both these phyla are known to contain antibiotic-producing species which might therefore play a protective role in these ant–plant systems.
Keywords: 16S pyrosequencing, Allomerus, fungus-growing ants, microbiome, Tetraponera
Introduction
Insect–bacterial symbioses are widespread in the environment (Moran 2006) and antibiotic-producing bacterial symbionts are often used to protect the host and/or their resources (Seipke et al. 2012a). In recent years, these symbioses have been explored as a potential source of new antibiotics as bacteria that have coevolved with their hosts might have evolved unique antibiotic-biosynthetic pathways (Poulsen 2010). Many insects (ants, termites, gall midges, and beetles) have evolved fungiculture and live in symbiosis with their fungal cultivar. In attine ants, it has been shown that mixed communities of antibiotic-producing bacteria (Phylum Actinobacteria) protect the ants and possibly their fungal cultivar against infection (Currie et al. 1999; Haeder et al. 2009; Sen et al. 2009; Barke et al. 2010; Schoenian et al. 2011; Mattoso et al. 2012).
Fungiculture has also been suggested to be widespread in plant–ants, which live in symbiosis with a host plant (Defossez et al. 2009). Recently discovered examples include Allomerus and Tetraponera species, which both use a single type of fungus, but for very different purposes. Allomerus spp. live in symbiosis with the Amazonian plant, Hirtella physophora, and cultivate a sooty mold (order Chaetothyriales) to use as a building material to carry out their unique mechanism of ambush hunting (Heil and McKey 2003; DeJean et al. 2005). These ants use their fungus to build a galleried structure along the stems and branches of their host plants in which they hide and await insect prey (DeJean et al. 2005; Ruiz-Gonzalez et al. 2010). The fungus may also act as a chemoattractant to prey insects (DeJean et al. 2005; Ruiz-Gonzalez et al. 2010). Tetraponera spp. live in symbiosis with the African plant, Acacia drepanolobium (swollen thorn acacia), which dominates the vegetation of the Kenyan Laikipia District (Young et al. 1997). The acacia defends itself against large- and medium-sized herbivores by a combination of long hollow thorns and by hosting four different ant species of the genera Tetraponera and Crematogaster (Young et al. 1997; Palmer et al. 2008). These ants have been observed cultivating a fungus (genus Chaetomium) inside their domatia and recent evidence suggests that they cultivate this fungus as food (Defossez et al. 2009). Remarkably, Tetraponera spp. also host nitrogen-fixing endosymbiotic bacteria, related to those found in the root nodules of legumes, which are proposed to fix nitrogen for their hosts (Van Borm et al. 2002).
The purpose of our study was to investigate the bacterial communities on the cuticles of fungus-growing Allomerus and Tetraponera ants and in particular to discover whether antibiotic-producing actinobacteria predominate in these communities as they do in attine ants. We previously reported that antibiotic-producing actinobacteria can be isolated from Allomerus spp. using culture-dependent techniques. However, selective isolation of these actinomycetes gave no indication as to their abundance on Allomerus workers or in their domatia and offered no evidence of a meaningful or specific interaction (Seipke et al. 2012c). In this study, we use culture-independent 16S rDNA 454-pyrosequencing to examine the bacterial communities associated with Allomerus and Tetraponera worker ants. We report that the cuticular microbiomes of Allomerus and Tetraponera spp. are dominated by Proteobacteria, but share a very similar distribution of bacterial phyla despite their geographic separation. At the genus level, the proteobacterial genera Erwinia and Serratia are dominant and both are known to include antibiotic-producing species. A single antibiotic-producing actinomycete (Streptomyces spp.) was found in the Allomerus samples and none were found associated with Tetraponera ants, suggesting that actinomycetes do not play an important role in this symbiosis. We conclude that if these plant–ants are using antibiotics to protect themselves or their fungal cultivars then they are most likely supplied by Erwinia or Serratia species.
