Abstract
Copper amine oxidases (CAOs) are responsible for the oxidative deamination of primary amines to their corresponding aldehydes. The CAO catalytic mechanism can be divided into two half-reactions: a reductive half-reaction, in which a primary amine substrate is oxidized to its corresponding aldehyde with the concomitant reduction of the organic cofactor 2,4,5-trihydroxyphenylalanine quinone (TPQ), and an oxidative half-reaction, in which reduced TPQ is re-oxidized with the reduction of molecular oxygen to hydrogen peroxide. The reductive half-reaction proceeds via Schiff base chemistry, in which the primary amine substrate first attacks the C5 carbonyl of TPQ, forming a series of covalent Schiff base intermediates. The X-ray crystal structures of copper amine oxidase-1 from the yeast Hansenula polymorpha (HPAO-1) in complex with ethylamine and benzylamine have been solved to resolutions of 2.18 and 2.25 Å, respectively. These structures reveal the two amine substrates bound at the back of the active site coincident with TPQ in its two-electron reduced aminoquinol form. Rearrangements of particular amino acid side chains within the substrate channel and specific protein-substrate interactions provide insight into substrate specificity in HPAO-1. These changes begin to account for this CAO’s kinetic preference for small, aliphatic amines over the aromatic amines or whole peptides preferred by some of its homologs.
Keywords: amine oxidase, TPQ, substrate specificity, substrate binding, amine, channel, copper
INTRODUCTION
The copper amine oxidase (CAO) family of enzymes, which are responsible for the oxidation of primary amines to their corresponding aldehydes, are found in nearly every aerobic organism, including bacteria, yeast, plants, fungi, and animals. This ubiquity is echoed in the wide range of primary amines that can act as CAO substrates, and consequently in the variety of biological functions attributed to these enzymes. In prokaryotes, CAOs allow for the use of primary amines as a sole carbon and/or nitrogen source during metabolism.1,2 The functions of eukaryotic CAOs are more complex, with proposed roles including the production of second messenger compounds involved in the cross-linking of cell wall components and wound healing in plants,3,4 and the uptake of inflammatory leukocytes within the vasculature of mammals.5-7 The presence of the extracellular CAO in adipocytes is also well documented, and while initially ascribed to a role in moderating glucose uptake,8 may also present a first line of defense against invading pathogens.9 The products generated during CAO catalysis can be highly cytotoxic, in particular formaldehyde, which has been implicated in protein crosslinking correlated with cellular damage in late-diabetic vascular complications and atherosclerosis.10
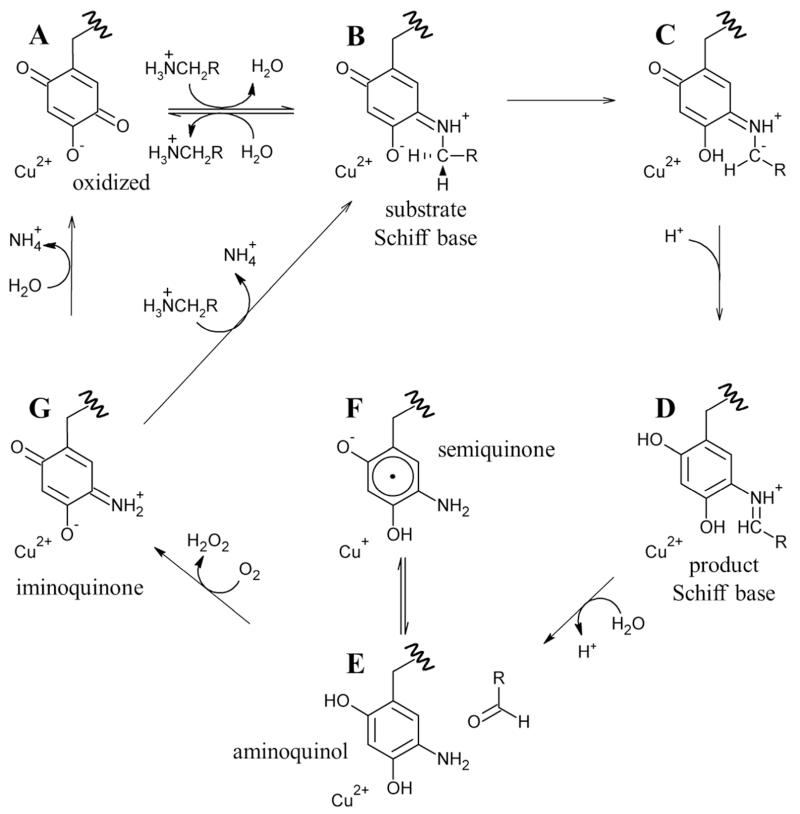
Catalysis in CAOs proceeds by means of a ping-pong mechanism: a reductive half-reaction in which a primary amine substrate is oxidized to its corresponding aldehyde with the concomitant reduction of the organic quinocofactor 2,4,5-trihydroxyphenylalanine quinone (TPQ), and an oxidative half-reaction in which TPQ is re-oxidized with the reduction of molecular oxygen to hydrogen peroxide and release of an ammonium ion:11
The reductive half-reaction begins with a nucleophilic attack on the C5 atom of oxidized TPQ by a primary amine (A in Scheme 1). This covalently links the two species to generate a substrate Schiff base intermediate (B in Scheme 1). An invariant aspartate residue abstracts a proton from the C1 atom of the substrate, which produces the product Schiff base (D in Scheme 1) by way of a carbanionic intermediate (C in Scheme 1). Rapid hydrolysis of the product Schiff base releases the corresponding aldehyde, leaving TPQ in a two-electron reduced aminoquinol form (E in Scheme 1) and marking the end of the reductive half-reaction.
Scheme 1.
Proposed CAO catalytic mechanism. RCH2NH3+ is representative of all primary amine substrates.
The oxidative half-reaction involves the re-oxidation of reduced TPQ coupled to the reduction of molecular oxygen to hydrogen peroxide. Following the release of the free aldehyde product at the end of the reductive half-reaction, the aminoquinol/Cu(II) couple exists in equilibrium with a semiquinone radical/Cu(I) couple (E and F in Scheme 1, respectively, with the position of this equilibrium varying from one CAO species to another and lying toward E in HPAO-1). The reduction of molecular oxygen generates an oxidized iminoquinone intermediate and hydrogen peroxide (G in Scheme 1). Finally, hydrolysis of the iminoquinone releases an ammonium ion and regenerates oxidized TPQ (A in Scheme 1). Alternatively, under steady state conditions where substrate levels are high, the iminoquinone is proposed to react with another primary amine, releasing an ammonium ion and forming the substrate Schiff base directly (B in Scheme 1).