Materials and Methods
Sample collection
Allomerus samples were collected in French Guiana from three inland sites, Basevie, Area 9, and Area 24, which all are close to the field station at Barrage de Petit Saut. Hirtella-hosted A. decemarticulatus colonies (HPAD) were sampled as well as Cordia-hosted A. octoarticulatus colonies (CNAO) and Hirtella-hosted A. octoarticulatus (HPAO) colonies. Five worker ants from each of the HPAD, CNAO, and HPAO samples were pooled and stored in 1 mL of 20% glycerol at −20°C, until processing. Allomerus decemarticulatus and A. octoarticulatus ants were distinguished morphologically by counting the number of antennal segments of representative worker ants under a stereomicroscope, 10 for A. decemarticulatus and eight for A. octoarticulatus. Tetraponera penzigi workers were provided by Naomi E. Pierce (Harvard, U.S.A.) and were sampled from Kitengela (S1o23′526′′ E36o49′108′′) and Ngong Hills (S1o26′946′′ E36o38′358′′) from southern Kenya (near Nairobi), as well as Mpala Road (N0o32′ E36o42′) from central Kenya (Laikipia district). An average of five worker ants were collected from each colony and were preserved in 1 mL of 20% glycerol.
Isolation of bacterial DNA, PCR, sequencing, and analysis
Bacteria present on worker ants stored in 20% glycerol were removed by vortexing and were subsequently trapped by passing the glycerol solution through a 0.45-μm filter. Total DNA was extracted from the filters using phenol–chloroform extraction followed by ethanol precipitation. The DNA was used as a template for PCR with the universal primers Gray28F 5′-GAGTTTGATCNTGGCTCAG and Gray519R 5′-GTNTTACNGCGGCKGCTG, which target variable regions V1–V3 in the 16S gene (Ishak et al. 2011b). A multiplex identification tag (11 nucelotides) was ligated to the 5′ and 3′ ends of 16S amplicons prior to 454-pyrosequencing (1/6 plate per sample) with a GS FLX sequencer (Roche, Hertfordshire, UK) using the GS FLX Titanium series chemistry kit at The Genome Analysis Centre (Norwich, U.K.). Raw data were deposited in the European Nucleotide Archive under accessions ERX177635 and ERX177636 for Tetraponera and Allomerus samples, respectively. The resulting 454 sequence reads were processed and analyzed using the software package Quantitative Insights Into Microbial Ecology v1.4.0 (http://qiime.org/index.html, Caporaso et al. 2010b) and the software packages contained within using the default parameters for each step unless otherwise specified. Reads that lacked the primer or possessed more than one primer mismatch, were too short (<200 nt), or contained a homopolymer run exceeding six nucleotides were removed. The remaining sequences were grouped into samples according to their 11-mer identifier and subsequently denoised using the denoise_wrapper.py script in QIIME (Reeder and Knight 2010). Denoised sequence reads were binned into operational taxonomic units (OTUs) using UCLUST (Edgar 2010) with shared identity threshold of 97%. Chimeric OTUs were identified by ChimeraSlayer (Haas et al. 2011) and discarded. One representative sequence from each OTU was aligned to the Greengenes core reference alignment (DeSantis et al. 2006) with PyNAST with a minimum percent identity of 75% (Caporaso et al. 2010a). Taxonomy was assigned using the Ribosomal Database Project (RDP) classifier 2.2 (Wang et al. 2007) with a minimum support threshold of 80%. Sequences belonging to OTUs designated as unclassified or that could not be assigned taxonomy beyond the domain Bacteria were removed and assumed to be derived from nonbacterial 16S sequences.
Results and Discussion
Allomerus samples were collected in French Guiana and Tetraponera samples were collected in Kenya (see Materials and Methods). Bacteria were washed off from ant cuticles using sterile 20% glycerol and bacterial DNA was extracted and used as template for PCR with universal primers Gray28F and Gray519R (Materials and Methods). The 16S amplicons that resulted were 454-pyrosequenced with a GS FLX sequencer (Roche) using the GS FLX Titanium series chemistry kit. Sequence data were quality filtered and taxonomy was assigned using the Quantitative Insights Into Microbial Ecology (QIIME) software package (Caporaso et al. 2010b). In total, 16,427 pyrotags (Allomerus) and 8388 pyrotags (Tetraponera) passed quality filtering and were assigned taxonomy.
The relative abundance of bacteria phyla on cuticles of Allomerus and Tetraponera ants is shown in Figure 1. Proteobacteria accounted for >75% of all pyrotags for both Allomerus and Tetraponera spp. Roughly 17% of pyrotags were classified as Actinobacteria (Allomerus) and Firmicutes (Tetraponera), while Firmicutes (1.8% of pyrotags) and Bacteroidetes (0.5% of pyrotags) were the least abundant phyla in Allomerus and Tetraponera cuticular microbiomes, respectively.