Despite well-conserved active site residues that carry out identical chemistry, members of the CAO family of enzymes are known to react with a wide variety of primary amine substrates, ranging from methylamine to whole proteins. Substrate preferences in CAOs vary by source and cellular location. For instance, CAOs isolated from Escherichia coli (ECAO) and Arthrobacter globiformis (AGAO) prefer primary aromatic monoamines such as phenylethylamine or tyramine.12 Two CAO paralogs isolated from the yeast Hansenula polymorpha (HPAO-1 and HPAO-2) demonstrate opposing preferences, with HPAO-1 preferring small aliphatic amines, such as ethylamine or methylamine, and HPAO-2 preferring the bulkier aromatic amine benzylamine.13 Vascular adhesion protein-1 (VAP-1), one of three functional CAO isoforms expressed in humans, is active against much larger amine substrates, and is proposed to regulate leukocyte adhesion and rolling along internal vascular surfaces during inflammation through its oxidation of membrane associated VAP-1 counter-receptors in endothelial cells.14 In contrast, VAP-1 that is localized to the outer plasma membrane of adipocytes recognizes a wide range of aliphatic and aromatic amines of variable size.9
Thus far in the study of substrate specificity in CAOs, there has been no structural investigation of a single CAO reduced by multiple physiological primary amine substrates, nor has an amine substrate been observed bound in the CAO active site. In order to explore the structural factors that influence substrate specificity in HPAO-1 as well as the interactions between HPAO-1 and amine substrates, crystals of the native protein have been anaerobically reduced with ethylamine or benzylamine before flash-freezing, and structures of the resulting complexes have been solved by X-ray crystallography. A comparison between HPAO-1 in complex with both amine substrates and the native enzyme provides insight into specific substrate-protein interactions involving residues located within the amine substrate channel and active site. These are considered within the context of steady state kinetic data available for the reaction of HPAO-1 with both amines.
MATERIALS AND METHODS
Protein expression and purification
Native HPAO-1 protein was heterologously expressed in Saccharomyces cerevisiae and purified as previously described.15
HPAO-1 crystallization and substrate soaks
Following purification, HPAO-1 protein was buffer-exchanged into 50 mM HEPES, pH 7.0, and concentrated to 13 mg/mL for crystallization. Crystals were grown by the hanging drop vapor diffusion method as described previously.16 The ratio of protein to crystallization solution (8-9% w/v polyethylene glycol 8000 in 0.22-0.25 M potassium phosphate, pH 6) in the hanging drops was 1:1 (6 μL total). Drops were seeded after 24 hours of equilibration using a streak-seeding technique and mature HPAO-1 crystals as seed donors. Cube-shaped crystals that were pink in color formed within 3-4 days.
Trays containing crystals of native HPAO-1 were brought into an anaerobic glove box (Belle Technology) and allowed to equilibrate for at least one week. Solutions used for crystal soaking and cryoprotection were brought into the glove box immediately after passing N2 gas over the container headspace, while stirring for >30 min in septa-covered bottles. Individual HPAO-1 crystals were soaked in an artificial crystallization solution containing either ethylamine hydrochloride or benzylamine hydrochloride. A range of amine concentrations and soak times were used for substrate soaks in order to optimize the diffraction quality of the crystals, as well as the occupancy of the amine substrate in the complex. All crystals were visually colorless after ~10 seconds following exposure to ethylamine or benzylamine; however crystals were left in the amine-supplemented solutions for longer periods than this to ensure that all residual O2 remaining in the crystallization drop had been consumed, and that any HPAO-1 turnover had terminated at the aminoquinol. The reduced HPAO-1 crystals were then soaked in a crystallization solution containing 25% glycerol mixed with well solution for ~5-10 seconds for cryoprotection before flash-freezing in N2(l). The conditions used to form the complexes reported here were 10 mM ethylamine hydrochloride for 2 hours, or 5 mM benzylamine hydrochloride for 5 minutes.
Single crystal UV/visible microspectrophotometry
HPAO-1 crystals that had been anaerobically reduced with ethylamine or benzylamine before flash-freezing were subjected to single crystal UV/visible spectroscopy before and after X-ray diffraction data collection, using a UV/visible single crystal microspectrophotometer from 4DX Systems (http://www.4dx.se/).
X-ray diffraction data collection, processing, and structure solution
X-ray diffraction data for anaerobic ethylamine- and benzylamine-treated HPAO-1 (designated EtAm-HPAO-1 and BzAm-HPAO-1, respectively) were collected from single crystals at 100K at the Advanced Photon Source, Argonne National Laboratory (beamline 23-ID, GM/CA-CAT; MARmosaic 300 CCD detector). Diffraction data were collected to resolutions of 2.18 and 2.25 Å for EtAm- and BzAm-HPAO-1, respectively, and processed using SCALEPACK and HKL2000.17
The EtAm- and BzAm-HPAO-1 crystals were found to be non-isomorphous with previously deposited structures of native HPAO-1. For phase determination of the BzAm-HPAO-1 data, molecular replacement was conducted using PHASER from the CCP4 suite, with a search model based on a polypeptide dimer from the previously deposited native HPAO-1 structure (PDB code 2OOV with solvent atoms, ligands, and the side chain of residue 405 (TPQ) removed from each protein chain).15,18,19 The structure of EtAm-HPAO-1 was then solved by difference Fourier, using the BzAm-HPAO-1 model with solvent atoms, ligands, and the side chain of residue 405 removed from each polypeptide chain for phasing in the CCP4 suite.19
X-ray crystal structure refinement
The initial coordinates for each structure were first subjected to rigid body refinement using REFMAC in the CCP4 suite.20 This was followed by one round of restrained refinement using automatically generated local non-crystallographic symmetry (NCS) restraints in REFMAC.20 Subsequent refinement cycles were done in the absence of NCS restraints. Water molecules were incorporated into the coordinates of the structures at positions with peaks >3.0σ in the corresponding 2Fo-Fc electron density maps. Multiple cycles of manual model building using COOT followed by refinement using REFMAC were completed based on peaks in the corresponding 2Fo-Fc and Fo-Fc electron density maps.20,21 Copper ions were assigned to the strongest peaks in the electron density maps, which correspond with the well-established copper binding site in CAOs.
When TPQ occupies an “off-copper” position in the native HPAO-1 structure, an ordered water molecule ligates the copper in an axial position.15 In all chains of the HPAO-1 substrate complexes, a water molecule was first modeled at this position based on strong peaks in the 2Fo-Fc and Fo-Fc electron density maps. After refinement, persistent positive peaks in the Fo-Fc difference electron density map in conjunction with very low B-factors (~2 Å2) indicated that water was electronically insufficient to account for the density at this position in chains B, C, D, and E of EtAm-HPAO-1 and chains A, B, E, and F of BzAm-HPAO-1. Although molecular oxygen, superoxide and peroxide cannot be distinguished at this resolution, the geometry and the Cu2+ oxidation state was only consistent with hydrogen peroxide, which has previously been visualized bound side-on to the copper in a crystal structure of ECAO.22 Therefore, hydrogen peroxide was systematically modeled at this position in the active site and subjected to test refinements. Test refinements against this alternate axial copper ligand and B-factor analysis were used to determine the identity of the appropriate species in each polypeptide chain of the two substrate-reduced datasets.
In all polypeptide chains of both substrate-HPAO-1 structures, the active site cofactor could be clearly modeled in its “off-copper” conformation, with no electron density suggesting that any proportion of the cofactor is present in its “on-copper” conformation. Based on single crystal UV/visible absorption spectra, which were bleached, the cofactor was modeled in its aminoquinol form in all polypeptide chains (using PDB residue TYQ).22
The structures of ethylamine and benzylamine were built into the appropriate polypeptide chains based on peaks in the 2Fo-Fc and Fo-Fc electron density maps, which indicated substrate bound in the back of the active site. Additional cycles of manual model building and refinement were carried out in all structures until peaks in the Fo-Fc electron density maps were at the level of noise.
RESULTS
Overall fold of the HPAO-1 substrate complexes
X-ray diffraction data for EtAm- and BzAm-HPAO-1 were collected and refined to resolutions of 2.18 and 2.25 Å, with final Rwork values of 0.150 (Rfree= 0.201) and 0.169 (Rfree = 0.230), respectively (Table 1).
Table 1.