Figure 1.
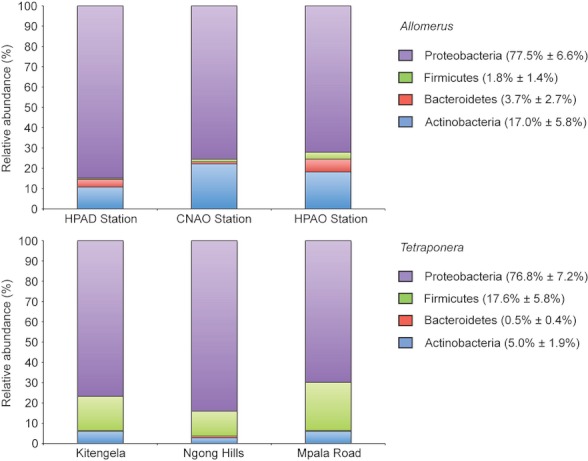
Relative pyrotag abundance of phyla in cuticular microbiomes of Allomerus and Tetraponera ants. The cuticles of both ant genera are dominated by Proteobacteria. We also report the average relative abundance ± the standard error for each phyla observed. For the Tetraponera graph, Cyanobacteria and candidate phylum TM7 are not shown but are present in average abundances of <0.01%.
Pyrotags were binned into OTUs sharing ≥97% identity and assigned taxonomy. The bacterial diversity at the phylum level is summarized in Table 1 and the relative abundance of these phyla are graphically depicted in Figure 2. Proteobacteria accounted for ∼58% of OTUs for Allomerus, and ∼50% of OTUs for Tetraponera on average. For Allomerus, OTU richness for the remaining phyla correlated perfectly to the pyrotag abundance displayed in Figure 1. However, for Tetraponera, despite accounting for only ∼5% of pyrotags, Actinobacteria possessed greater bacterial diversity (an average of 20 OTUs) compared with Firmicutes, which accounted for ∼17% of pyrotags, but only consisted of 11.7 OTUs on average.
Table 1.
Richness of operational taxonomic units (OTUs) for Allomerus and Tetraponera ant cuticular microbiomes
| Phylum | Allomerus | Tetraponera | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HPAD Station | CNAO Station | HPAO Station | Avg | StErr | Kitengela | Ngong Hills | Mpala Road | Avg | StErr | |
| Actinobacteria | 29 | 35 | 45 | 36.3 | 8.1 | 26 | 23 | 23 | 20.0 | 2.0 |
| Bacteroidetes | 11 | 11 | 11 | 11.0 | 0.0 | 2 | 3 | 2 | 2.3 | 0.6 |
| Firmicutes | 5 | 7 | 12 | 8.0 | 3.6 | 13 | 16 | 10 | 11.7 | 2.5 |
| Proteobacteria | 80 | 72 | 74 | 75.3 | 4.2 | 42 | 42 | 34 | 39.0 | 4.4 |
| Total | 125 | 125 | 142 | 130.7 | 9.8 | 83 | 84 | 69 | 78.7 | 8.4 |
OTUs comprise pyrotags sharing ≥97% nucleotide identity. HPAD, Hirtella-hosted A. decemarticulatus; CNAO, Cordia-hosted A. octoarticulatus; HPAO, Hirtella-hosted A. octoarticulatus; Avg, average; StErr, standard error.
Figure 2.
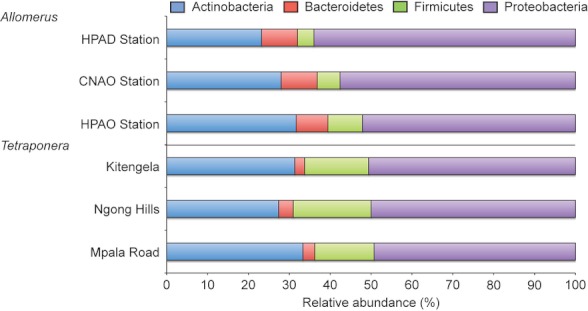
Relative operational taxonomic unit (OTU) richness of bacterial phyla in cuticular microbiomes of Allomerus and Tetraponera ants. For Allomerus, two Proteobacteria OTUs that could not be assigned to a class were omitted from the analysis.
Strikingly, of the OTUs classified as Actinobacteria, Allomerus ants possessed only a single OTU (0.06% relative abundance of pyrotags for the CNAO sample) corresponding to a known antibiotic-producing actinomycete genus (Streptomyces) and the Tetraponera sample did not possess any OTUs classified as antibiotic-producing Actinobacteria. This starkly contrasts with the cuticular microbiome of attine ants, which is dominated by antibiotic-producing actinomycetes Pseudonocardia and Amycolatopsis spp. (Sen et al. 2009; Ishak et al. 2011a; Anderson et al., 2011), and is also dissimilar from the microbiome of fire ant workers, which are dominated by Spiroplasma and Nocardia spp. (Ishak et al. 2011b).
As Proteobacteria was the dominant phyla identified both in terms of pyrotag abundance and OTU richness, we analyzed the diversity of Proteobacteria at the level of class (Fig. 3). γ-Proteobacteria was the most abundant class, accounting for 68% and 91% of pyrotags, and 49% and 61% of OTU richness for Allomerus and Tetraponera ants, respectively (Table S1). The decrease in relative abundance of γ-proteobacteria pyrotags versus OTU richness suggests that a few taxa account for the majority of Proteobacteria pyrotags. In order to further analyze this possibility, we tabulated OTUs with a pyrotag relative abundance ≥1% (Table 2). Bacterial types at greater than 84% abundance in the microbial community associated with Allomerus ants is represented by 36 species, with a single Erwinia species (29.2%), two Ochrobactrum species (10.6%), and a single Serratia species (10.2%) comprising the most abundant taxa. Likewise, greater than 84% of the microbial community associated with the cuticles of Tetraponera ants is represented by just 15 genera, with two Serratia species being the most abundant (53.6%). Rarefaction plots using 97% shared identity (i.e., species-level classification) (Fig. S1) indicated that our sampling captured the majority of the bacterial diversity at this taxonomic level associated with cuticles of Allomerus and Tetraponera ants, though it is likely we undersampled rare bacterial genera.
Figure 3.
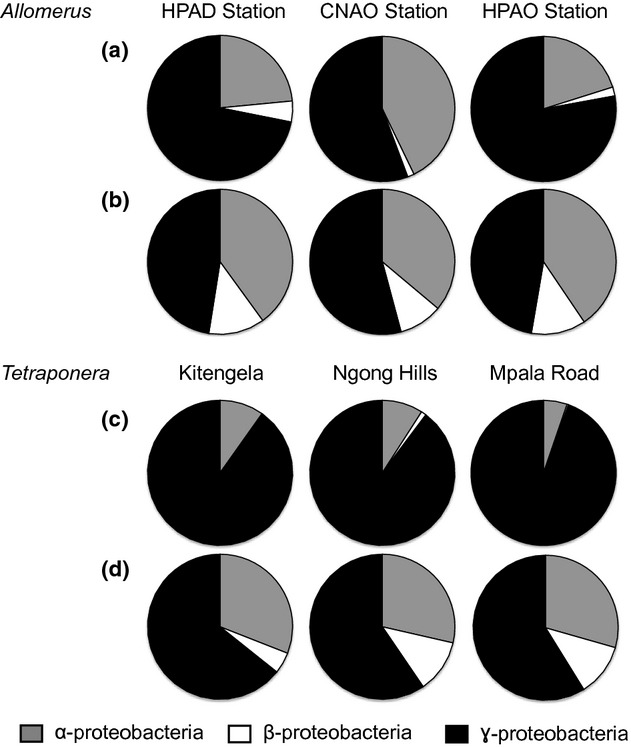
The diversity of Proteobacteria within Allomerus and Tetraponera cuticular microbiomes. (A and C) Relative abundance of pyrotags from proteobacterial classes; (B and D) Relative abundance of operational taxonomic units (OTUs) (97% identity) from proteobacterial classes. The distribution and average relative abundance of Proteobacteria pyrotags and OTUs is enumerated in Table S1.
Table 2.