X-ray data collection, processing, and refinement statistics for the ethylamine- and benzylamine-HPAO-1 complexes.
| Data collection and processing statistics | ||
| substrate | ethylamine | benzylamine |
| beamline | 23-ID, GM/CA-CAT | 23-ID, GM/CA-CAT |
| space group | P21 | P21 |
| unit cell dimensions a × b × c, (Å); β (°) | 104.4, 232.8, 105.1; 96.7 | 104.2, 233.7, 105.1; 96.6 |
| no. of molecules in the unit cell, Z | 3 | 3 |
| resolution (Å)a | 50.00-2.18 (2.22-2.18) | 50.00-2.25 (2.29-2.25) |
| no. of unique reflections | 257,277 | 232,595 |
| completeness (%)a | 100.0 (100.0) | 98.8 (97.7) |
| Rmerge(%)a,b | 0.108 (0.457) | 0.089 (0.324) |
| I/σ(I) | 10.0 (1.7) | 9.8 (1.8) |
| redundancya | 3.8 (3.6) | 4.3 (4.1) |
| Structure refinement statistics | ||
| refinement resolution range (Å)a | 49.0-2.18 (2.24-2.18) | 48.9-2.25 (2.31-2.25) |
| no. of reflections in the working seta | 244,276 (16,892) | 220,912 (16,116) |
| no. of reflections in the test seta | 12,922 (875) | 11,654 (793) |
| Rwork(%)a,c | 0.150 (0.243) | 0.169 (0.258) |
| Rfree(%)a,d | 0.201 (0.296) | 0.230 (0.332) |
| rmsd from ideal geometry | ||
| bond lengths (Å) | 0.016 | 0.023 |
| bond angles (°) | 1.59 | 1.95 |
| Ramachadran plot | ||
| energetically favored regions (%) | 95.2 | 94.6 |
| allowed regions (%) | 4.2 | 4.7 |
| outliers (%) | 0.6 | 0.7 |
| Overall average B factor (Å2) | 33.3 | 22.6 |
| Cruickshank’s DPI (Å) | 0.19 | 0.23 |
Numbers in parentheses refer to the highest-resolution shell.
Rmerge =Σhkl Σi|Ihkl,i - 〈Ihkl〉|/ΣhklΣi Ihkl,i, where I is the observed intensity and 〈I〉 is the average intensity for multiple measurements.
Rwork = Σ||Fo/-|Fc||/Σ|Fo|, where |Fo| is the observed structure factor amplitude and |Fc| is the calculated structure factor amplitude for 95% of the data used in refinement.
Rfree based on 5% of the data excluded from refinement.
Both substrate-reduced HPAO-1 datasets were non-isomorphous with native HPAO-1 (Table S1). Consequently, molecular replacement was used to phase the BzAm-HPAO-1 data. The resulting BzAm-HPAO-1 structure was then used to phase the EtAm-HPAO-1 data using difference Fourier techniques. Both substrate-reduced HPAO-1 structures contain six polypeptide chains comprising three physiological dimers within the asymmetric unit (ASU) in space group P21. The overall fold is nearly identical to that of the native enzyme, with superimposition of corresponding main chain atoms with those of native HPAO-1 (PDB code 2OOV) yielding root-mean-square deviations (rmsd) of 0.36 and 0.43 Å for the EtAm- and BzAm-HPAO-1 complexes, respectively.15 Any significant structural differences between the substrate-reduced HPAO-1 and native HPAO-1 structures are limited to areas around the active site and substrate channel of the enzyme.
Substrate binding in the HPAO-1 active site
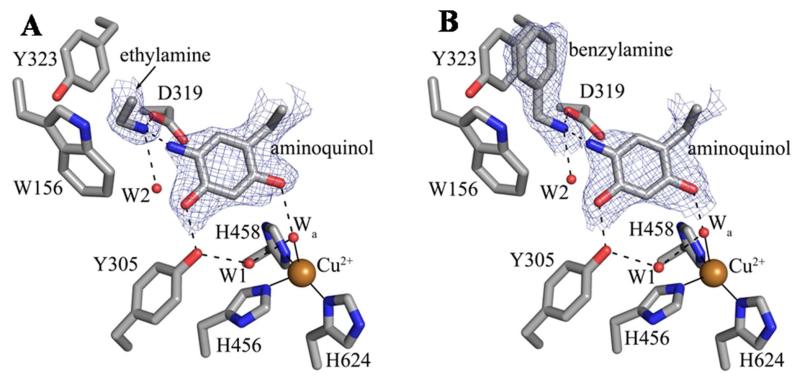
Peaks in the 2Fo-Fc and Fo-Fc electron density maps immediately following phase calculation and rigid body refinement indicated the presence of substrate amine bound in the back of the active site in all protein chains located in the ASU of both structures (from here on, all values are reported as the range observed across the six polypeptides in the ASU of each structure). In all six polypeptide chains of both substrate-reduced HPAO-1 structures, the terminal functional group of the bound molecule is involved in three hydrogen bonding interactions with an ordered water molecule (W2), the side chain of Asp319, and the N5 atom of the aminoquinol (Figure 1). This identifies it as substrate amine, as the aldehyde oxygen of product would only be able to participate in two hydrogen bonds. The geometry suggested by the bound species’ electron density also supported the amine substrate assignments: in the case of EtAm-HPAO-1, the angle between the methyl carbon, methylene carbon, and amine nitrogen ranges from 104-111° (aldehyde ~120°), and in the BzAm-HPAO-1 structure, it was clear that the terminal functional group is non-coplanar with the phenyl ring by 18-40° (benzaldehyde is fully conjugated and coplanar). Additionally, the hydrogen bond geometries and distances between the amine substrates and Asp319/aminoquinol support the bound amine being uncharged; a requirement for initial Schiff base formation between substrate and TPQ (Scheme 1).
Figure 1.
Active sites of the (A) ethylamine- and (B) benzylamine-containing HPAO-1 complexes. Residues and substrate species from polypeptide chain D of both structures are drawn in stick and colored by atom type (carbon, grey). Water molecules are drawn as small red spheres and copper ions are drawn as gold spheres. Hydrogen bonds are indicated by dashed lines, and metal-ligand interactions are indicated by solid lines. Electron density from the 2Fo-Fc electron density maps contoured at 1σ is drawn as blue mesh.
In both EtAm- and BzAm-HPAO-1, the substrates are bound with their non-polar R groups in a hydrophobic pocket (formed by Tyr323 and Trp156) adjacent to the aminoquinol and their amine groups oriented toward atom C5 of the cofactor (the site of nucleophilic attack during catalysis) (Figure 1). In the EtAm-HPAO-1 refined model, all six polypeptide chains within the ASU contain ethylamine modeled at a single binding site (Figure 1A). However, positive peaks in the Fo-Fc electron density map (+5.5-7σ) located adjacent to this species in all crystallographic active sites suggest a partially-occupied second ethylamine binding site that is sterically exclusive to the modeled substrate molecule (Table S2, Figure S2). This indicates positional mobility associated with ethylamine binding; however, the electron density is not clear enough to model this alternate position with confidence.