Genera of OTUs associated with Allomerus and Tetraponera ant cuticles
| Allomerus | Phylum or class | HPAD Station | CNAO Station | HPAO Station | Avg | StErr |
|---|---|---|---|---|---|---|
| Quality-filtered pyrotags | 5312 | 5661 | 5454 | |||
| Genus | ||||||
| Actinomycetales1 | Actinobacteria | 0.6 | – | 1.1 | 0.6 | 0.6 |
| Actinomycetales1 | Actinobacteria | – | 1.1 | 1.0 | 0.7 | 0.6 |
| Microbacterium | Actinobacteria | 0.3 | 8.9 | – | 3.1 | 5.1 |
| Actinomycetales1 | Actinobacteria | 0.1 | 1.1 | 0.2 | 0.5 | 0.6 |
| Microbacterium | Actinobacteria | 6.3 | 1.2 | 8.0 | 5.2 | 3.5 |
| Brevibacterium | Actinobacteria | – | 3.4 | 0.6 | 1.3 | 1.8 |
| Actinomycetales1 | Actinobacteria | 0.9 | 2.0 | 1.5 | 1.4 | 0.5 |
| Sphingobacterium | Bacteroidetes | 0.3 | 0.1 | 1.2 | 0.5 | 0.6 |
| Sphingobacterium | Bacteroidetes | – | – | 2.6 | 0.9 | 1.5 |
| Sphingobacterium | Bacteroidetes | 1.6 | 0.4 | 0.6 | 0.9 | 0.7 |
| Bacillus | Firmicutes | 0.5 | 0.1 | 1.9 | 0.8 | 1.0 |
| Staphylococcus | Firmicutes | <0.1 | 1.1 | 0.4 | 0.5 | 0.6 |
| Bosea | α-proteobacteria | 0.8 | 0.1 | 1.8 | 0.9 | 0.8 |
| Devosia | α-proteobacteria | – | <0.1 | 1.1 | 0.4 | 0.7 |
| Rhizobium | α-proteobacteria | 1.3 | 0.2 | 0.2 | 0.6 | 0.7 |
| Phyllobacteriaceae2 | α-proteobacteria | 1.1 | <0.1 | – | 0.4 | 0.6 |
| Bradyrhizobiaceae2 | α-proteobacteria | 0.8 | 0.1 | 1.3 | 0.7 | 0.6 |
| Rhizobiales1 | α-proteobacteria | 2.7 | 3.5 | 2.5 | 2.9 | 0.5 |
| Ochrobactrum | α-proteobacteria | 2.6 | 10.4 | 1.3 | 4.8 | 4.9 |
| Ochrobactrum | α-proteobacteria | 4.6 | 12.4 | 0.2 | 5.8 | 6.2 |
| Bosea | α-proteobacteria | 1.7 | 2.4 | 1.3 | 1.8 | 0.6 |
| Rhizobium | α-proteobacteria | 0.9 | <0.1 | 1.5 | 0.8 | 0.7 |
| Bradyrhizobiaceae2 | α-proteobacteria | 0.7 | 1.6 | 0.3 | 0.9 | 0.6 |
| Comamonas | β-proteobacteria | 1.4 | – | – | 0.5 | 0.8 |
| Shinella | β-proteobacteria | 0.3 | 0.3 | 1.1 | 0.6 | 0.4 |
| Shinella | β-proteobacteria | 1.0 | 0.1 | 0.1 | 0.4 | 0.5 |
| Enterobacteriaceae2 | γ-proteobacteria | 0.8 | 0.3 | 1.6 | 0.9 | 0.7 |
| Stenotrophomonas | γ-proteobacteria | <0.1 | 1.0 | 0.6 | 0.5 | 0.5 |
| Serratia | γ-proteobacteria | 12.5 | 15.6 | 1.4 | 9.8 | 7.5 |
| Stenotrophomonas | γ-proteobacteria | 1.9 | 4.4 | 0.9 | 2.4 | 1.8 |
| Enterobacteriaceae2 | γ-proteobacteria | 0.1 | 4.6 | <0.1 | 1.6 | 2.6 |
| Enterobacteriaceae2 | γ-proteobacteria | 1.1 | 0.5 | 1.6 | 1.1 | 0.5 |
| Enterobacteriaceae2 | γ-proteobacteria | <0.1 | 0.5 | 1.1 | 0.5 | 0.5 |
| Erwinia | γ-proteobacteria | 38.6 | 5.3 | 43.7 | 29.2 | 20.8 |
| Enterobacteriaceae2 | γ-proteobacteria | 1.1 | 1.1 | 1.9 | 1.4 | 0.5 |
| Enterobacter | γ-proteobacteria | 0.4 | 3.7 | 0.4 | 1.5 | 1.9 |
| Total | 86.9 | 87.5 | 84.8 | |||
| Tetraponera | Kitengela | Ngong Hills | Mpala Road | |||
|---|---|---|---|---|---|---|
| Quality-filtered pyrotags | 2709 | 2601 | 3078 | |||
| Genus | ||||||
| Curtobacterium | Actinobacteria | 1.0 | 0.3 | 0.3 | 0.5 | 0.4 |
| Microbacteriaceae2 | Actinobacteria | 1.6 | 0.8 | 1.8 | 1.4 | 0.5 |
| Actinomycetospora | Actinobacteria | <0.1 | 0.1 | 1.4 | 0.5 | 0.8 |
| Staphylococcus | Firmicutes | – | 0.5 | 19.8 | 6.7 | 11.3 |
| Geobacillus | Firmicutes | 10.2 | 8.2 | 2.8 | 7.1 | 3.8 |
| Weissella | Firmicutes | 4.4 | 0.9 | – | 1.8 | 2.3 |
| Rhizobiales1 | α-proteobacteria | 1.6 | 0.8 | 0.4 | 0.9 | 0.6 |
| Sphingomonadaceae2 | α-proteobacteria | 0.7 | 5.3 | – | 2.0 | 2.9 |
| Brevundimonas | α-proteobacteria | 1.3 | 0.1 | 0.4 | 0.6 | 0.6 |
| Sphingobium | α-proteobacteria | <0.1 | – | 1.1 | 0.4 | 0.6 |
| Serratia | γ-proteobacteria | 1.9 | 3.5 | 3.3 | 2.9 | 0.9 |
| Serratia | γ-proteobacteria | 27.6 | 64.8 | 59.6 | 50.7 | 20.2 |
| Enterobacteriaceae2 | γ-proteobacteria | 2.7 | – | – | 0.9 | 1.5 |
| Enterobacteriaceae2 | γ-proteobacteria | 30.7 | – | <0.1 | 10.2 | 17.7 |
| Halomonas | γ-proteobacteria | 0.8 | 1.0 | 0.2 | 0.7 | 0.4 |
| Total | 84.4 | 86.2 | 91.1 | |||
Only OTUs comprising ≥1% relative pyrotag abundance are shown. Also shown is the average percent abundance (Avg) and standard error (StErr) of taxa. Taxonomy at the level of class is shown for Proteobacteria. OTU, operational taxonomic unit; HPAD, Hirtella-hosted A. decemarticulatus; CNAO, Cordia-hosted A. octoarticulatus; HPAO, Hirtella-hosted A. octoarticulatus.
The 1order or 2family is reported in instances when a genus-level classification could not be made.
The high abundance of Erwinia spp. in the Allomerus samples may not be surprising, because this genus of bacteria are abundant epiphytic phytopathogens that cause disease on crop plants and crop trees (Starr and Chatterjee 1972). The γ-proteobacteria Erwinia species associated with Allomerus ants is possibly carryover from the close association with their plant host. The α-proteobacterium Ochrobactrum and γ-proteobacteria Serratia spp. are well-known insect symbionts (both harmful and beneficial) and more than 70 species of insects are susceptible to infections by Serratia spp. (Bucher 1963; Grimont and Grimont 1978). One highly coevolved species (Candidatus Serratia symbiotica) is an important endosymbiont of the aphid, Cinara cedri (Lamelas et al. 2008). It is notable that Tetraponera ant samples from three separate sites were dominated by Serratia species with an abundance as high as 64.8% in the Ngong Hills sample. In addition to their roles as plant and insect pathogens or symbionts, Erwinia and Serratia spp. are also antibiotic producers (Wilf and Salmond 2012), which suggests that Allomerus and Tetraponera ants may use these taxa for protection. Certainly, the dominance of Serratia species on the Tetraponera worker ants is reminiscent of the levels of abundance of actinomycetes in attine ants (Sen et al. 2009; Anderson et al. 2011; Ishak et al. 2011a).