The benzylamine molecules in all six ASU polypeptide chains of the BzAm-HPAO-1 complex are present in only one position at the back of the active site (Figure 1B). Benzylamine is positioned such that its phenyl ring is involved in a π-π stacking interaction with Tyr323, as well as a perpendicular π-π interaction with Trp156. The distances between atoms in the phenyl rings of benzylamine and Tyr323 are ~3.5-4.0 Å, which is consistent with a stabilizing aromatic interaction. Likewise, the position of benzylamine relative to the side chain of Trp156 is similar to that seen in other examples of perpendicular aromatic stabilization, with the centers of the phenyl ring in benzylamine and the six-membered ring of Trp156 located 5.4-5.9Å apart.20,21
Active site features of the substrate-HPAO-1 complexes
Copper binding
The HPAO-1 active site, formed by residues from domains D3 and D4, lies deeply buried within the protein interior (Figure S1). In all polypeptide chains within the ASU of both substrate-reduced HPAO-1 structures, copper is bound at full occupancy by four ligands, which include either a water molecule or hydrogen peroxide at an axial position, as well as the imidazole groups from a conserved trio of histidine residues at distances of 2.0-2.1, 1.9-2.2, and 1.9-2.1 Å for His456, His458, and His624, respectively (Figure 1). Additionally, strong electron density indicated an equatorial water molecule acting as a fifth copper ligand in some HPAO-1 chains in the ASUs (chains C and E of the EtAm-HPAO-1 structure at distances of 2.5 and 3.0 Å, respectively, and chain C of the BzAm-HPAO-1 structure at a distance of 2.6 Å). The equatorial water ligand is known to be labile, and the ability to model it varies across CAO crystal structures and within crystal forms.
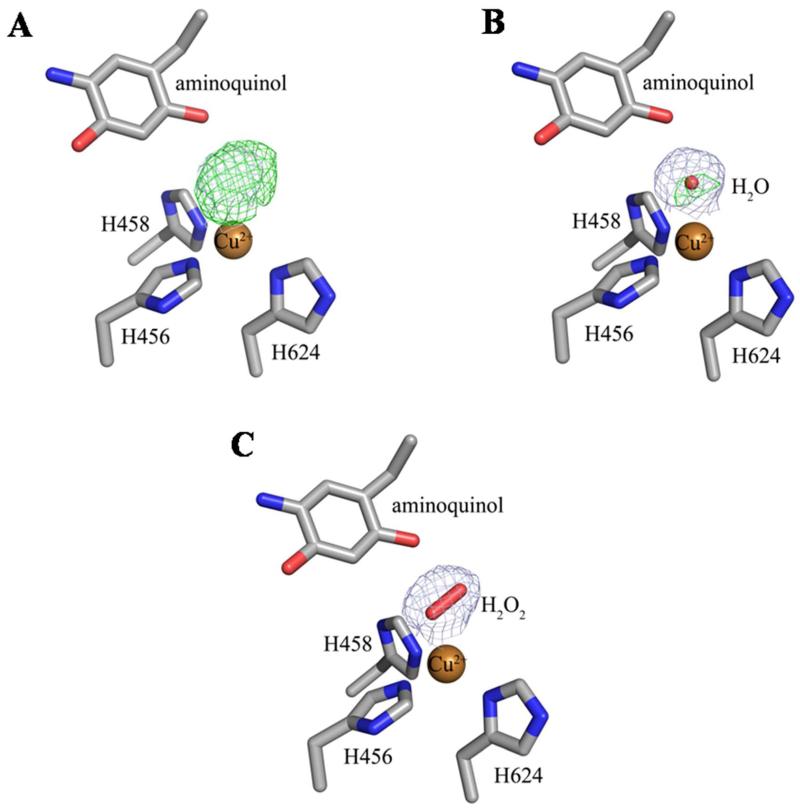
In the structure of native HPAO-1, a water molecule ligates the bound copper in an axial position when TPQ is in the “off-copper” conformation, and indirectly links the O2 atom of TPQ and the bound copper through hydrogen bonding and metal coordination. However, in chains A, B, E and F of both the EtAm- and BeAm-HPAO-1 structures, water is electronically insufficient to account for the electron density at this position (Figure 2). Hydrogen peroxide refined well in these chains, indicating that low levels of oxygen had been present during crystal soaking, handling, and freezing.22 The fact that the same chains are involved in both structures suggests that H2O2 retention in the active sites is at least partially due to crystal packing.
Figure 2.
Structures of the active site of chain E of the ethylamine-HPAO-1 complex with (A) no model, (B) water (Wa), or (C) hydrogen peroxide (H2O2) modeled as the axial copper ligand. Residues in all panels and the peroxide in panel (C) are drawn in stick and colored by atom type (carbon, grey). Copper ions in all panels are drawn as gold spheres and the water molecule in panel (B) is drawn as a small red sphere. The 2Fo-Fc electron density maps after refinement against the corresponding species are drawn as a blue mesh and contoured at 1σ. The Fo-Fc electron density maps after refinement are drawn as a green mesh and contoured at +3.5σ.
In the EtAm-HPAO-1 structure, water is bound 2.6-2.7 Å from the copper in chains C and D, and a side-on hydrogen peroxide is bound with both oxygen atoms 2.5-3.0 Å from the copper in chains A, B, E, and F (water, Wa in Figure 1A; H2O2, Figure 2; Tables S2, S3). In the benzylamine-HPAO-1 complex, hydrogen peroxide was bound with both oxygen atoms at a distance of 2.7-3.3 Å from the copper, with water ligating the active site copper in an axial position at a distance of 2.7-2.8 Å in chains C and D, which is similar to that of the native enzyme (Wa in Figure 1B; Tables S2, S3).
Electronic form of the cofactor in the substrate-HPAO-1 complexes
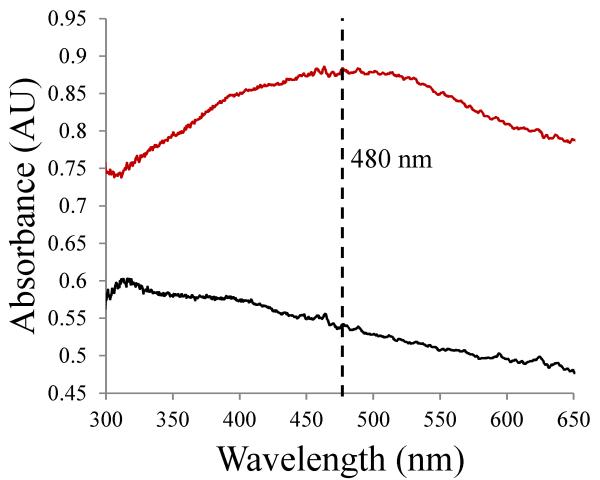
The active site cofactor could be clearly modeled in its “off-copper” conformation in all six polypeptide chains of both structures. In the native enzyme, the “off-copper” conformation, which represents a catalytically productive state, denotes TPQ with its C5 carbonyl directed toward the substrate entry channel. In this position, TPQ is not a direct ligand to the copper, but instead interacts with copper through an axial water molecule. No electron density was found to indicate the presence of cofactor in its unproductive “on-copper” conformation, in which the O4 atom is a direct ligand to the copper. This contrasts with the native HPAO-1 structure, in which four of six polypeptide chains in the ASU contain a mixture of “off-copper” and “on-copper” TPQ conformations.15 Single crystal UV/visible spectroscopy of crystals anaerobically treated with ethylamine or benzylamine showed only a small shoulder feature at ~310 nm, but were otherwise bleached as compared to spectra of the native enzyme (Figure 3). This is consistent with the presence of cofactor in its two-electron reduced aminoquinol form rather than either the semiquinone or iminoquinone (semiquinone: λmax ~360, 435, 460 nm; protonated iminoquinone: λmax ~350 nm; deprotonated iminoquinone: λmax ~450 nm).
Figure 3.
UV/visible spectra showing HPAO-1 crystals before (red curve) and after (black curve) exposure to ethylamine.
Changes in the HPAO-1 substrate channel associated with exposure to amine substrates
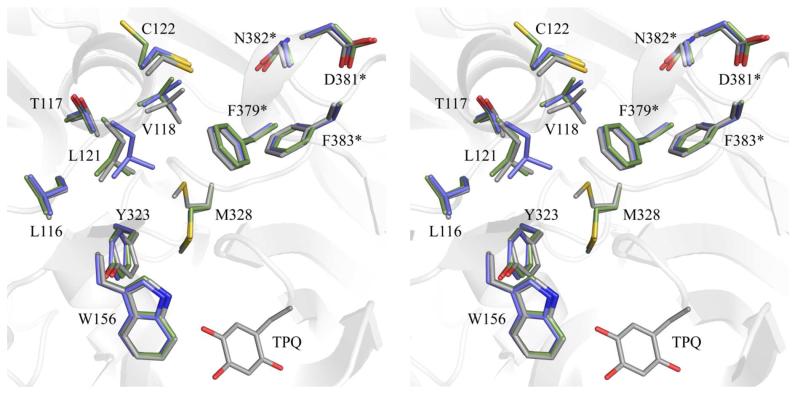
The structural changes in HPAO-1 following anaerobic treatment with both amine substrates are localized to a conserved channel that leads from the enzyme surface to the deeply buried CAO active site (a distance of ~18 Å in HPAO-1).16 The CAO amine substrate channel is defined by a series of secondary structural elements at different depths relative to the enzyme surface. The entrance to this channel is delineated by a surface-exposed helix which lies across its edge.25 Ethylamine or benzylamine binding resulted in the movements of residues Leu116-Cys122 located within this helix away from the center of the amine substrate channel (Figure 4).
Figure 4.
Stereoview of structural changes in the HPAO-1 amine substrate channel following substrate binding. Residues are drawn in stick and colored by structure and by atom type (native HPAO-1: carbon, grey; ethylamine-HPAO-1: carbon, green; benzylamine-HPAO-1: carbon, blue). TPQ from native HPAO-1 is drawn in stick and colored by atom type (carbon, grey). Asterisks indicate side chains which are derived from the other monomer of the homodimer. The overall fold of native HPAO-1 is drawn as a semi-transparent grey cartoon.
Lying closer to the deeply-buried HPAO-1 active site, the side chain of residue Met328 has rotated ~120° around its Cβ-Cγ bond in the EtAm- and BzAm-HPAO-1 complexes, which further expands the width of the substrate channel relative to that of the native enzyme (Figure 4). Additionally, the side chain of Tyr323, which has been proposed to influence substrate specificity in HPAO-1, is rotated ~30° away from the active site in the EtAm- and BzAm-HPAO-1 complexes relative to its position in native HPAO-1 (Figure 4).13
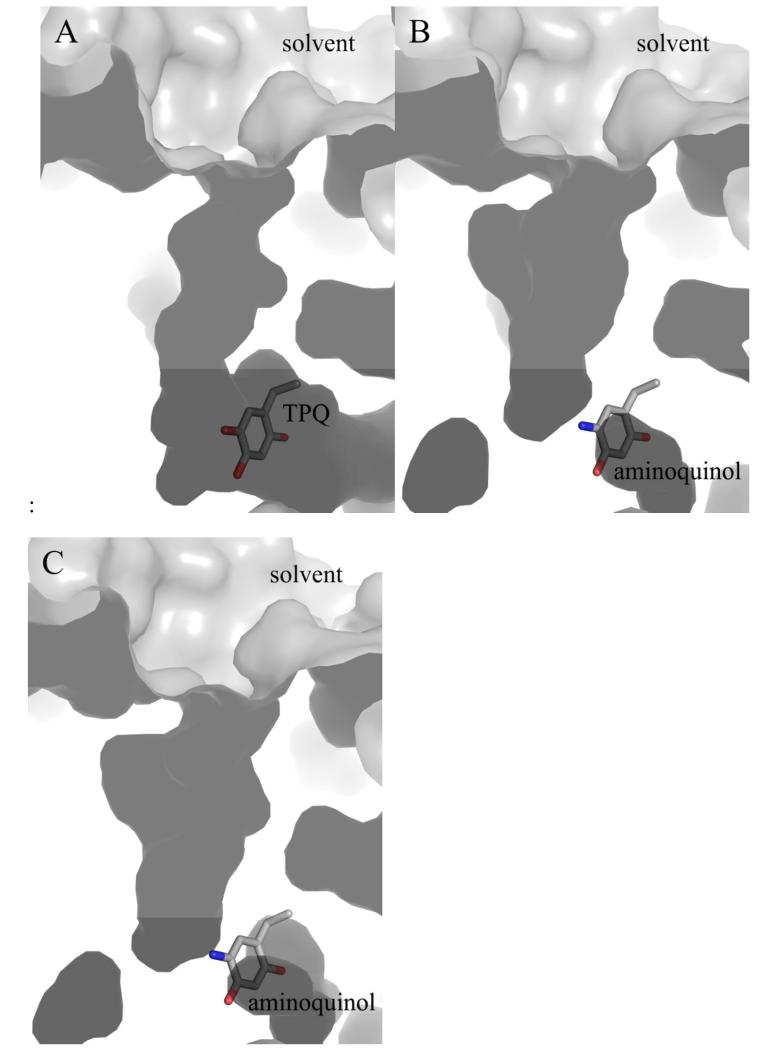
The back wall of the active site of one CAO monomer is partially formed by residues from domain D4 of the other monomer in the protein dimer. These residues are derived from a loop connecting a pair of β-strands that pack tightly against the other monomer to form a β-hairpin “arm” (red in Figure S1). In the structures of HPAO-1 in complex with ethylamine and benzylamine, the side chains of residues Phe379*, Asp381*, Asn382*, and Phe383* from this loop (* = from the other polypeptide in the dimer) have shifted away from the center of the substrate channel (Figure 4). This movement, in conjunction with movements of the specific amino acid side chains mentioned previously, results in a general broadening of the amine substrate channel in the EtAm- and BzAm-HPAO-1 complexes (Figure 5).
Figure 5.
Cut-through of the surface of the substrate entry channel in (A) native HPAO-1, (B) ethylamine-HPAO-1, and (C) benzylamine-HPAO-1. Structures are drawn as a molecular surface and colored grey. TPQ and aminoquinol moieties are drawn in stick and colored by atom type (carbon, grey).