Given that we previously isolated six actinobacterial species from Allomerus ants (one Amycolatopsis spp. and five Streptomyces spp.) and four of the isolates produced antifungal compounds (Seipke et al. 2012c), it is somewhat surprising that antibiotic-producing actinobacteria are not abundant on Allomerus ants. Additionally, even though our sampling suggested the absence of antibiotic-producing actinobacteria within the cuticular microbiome of Tetraponera ants, we could readily isolate antibiotic-producing actinobacteria from the ants used in this study, including three Streptomyces spp. and three Saccharopolyspora spp, five of which have antifungal activity (Table S2). In fact, this highlights the caution with which culture-dependent studies should be treated as our own approach was biased toward the selective isolation of actinomycetes, which have a “hairy” colony morphology due to their filamentous growth and are easy to identify by eye on agar plates. The culture-independent data suggests that filamentous actinomycete bacteria have a rare abundance on Allomerus and Tetraponera spp. as most of the species observed in culturing studies were not detected by deep sequencing the same samples (Materials and Methods). As noted above, however, it is likely we under-sampled rare bacterial genera.
Our direct sequencing approach suggests that it is unlikely that Allomerus and Tetraponera species use actinomycetes for protection and instead suggest that antibiotic-producing Proteobacteria might perform this role. This would make sense as these plant–ants rarely, if ever, leave their host plants and do not come into contact with the soil where actinomycete species commonly reside. Alternatively, they will come into frequent contact with Erwinia and Serratia spp. because they are common symbionts of plants and insects and so these bacteria may have colonized or been recruited to the plant–ant cuticles. It is also possible that the ants do not need protection from antibiotic-producing bacteria, for example, they are less exposed to pathogens because they do not leave their host plants and do not bring exogenous material (e.g., leaf matter) into the nest; or they may have evolved efficient grooming techniques that are sufficient to limit spread of pathogens in the nest and they may produce their own antimicrobial compounds and these are sufficient to protect against infection.
Conclusions
Our study supports the hypothesis that plant–ants like Allomerus and Tetraponera are colonized by very different bacterial phyla compared with soil-dwelling ants like the attines. It seems likely that attine ants recruited antibiotic-producing actinomycete bacteria from the soil and evolved a symbiosis in which they use these bacteria for protection. We propose that a similar scenario might be true for plant–ants but in their case they used antibiotic-producing bacteria commonly found on their host plants or prey insects, notably Erwinia and Serratia spp. Certainly it is striking that worker ants of both ant–plant systems are dominated by the same bacterial phyla and future work will be aimed at understanding the role that these bacterial species play in ant–plant–fungal symbioses.
Acknowledgments
We thank Naomi Pierce for providing Tetraponera samples and Jérôme Orivel for assisting J. B. with collection of Allomerus samples. We also thank Heather Ishak and Ulrich Mueller for helpful discussions concerning this work. 454-pyrosequencing was supported by a Capacity and Capability Challenge Programme (project number CCC-1-12) with The Genome Analysis Centre in Norwich, U.K. Ryan F. Seipke was supported through the MRC Milstein award G0801721 and NERC grant NE/J01074X/1. Jörg Barke was supported by a UEA-funded Ph.D. studentship.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Rarefaction curves (97% shared identity) for Allomerus (HPAO Station, HPAD Station, CNAO Station) and Tetraponera (Ngong Hills, Kitengela, Mpala Road) ant samples.
Table S1. Distribution of Proteobacteria within cuticular microbiomes Allomerus and Tetraponera ants.
Table S2. 16S identification of the antibiotic-producing Actinobacteria genera from Tetraponera ants.