DISCUSSION
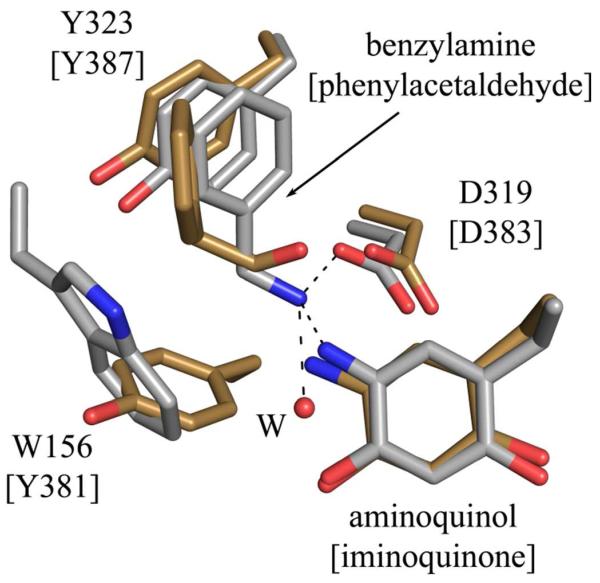
The retention of aldehyde product in CAO has been observed in ECAO crystals aerobically undergoing turnover with 2-phenylethylamine, which revealed a steady-state structure in which iminoquinone was present, product aldehyde was found at the back of the amine substrate channel and hydrogen peroxide bound at the copper (PDB code 1D6Z) (Figure 6).22 It was proposed that in solution, dynamic motions of domains D3 and D4, which both contribute residues to the substrate channel, are key to product dissociation from the ECAO active site. Intermolecular contacts appear to slow aldehyde release in crystallo and disrupt the ping-pong kinetics normally seen in solution, resulting in the accumulation of product aldehyde and hydrogen peroxide in the active site. In HPAO-1, the presence of hydrogen peroxide suggests that its release is also slowed in at least some of the active sites in crystallo. In both HPAO-1 substrate-reduced structures, one dimer of the ASU contains axially ligated water molecules, as would be expected in solution under equivalent conditions. Even with trace oxygen initially present, the enzyme should have turned over until the oxygen was exhausted and then halted at the substrate-reduced aminoquinol with axial water. Therefore, crystal packing must be playing a role, although it is unclear why the retention of H2O2 occurs in two dimers, while the third exhibits the expected active site components after anaerobic reduction. The broadening of the substrate channel occurs in all chains and the trimer of dimers in the ASU involves interactions between the D2 and D3 domains of all monomers, while the interactions with symmetry mates is similar in each physiological dimer, with one monomer having more extensive interactions than the other.
Figure 6.
Overlay of the active sites of HPAO-1 in complex with benzylamine (chain D) and ECAO in complex with product phenylacetaldehyde and hydrogen peroxide (chain A, PDB code 1D6U).22 Residues are drawn in stick and colored by atom type (HPAO-1: carbon, grey; ECAO: carbon, gold). Residues labels are those of HPAO-1, with labels for the ECAO structure in brackets.
The CAO family of enzymes possesses only ~20-40% sequence identity, but displays a high level of structural similarity regardless of enzyme source, particularly in the overall fold and the residues which perform chemistry at the enzyme active site. These conserved residues include Tyr305, which stabilizes the position of TPQ during both biogenesis and catalysis; three histidine residues that ligate the active site copper ion; and an invariant aspartate residue (Asp319) that acts as a general catalytic base (Figure 1). Given the wide variety of functional roles played by CAOs depending on source and cellular location, substrate selectivity is critical and appears to be controlled by several factors. Not surprisingly, the overall shape and size of a CAO’s amine substrate channel correlates with its preferred substrates. In general, CAOs that display the highest activity against bulkier or branched amines contain a correspondingly broad and solvent-exposed substrate channel, while those that show a preference for smaller amines have a more restricted substrate channel. The crystal structures of CAOs from Aspergillus nidulans (ANAO) and Pichia pastoris (PPLO), for example, reveal amine substrate channels so wide that there is unimpeded access to the active site from bulk solvent.25,27 This correlates well with the documented ability of these CAOs to deaminate peptidyl lysine residues. In addition, the active site channel in human VAP-1 resembles the wide funnel shape observed in the PPLO and ANAO structures and is large enough to accommodate peptide or protein ligands, including counter-receptors located on the surface of leukocytes.28,29 In contrast, the HPAO-1 substrate channel is relatively narrow compared to some of its CAO counterparts. This reduces the number of orientations with which a substrate can approach the active site, thus the likelihood of binding and subsequent catalysis with a bulky substrate such as benzylamine is lessened.
The structures of HPAO-1 in complex with two different amine substrates provide insight into specific substrate-protein interactions preceding the nucleophilic attack on atom C5 of TPQ that initiates catalysis. Non-mammalian CAOs contain an aromatic residue (typically a Phe or Tyr) at the intersection of the enzyme active site and the amine substrate channel. This residue has been proposed to adopt two conformations through the rotation of its Cβ-Cγ bond and either allow or block substrate access to the active site. In the two CAOs that have been structurally characterized from Hansenula polymorpha, however, this so-called “gating residue” is a tryptophan, and has only been observed in an “open” conformation. The HPAO-1 and HPAO-2 Trp residues are derived from β-sheet 2.1 of domain D3, one of two small α/β domains N-terminal to the larger catalytic domain D4. Despite the highly conserved overall fold among members of the CAO family of enzymes, in all other structurally characterized CAOs the aromatic gating residues originate from a loop connecting the two β-sheet sandwich motifs in domain D4.
In CAOs that efficiently oxidize aromatic amines such as ECAO or AGAO, the gating residue has been proposed to facilitate reaction with substrates such as 2-phenylethylamine or tyramine through aromatic interactions.31 This is thought to assist in aligning aromatic substrates such that their amine groups are poised for nucleophilic attack on the C5 atom of TPQ at an optimal distance and angle. A comparison of the hydrophobic region near the cofactor’s C5 carbonyl in BzAm-HPAO-1 with that of product-bound ECAO reveals that in HPAO-1 the location of this pocket is shifted relative to TPQ and the catalytic base, where the initial nucleophilic attack and subsequent proton abstraction step take place (Figure 6). In addition to potential interactions with residue Trp156 in HPAO-1, which is positioned for a perpendicular π-π interaction with an aromatic substrate, Tyr323 is well-positioned for a stacked π-π interaction with an aromatic substrate as well (Figure 6).
A comparison of the steady-state kinetic parameters kcat and kcat/Km for the HPAO-1-catalyzed oxidation of ethylamine and benzylamine reveals that HPAO-1 is an effective ethylamine oxidase, but does not efficiently oxidize benzylamine (Table 2).13,28 Ethylamine is the most efficient known substrate for HPAO-1. The values of kcat and kcat/Km for the reaction with benzylamine are reduced ~300-fold and ~580-fold, respectively, relative to those corresponding to the oxidation of ethylamine. Data describing the kinetic isotope effect for the reaction with benzylamine are available, and the values for Dkcat and Dkcat/Km(S) (5.9 ± 0.7 and 3 ± 1, respectively) indicate that proton abstraction and substrate binding are similarly rate-limiting during the reaction with benzylamine.13
Table 2.
Steady state kinetic parameters for the oxidation of ethylamine or benzylamine by HPAO-1.
The interaction between two aromatic species, such as the phenyl ring of benzylamine with residues Trp156 or Tyr323 in HPAO-1, is associated with a small but favorable change in free energy.23,24 These interactions could potentially influence the efficiency of catalysis with an aromatic substrate, depending on the angle and distance between the amine group of the substrate and TPQ. In HPAO-1, the steps known to be rate-limiting when benzylamine is the amine substrate (substrate binding and proton abstraction) imply that the residues in the hydrophobic pocket are not optimally positioned for an efficient nucleophilic attack on the C5 atom by an aromatic substrate, and that the substrate Schiff base formed with benzylamine is not well positioned relative to Asp319, hindering proton abstraction. This is consistent with the observation that the electron density for bound benzylamine supports the presence of substrate in only one conformation. In contrast, peaks in the electron density of the Fo-Fc map of the ethylamine-HPAO-1 complex indicate that this substrate binds in at least two positions within the active site, consistent with benzylamine being more restricted in the orientations it can adopt compared to ethylamine (Figure S2). This structural observation is supported by the approximately two-fold lower Km value for the reaction of HPAO-1 with benzylamine as opposed to ethylamine (0.38 mM vs. 0.73 mM, respectively).10,25 In the steady state crystal structure of ECAO, an efficient aromatic monoamine oxidase, the observed position of the phenyl ring of product phenylacetaldehyde is significantly different from that observed for benzylamine in HPAO-1, although no structure for a 2-phenylethylamine substrate complex is available for ECAO (Figure 6).22 Furthermore, the shape and location of the hydrophobic pocket adjacent to TPQ where substrate binds is not optimal for accommodating Schiff base intermediates formed during catalysis with benzylamine.13 The product Schiff base formed during this reaction is fully conjugated and thus planar, further exacerbating the situation with benzylamine.