References
- Anderson SB, Hughes DP, Boomsma JJ. Copenhagen, Denmark: Graduate School of Science, Faculty of Science, University of Copenhagen; 2011. Dynamics of ant-microbial interactions: coevolution along the parasitism-mutualism continuum. Ph.D. diss. [Google Scholar]
- Barke J, Seipke RF, Grüschow S, Heavens D, Drou N, Bibb MJ, et al. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010;8:109. doi: 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher GE. Nonsporulating bacterial pathogens. In: Steinhaus EA, editor. Insect pathology: an advanced treatise. Vol. 2. New York: Academic Press; 1963. pp. 117–147. [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010a;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010b;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-produce bacteria to control garden parasites. Nature. 1999;398:701–705. [Google Scholar]
- Defossez E, Selosse MA, Dubois MP, Mondolot L, Faccio A, Djieto-Lordon C, et al. Ant-plants are fungi: a new threeway symbiosis. New Phytol. 2009;182:942–949. doi: 10.1111/j.1469-8137.2009.02793.x. [DOI] [PubMed] [Google Scholar]
- DeJean A, Solano PJ, Ayroles J, Corbara B, Orivel J. Arboreal ants build traps to capture prey. Nature. 2005;434:973. doi: 10.1038/434973a. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Grimont PAD, Grimont F. The genus Serratia. Annu. Rev. Microbiol. 1978;32:221–248. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeder SR, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, McKey D. Protective ant-plant interactions as model systems in ecological and evolutionary research. Annu. Rev. Ecol. Evol. Syst. 2003;34:425–453. [Google Scholar]
- Ishak HD, Miller JL, Sen R, Dowd SE, Meyer E, Mueller UG. Microbiomes of ant castes implicate new microbial roles in the fungus-growing ant Trachymyrmex septentrionalis. Sci. Rep. 2011a;1:204. doi: 10.1038/srep00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, et al. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb. Ecol. 2011b;61:821–831. doi: 10.1007/s00248-010-9793-4. [DOI] [PubMed] [Google Scholar]
- Lamelas A, Perez-Brocal V, Gomez-Valero L, Gosalbes MJ, Moya A, Latorre A. Evolution of the secondary symbiont “Candidatus serratia symbiotica” in aphid species of the subfamily lachninae. Appl. Environ. Microbiol. 2008;74:4236–4240. doi: 10.1128/AEM.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoso TC, Moreira DD, Samuels RI. Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biol. Lett. 2012;8:461–464. doi: 10.1098/rsbl.2011.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. Symbiosis. Curr. Biol. 2006;16:R866–R871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Palmer TM, Stanton NL, Young TP, Goheen JR, Pringle RM, Karban R. Breakdown of an ant-plant mutualism follows the loss of large herbivores from an African savanna. Science. 2008;319:192–195. doi: 10.1126/science.1151579. [DOI] [PubMed] [Google Scholar]
- Poulsen M. Biomedical exploitation of the fungus-growing ant symbiosis. Drug News Perspect. 2010;23:203–210. doi: 10.1358/dnp.2010.23.3.1489981. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gonzalez MX, Male P-JG, Leroy C, DeJean A, Gryta H, Jargeat P, et al. Specific, non-nutritional association between an ascomycete fungus and Allomerus plant-ants. Biol. Lett. 2010;7:475–479. doi: 10.1098/rsbl.2010.0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenian IM, Spiteller M, Ghaste M, Wirth R, Herz H, Spiteller D. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc. Natl. Acad. Sci. USA. 2011;108:1955–1960. doi: 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke RF, Kaltenpoth M, Hutchings MI. Streptomyces as symbionts: a emerging and widespread theme? FEMS Microbiol. Rev. 2012a;36:862–876. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- Seipke RF, Grüschow S, Goss RJM, Hutchings MI. Isolating antifungals form fungus-growing ant symbionts using a genome-guided chemistry approach. Methods Enzymol. 2012b;517:47–70. doi: 10.1016/B978-0-12-404634-4.00003-6. [DOI] [PubMed] [Google Scholar]
- Seipke RF, Barke J, Ruiz-Gonzalez MX, Orivel J, Yu DW, Hutchings MI. Fungus-growing Allomerus ants are associated with antibiotic-producing actinobacteria. Antonie Van Leeuwenhoek. 2012c;101:443–447. doi: 10.1007/s10482-011-9621-y. [DOI] [PubMed] [Google Scholar]
- Sen R, Ishak HD, Estrada E, Dowd SE, Hong E, Mueller UG. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. USA. 2009;106:17805–17810. doi: 10.1073/pnas.0904827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr MP, Chatterjee AK. The genus Erwinia: enterobacteria pathogenic to plants and animals. Annu. Rev. Microbiol. 1972;26:389–426. doi: 10.1146/annurev.mi.26.100172.002133. [DOI] [PubMed] [Google Scholar]
- Van Borm S, Buschinger A, Boomsma JJ, Billen J. Tetraponera ants have gut symbionts related to nitrogen-fixing root-nodule bacteria. Proc. Biol. Sci. 2002;269:2023–2027. doi: 10.1098/rspb.2002.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilf NM, Salmond GP. The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology. 2012;158:648–658. doi: 10.1099/mic.0.055780-0. [DOI] [PubMed] [Google Scholar]
- Young TP, Stubblefield CH, Isbell LA. Ants on swollen-thorn acacias: species coexistence in a simple system. Oecologia. 1997;109:98–107. doi: 10.1007/s004420050063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