A recent study of substrate specificity of HPAO-1 and a paralogous CAO (HPAO-2) emphasized the importance of the orientation of substrates and catalytic intermediates relative to residues in the active site.13 HPAO-2 is known to prefer aromatic amines over the small aliphatic amines preferred by HPAO-1. It was proposed that an amine’s position before its nucleophilic attack on TPQ and the ability of different CAOs to accommodate subsequent Schiff base intermediates are critical to substrate specificity. This is primarily controlled by second sphere residues within the substrate channel,13 which in HPAO-1 include the gating residue Trp156 as well as Tyr323. Because each CAO’s ability to accommodate catalytic intermediates varies, the identities and positions of these second sphere residues likely influence how complementary each CAO active site is to the bulky Schiff base species formed with an aromatic amine.
Following ethylamine or benzylamine binding, the HPAO-1 active site becomes more accessible due to movements of specific amino acid side chains at different depths relative to the enzyme surface that widen the substrate entry channel. These include a surface-exposed helix (Leu116-Cys122); residues Tyr323 and Met328; and residues Phe379*, Asp381*, Asn382*, and Phe383* from the other monomer in the dimer, all of which move further from the active site following substrate binding (Figure 4). Within these changes are residues that differ between HPAO-1 and -2 (Tyr323, Met328).13 These were proposed to impact substrate specificity, but other identified residues are essentially superimposable, at least within the context of the EtAm- and BzAm-HPAO-1 structures. An investigation of structural changes in AGAO associated with the formation of substrate Schiff base analogs with three aromatic hydrazine inhibitors has recently been conducted.32 The effects associated with the binding of these inhibitors include movements in the side chains of residues Phe105, Tyr302, and Leu358* in AGAO, resulting in an “induced;fit” effect.32 These residues are homologous with those found to move upon binding of the physiological substrates ethylamine and benzylamine in HPAO-1 (Phe105, Tyr302, and Leu358* in AGAO are sequentially equivalent to Leu121, Tyr323, and Arg380* in HPAO-1, respectively). In addition, docking studies conducted with a truncated, soluble form of VAP-1 (sVAP-1) have implicated residues Phe389 and Tyr384 as important in determining substrate specificity due to predicted interactions with bound aromatic substrates.33 Phe389 in sVAP-1 corresponds sequentially to Tyr323 in HPAO-1. However, although it is derived form a different part of the primary sequence, the side chain of Tyr384 in sVAP-1 occupies the same space near the active site as Trp156 in HPAO-1. In AGAO ethylamine is a poor substrate, and crystallographic data for an ethylamine-derived substrate Schiff base revealed a disordered adduct, and no associated structural changes within the substrate channel.34
In addition, C-H bond activation in CAOs has been determined to occur via hydrogen tunneling,35 which is sensitive to subtle changes in donor-acceptor distances as tunneling requires precise overlap between donor/acceptor wave functions.36 The small variations in the positions of ethylamine and benzylamine in the CAO active site may influence C-H bond activation by disrupting the close distance between donor and acceptor required for hydrogen tunneling.
In closing, a structural analysis of HPAO-1 in complex with two kinetically-distinct amine substrates has identified specific amino acid residues lining the amine substrate channel that move upon exposure to substrate. These widen the channel at different depths from the buried CAO active site relative to the substrate channel in the native enzyme. For the first time substrate amine has been visualized bound within a reduced CAO active site, and the conformations of specific amino acid side chains which interact with the substrate prior to its nucleophilic attack on TPQ appear to influence substrate specificity. This is primarily due to differences in the efficiency with which primary amines can interact with catalytically-relevant residues in the CAO active site. Species-specific variations in the positions of these side chains and their ability to accommodate catalytic intermediates likely play a role in substrate selection, whilst subtle differences in donor/acceptor distance may influence the efficiency of C-H activation during tunneling, helping generate the broad substrate specificity seen across the CAO family of enzymes.
Supplementary Material
ACKNOWLEDGEMENTS
Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute. X-ray data were collected at GM/CA-CAT at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. We thank Ed Hoeffner for Kahlert Structural Biology Laboratory support at The University of Minnesota.
Footnotes
This work was supported by National Institutes of Health Grants (GM66569 to C.M.W., and GM039326 to J.P.K.), Minnesota Medical Foundation Grant 3714-9221-06, Office of the Dean of the Graduate School of the University of Minnesota Grant 21087, a 3M Science and Technology fellowship to V.J.K., a University of Minnesota UROP grant to C.J.S., and a Minnesota Partnership for Biotechnology and Medical Genomics Grant SPAP-05-0013-P-FY06 to C.M.W.
Coordinates and structure factor data for the reported structures have been deposited in the Protein Data Bank with accession code 4EV2 for ethylamine-reduced HPAO-1, and 4EV5 for benzylamine-reduced HPAO-1.
Abbreviations: CAO, copper amine oxidase; HPAO-1, copper amine oxidase from Hansenula polymorpha which prefers small aliphatic amines; TPQ, 2,4,5-trihydroxyphenylalanine quinone; EtAm-HPAO-1, HPAO-1 in complex with ethylamine; BzAm-HPAO-1, HPAO-1 in complex with benzylamine; ASU, crystallographic asymmetric unit; NCS, non-crystallographic symmetry; ECAO, copper amine oxidase from Escherichia coli; AGAO, copper amine oxidase from Arthrobacter globiformis; ANAO, copper amine oxidase from Aspergillus nidulans; PPLO, amine oxidase from Pichia pastoris
SUPPORTING INFORMATION AVAILABLE
This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Parrott S, Jones S, Cooper RA. 2-Phenylethylamine catabolism by Escherichia coli K12. J. Gen. Microbiol. 1987;133:347–351. doi: 10.1099/00221287-133-2-347. [DOI] [PubMed] [Google Scholar]
- 2.Hacisalihoglu A, Jongejan JA, Duine JA. Distribution of amine oxidases and amine dehydrogenases in bacteria grown on primary amines and characterization of the amine oxidase from Klebsiella oxytoca. Microbiology. 1997;143:505–512. doi: 10.1099/00221287-143-2-505. [DOI] [PubMed] [Google Scholar]
- 3.Angelini R, Cona A, Federico R, Fincato P, Tavladoraki P, Tisi A. Plant amine oxidases “on the move”: an update. Plant Physiol. Biochem. 2010;48:560–564. doi: 10.1016/j.plaphy.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Tisi A, Angelini R, Cona A. Wound healing in plants: cooperation of copper amine oxidase and flavin-containing polyamine oxidase. Plant Signaling Behav. 2008;3:204–206. doi: 10.4161/psb.3.3.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntire WS. Newly discovered redox cofactors: possible nutritional, medical, and pharmacological relevance to higher animals. Annu. Rev. Nutr. 1998;18:145–177. doi: 10.1146/annurev.nutr.18.1.145. [DOI] [PubMed] [Google Scholar]
- 6.Salmi M, Yegutkin GG, Lehvonen R, Koskinen K, Salminen T, Jalkanen S. A cell surface amine oxidase directly controls lymphocyte migration. Immunity. 2001;14:265–276. doi: 10.1016/s1074-7613(01)00108-x. [DOI] [PubMed] [Google Scholar]
- 7.Enrique-Tarancon G, Marti L, Morin N, Lizcano JM, Unzeta M, Sevilla L, Camps M, Palacin M, Testar X, Carpene C, Zorzano A. Role of semicarbazide-sensitive amine oxidase on glucose transport and GLUT4 recruitment to the cell surface in adipose cells. J. Biol. Chem. 1998;273:8025–8032. doi: 10.1074/jbc.273.14.8025. [DOI] [PubMed] [Google Scholar]
- 8.Stolven CM, Madanat R, Marti L, Kari S, Yegutkin GG, Sariola H, Zorzano A, Jalkanen S. Semicarbazide-sensitive amine oxidase overexpression has dual consequences: insulin mimicry and diabetes-like complications. FASEB J. 2004;18:702–704. doi: 10.1096/fj.03-0562fje. [DOI] [PubMed] [Google Scholar]
- 9.Shen SH, Wertz DL, Klinman JP. Implications for functions of the ectopic adipocyte copper amine oxidase (AOC3) from purified enzyme and cell-based kinetic studies. Plos One. 2012;7:1–11. doi: 10.1371/journal.pone.0029270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obata T. Diabetes and semicarbazide-sensitive amine oxidase (SSAO) activity: a review. Life Sci. 2006;79:417–422. doi: 10.1016/j.lfs.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Klema VJ, Wilmot CM. The role of protein crystallography in defining the mechanisms of biogenesis and catalysis in copper amine oxidase. IJMS. 2012;13:5375–5405. doi: 10.3390/ijms13055375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, Corner AS, Phillips SE, McPherson MJ, Knowles PF. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 Å resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 13.Chang CM, Klema VJ, Johnson BJ, Mure M, Klinman JP, Wilmot CM. Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha. Biochemistry. 2010;49:2540–2550. doi: 10.1021/bi901933d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kivi E, Elima K, Aalto K, Nymalm Y, Auvinen K, Koivunen E, Otto DM, Crocker PR, Salminen TA, Salmi M, Jalkanen S. Human Siglec-10 can bind to vascular adhesion protein-1 and serves as its substrate. Blood. 2009;114:5385–5392. doi: 10.1182/blood-2009-04-219253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson BJ, Cohen J, Welford RW, Pearson AR, Schulten K, Klinman JP, Wilmot CM. Exploring molecular oxygen pathways in Hansenula polymorpha copper-containing amine oxidase. J. Biol. Chem. 2007;282:17767–17776. doi: 10.1074/jbc.M701308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Chen L, Cai D, Klinman JP, Mathews FS. Crystallographic study of yeast copper amine oxidase. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;53:364–370. doi: 10.1107/S0907444997000814. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.McCoy A, Grosse-Kunstleve RW, Adams PD, Winn M, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol.Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol.Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Wilmot CM. Visualization of dioxygen bound to copper during enzyme catalysis. Science. 1999;286:1724–1728. doi: 10.1126/science.286.5445.1724. [DOI] [PubMed] [Google Scholar]
- 23.Tsuzuki S, Honda K, Azumi R. Model chemistry calculations of thiophene dimer interactions: origin of pi-stacking. J. Am.Chem. Soc. 2002;124:12200–12209. doi: 10.1021/ja0204877. [DOI] [PubMed] [Google Scholar]
- 24.Tsuzuki S, Honda K, Uchimaru T, Mikami M, Tanabe K. Origin of attraction and directionality of the pi/pi interaction: model chemistry calculations of benzene dimer interaction. J. Am. Chem. Soc. 2002;124:104–112. doi: 10.1021/ja0105212. [DOI] [PubMed] [Google Scholar]
- 25.McGrath AP, Mithieux SM, Collyer CA, Bakhuis JG, van den Berg M, Sein A, Heinz A, Schmelzer C, Weiss AS, Guss JM. Structure and activity of Aspergillus nidulans copper amine oxidase. Biochemistry. 2011;50:5718–5730. doi: 10.1021/bi200555c. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Z, Davidson VL. Identification of a new reaction intermediate in the oxidation of methylamine dehydrogenase by amicyanin. Biochemistry. 1999;38:4862–4867. doi: 10.1021/bi982939r. [DOI] [PubMed] [Google Scholar]
- 27.Duff AP, Cohen AE, Ellis PJ, Kuchar JA, Langley DB, Shepard EM, Dooley DM, Freeman HC, Guss JM. The crystal structure of Pichia pastoris lysyl oxidase. Biochemistry. 2003;42:15148–15157. doi: 10.1021/bi035338v. [DOI] [PubMed] [Google Scholar]
- 28.Arienne TT, Nymalm Y, Kidron H, Smith DJ, Pihlavisto M, Salmi M, Jalkanen S, Johnson MS, Salminen TA. Crystal structure of the human vascular adhesion protein-1: unique structural features with functional implications. Protein Sci. 2005;14:1964–1974. doi: 10.1110/ps.051438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmi M, Yegutkin GG, Lehvonen R, Koskinen K, Salminen TA, Jalkanen S. A cell surface amine oxidase directly controls lymphocyte migration. Immunity. 2001;14:265–276. doi: 10.1016/s1074-7613(01)00108-x. [DOI] [PubMed] [Google Scholar]
- 30.Hevel JM, Mills SA, Klinman JP. Mutation of a strictly conserved, active-site residue alters substrate specificity and cofactor biogenesis in a copper amine oxidase. Biochemistry. 1999;38:3683–3693. doi: 10.1021/bi982199m. [DOI] [PubMed] [Google Scholar]
- 31.Kurtis CR, Knowles PF, Parsons MR, Gaule TG, Phillips SE, McPherson MJ. Tyrosine 381 in E. coli copper amine oxidase influences substrate specificity. J. Neural. Transm. 2011;118:1043–1053. doi: 10.1007/s00702-011-0620-y. [DOI] [PubMed] [Google Scholar]
- 32.Murakawa T, Hayashi H, Taki M, Yamamoto Y, Kawano Y, Tanizawa K, Okajima T. Structural insights into the substrate specificity of bacterial copper amine oxidase obtained by using irreversible inhibitors. J. Biochem. 2012;151:167–178. doi: 10.1093/jb/mvr125. [DOI] [PubMed] [Google Scholar]
- 33.Elovaara H, Kidron H, Parkash V, Nymalm Y, Bligt E, Ollikka P, Smith DJ, Pihlavisto M, Salmi M, Jalkanen S, Salminen TA. Identification of two imidazole binding sites and key residues for substrate specificity in human primary amine oxidase AOC3. Biochemistry. 2011;50:5507–5520. doi: 10.1021/bi200117z. [DOI] [PubMed] [Google Scholar]
- 34.Taki M, Murakawa T, Nakamoto T, Uchida M, Hayashi H, Tanizawa K, Yamamoto Y, Okajima T. Further insight into the mechanism of stereoselective proton abstraction by bacterial copper amine oxidase. Biochemistry. 2008;47:7726–7733. doi: 10.1021/bi800623f. [DOI] [PubMed] [Google Scholar]
- 35.Grant KL, Klinman JP. Evidence that both protium and deuterium undergo significant tunneling in the reaction catalyzed by bovine serum amine oxidase. Biochemistry. 1989;28:6597–6605. doi: 10.1021/bi00442a010. [DOI] [PubMed] [Google Scholar]
- 36.Klinman JP. An integrated model for enzyme catalysis emerges from studies of hydrogen tunneling. Chem. Phys. Lett. 2009;471:179–193. doi: 10.1016/j.cplett.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.